Insights into the epimerization activities of RaCE and pAGE: the quantum mechanics/molecular mechanics simulations†
Yulai Zhanga,
Qingchuan Zheng*ab,
Jilong Zhanga and
Hongxing Zhanga
aState Key Laboratory of Theoretical and Computational Chemistry, Institute of Theoretical Chemistry, Jilin University, Changchun 130023, People's Republic of China. E-mail: zhengqc@jlu.edu.cn
bKey Laboratory for Molecular Enzymology and Engineering of the Ministry of Education, Jilin University, Changchun 130023, People's Republic of China
First published on 20th November 2015
Abstract
Ruminococcus albus cellobiose 2-epimerase (RaCE) and N-acetyl-D-glucosamine 2-epimerase from porcine kidney (pAGE) belong to the AGE superfamily and have a detectable AGE activity. Interestingly, RaCE and pAGE share a set of conserved residues and have a similar catalytic mechanism, although they exhibit different substrate specificities, in that RaCE reacts with unmodified oligosaccharides and pAGE reacts with modified monosaccharides. In this study, the reaction mechanisms of RaCE and pAGE were theoretically calculated by using the quantum mechanics/molecular mechanics method based on the umbrella sampling simulations. The simulation results not only characterize the detailed mechanism of enzyme actions, but also reveal the difference between the catalytic mechanism of RaCE and pAGE. The energy barriers of the two reactions indicate that the activity of reprotonation in the epimerization activities is much higher than that of the deprotonation. Additionally, we obtained the structural information of the transition states of deprotonation and reprotonation and the stable intermediate states and analyzed the enzyme function of lowering the activation energy. Our work can provide important information to understand the catalytic mechanism of the AGE family.
Introduction
Cellobiose 2-epimerase (CE, EC 5.1.3.11) is an enzyme that catalyzes the isomerization of carbohydrate, and its main function is to catalyze reversible epimerization of the D-glucose moiety at the reducing end of oligosaccharides linked by β-1,4-glycosidic linkages and generated D-mannose, such as cellobiose, lactose, and 4-O-β-D-mannosyl-D-glucose.1,2 With most sugar epimerase difference, which act on substrate was activated phosphate groups or nucleotide phosphate group modified and catalyzed isomerization of non anomeric hydroxyl, CE catalyzes the epimerization of unmodified sugars at the C2 position.1–3CE, which was first found in the ruminal anaerobic bacterium, Ruminococcus albus, is the only enzyme responsible for epimerization of the 2-OH group of non-modified oligosaccharide.4–7 CE has an important role in the metabolism of mannan that can convert β-1,4-mannobiose into 4-O-β-D-mannosyl-D-glucose (Man-Glc) and also convert lactose into epilactose (4-O-β-D-galactosyl-D-mannose) that is a non-digestible oligosaccharide. Epilactose can enhance proliferation of beneficial bacteria, which enhances absorption of minerals and reduces a risk factor for colon cancer.8–10 Furthermore, epilactose is useful for medical applications and functional foodstuff, etc.11,12
CE, N-acetyl-glucosamine 2-epimerases (EC 5.1.3.8, AGEs) and aldose–ketose isomerase YihS belong to the AGE superfamily.1,2 The amino acid sequences of CEs show weak similarity to AGE and aldose–ketose isomerases YihS, while the three-dimensional structures of CEs are significantly similar to these of N-acetyl-glucosamine 2-epimerases (EC 5.1.3.8, AGEs) and aldose–ketose isomerase YihS with respect to a common scaffold, an (α/α)6 barrel.1,2,13–15 A previous report showed that CE and AGE have a detectable AGE activity,1–3,14–16 but YihS shows no AGE activity.13 AGE catalyzes the reversible epimerization between N-acetyl-D-glucosamine (GlcNAc) and N-acetyl-D-mannosamine (ManNAc).14,15 Interestingly, although they exhibit different substrate specificities, which CE reacts with unmodified oligosaccharides and AGE reacts with modified monosaccharides, CE and AGE share a set of conserved residues forming the catalytic core and have a similar catalytic mechanism that the catalytic activity both proceed through ring opening, deprotonation/reprotonation, carbon–carbon bond rotation, and ring closure. Recently, a number of crystal structures of CE and AGE have been reported.1,2,14,15 The crystal structure of N-acetyl-D-glucosamine 2-epimerase from porcine kidney (pAGE)14 and Anabaena sp. CH1 (aAGE)15 have been determined and proposed that two key histidine residues (pAGE-His248 and pAGE-His382) for reversible conversion. The crystal structure of cellobiose 2-epimerase from Ruminococcus albus (RaCE)1 and Rhodothermus marinus (RmCE)2 have been determined, and Fujiwara et al.1,2 proposed that the “triplet histidine center” (RaCE-His184, RaCE-His243 and RaCE-His374) is responsible for the catalytic reaction. It is noteworthy that the crystal structures of Rhodothermus marinus CE (RmCE) in the apo form, and complexes with 4-O-β-D-glucopyranosyl-D-mannose, epilactose, or cellobiitol were resolved by Fujiwara et al.2 These crystal structures have important significance for understanding the details of the catalytic mechanism of CE. However, they do not reveal the high energy transition states of deprotonation and reprotonation and the stable intermediate states. Transition state structure is central to understanding catalysis, because enzymes function by lowering activation energy. Therefore, it is vital to characterize the mechanism of CE and AGE through computational approaches because no physical or spectroscopic method is available to directly observe the structure of the transition state for enzymatic reactions.
In this article, we mainly focus on the deprotonation and reprotonation process and the proposed mechanisms for the epimerization reactions by RaCE and pAGE were depicted in Fig. 1. To the best of our knowledge, the detailed mechanism of AGE activity has not yet been addressed by any theoretical studies. Our current study is aiming to discover the epimerization activities of RaCE and pAGE with the classic molecular dynamics (MD) combined with the quantum mechanics/molecular mechanics (QM/MM) umbrella sampling methods. As we all know, the QM/MM method provides an accurate and efficient energetic description of complex chemical and biological systems, leading to significant advances in the understanding of chemical reactions in solution and in enzymes.17–19 The simulation results present the estimated conformations and energies of the important sites during the catalysis, which show good consistency with the experimental data and may be helpful for further experiments of the AGE family.
 | ||
| Fig. 1 The proposed mechanisms of the epimerization reactions by RaCE and pAGE. The residues involved in the reactions and the names of some atoms used in calculations are labeled in this figure. | ||
Materials and methods
Preparation of the structure
The atomic coordinates and the structure factors for the native RaCE enzyme and pAGE enzyme were deposited in the Protein Data Bank with PDB codes 3VW5 and 1FP3, respectively. In order to take into account the efficiency and accuracy of the calculations, we built two models. One model is the complex of RaCE and D-mannose (MAN) (RaCE/MAN), which was built by superimposing the crystal structure of RmCE/4-O-β-D-glucopyranosyl-D-mannose complex (PDB code: 3WKG) on that of RaCE (PDB codes: 3VW5), then deleting the redundant glucopyranosyl and the protein structure in 3WKG. This complex was chosen as the initial structure of RaCE/MAN complex. The other is the complex of pAGE and N-acetyl-D-mannosamine (ManNAc) (pAGE/ManNAc) was built like RaCE/MAN complex. The protonation state of all titratable residues of the full protein were determined according to the PROPKA on-line toolkit20 (http://propka.ki.ku.dk/). It is noteworthy that the protonation states of some residues in active site were determined according to the experimental results.1,2,13–15 The RaCE-His243 (pAGE-His248) was single-protonated at the δ site according to the proposed mechanism. The RaCE-His184 and the RaCE-His374 (pAGE-His382) were double-protonated. All missing hydrogen atoms of the proteins were added using the LEaP module in the AMBER 12 package.21 The ff99SB force field22 was applied to produce the parameters for the protein. The general Amber force field (GAFF)23 was used to obtain the force field parameters for the MAN and ManNAc. Finally, an appropriate number of sodium counterions were placed in the both complex system, which were solvated in an octahedral periodic box of TIP3P water molecules with a minimum distance of 8.0 Å between the outermost protein atoms and the walls of the simulation box.Molecular dynamics simulations
The RaCE/MAN and pAGE/ManNAc were prepared for the classical MD simulations. Two steps of energy minimizations were performed to the systems, using the sander module of AMBER 12.24 First, all the water molecules and counterions were minimized with 5000 steps of steepest descent followed by 5000 steps of conjugate gradient. Then the whole system was minimized with the same process to remove the bad contacts. The system was gradually heated up to 300 K in the NVT ensemble, with velocity of 1 K ps−1. Then, it reached equilibration after another 200 ps simulation. In the meantime, weak restrains were performed on the Cα atoms of the protein and all the atoms of the ligand during the first two processes to ensure the accomplishment of the stabilization. Finally, 10 ns MD simulation was carried out with the NPT ensemble without restrains. A SHAKE algorithm25 was applied to constrain all bonds involving hydrogen atoms, and particle mesh Ewald (PME) method26 was used to calculate the electrostatic interactions with a cutoff value of 10 Å. The time step was set to 1 fs. All figures of the complex conformations in this contribution were generated by PyMOL.27Umbrella sampling
Umbrella sampling28–32 is a powerful algorithm to rapidly explore and accurately determine the free energy landscape of a system. Umbrella sampling simulation can drive the enzyme system over the energy barrier by harmonically restraining the reaction coordinate to a series of windows. The program AMBER coupled with the Weighted Histogram Analysis Method (WHAM)30 was applied to perform the QM/MM umbrella sampling simulations, using the structure of the equilibrated complex system as the start point. In RaCE/MAN complex, the QM part of the umbrella sampling simulations included the substrate (D-mannose), residue His184, His243 and His374. The rest of the complex system was the MM part. Additionally, in the umbrella sampling simulations, we cut through a carbon–carbon bond (Cα–Cβ) on the QM/MM frontier.33 Three hydrogen link atoms were introduced along the covalent bonds crossing the boundary between the QM and the MM regions, to satisfy the valence requirements of the QM fragments. The reaction coordinates were taken as the distance difference of between the bonds being broken/formed in the two steps, respectively. In general, the coordinate that describes the transformation of the system from reactants to products is a collective coordinate. Therefore, in the first step (deprotonation), the distance difference of between C2–H2 and H2–NE2 (R(C2–H2)–R(H2–NE2)) was set as the reaction coordinate. The distance difference of between NE2–HE2 and HE2–C2 (R(NE2–HE2)–R(HE2–C2)) was set as the reaction coordinate of the second step (reprotonation). The atom names are shown labeled in Fig. 1A. For each window on the reaction coordinate (ranging from −1.60 to 1.40, from −2.30 to 3.00 in steps of 0.05 Å), 50 ps sampling was performed with a force constant of 500.0 kcal mol−1 Å−2. In a similar manner, in pAGE/ManNAc complex, the QM part of the umbrella sampling simulations included the substrate (ManNAc), residue His248 and residue His382. Then, the first step of the epimerization reaction was calculated. The length of R(C2–H2)–R(H2–NE2) was set as the reaction coordinate of the first step, the length of R(NE2–HE2)–R(HE2–C2) was set as the reaction coordinate of the second step. The atom names are shown labeled in Fig. 1B. For each window on the reaction coordinate (ranging from −1.90 to 1.60, from −2.70 to 4.00 in steps of 0.05 Å), 50 ps sampling was performed with a force constant of 500.0 kcal mol−1 Å−2. Each subsequent simulation was started from the last frame of the previous run, using AM1/ff99SB of the AMBER simulation package. Semi-empirical methods, such as AM134 and so on, are fast and widely-used methods in the QM/MM calculations. After getting the distribution of the reaction coordinate of each window, the WHAM was used to obtain the potential of mean force (PMF) by eliminating the part of the harmonic umbrella potential. Along every single reaction path, the two points of minimum energy were set as the reactant state (R) and product state (P), the point of highest energy was set as the transition state (TS).Results and discussion
MD simulation of the RaCE/MAN complex and the pAGE/ManNAc complex
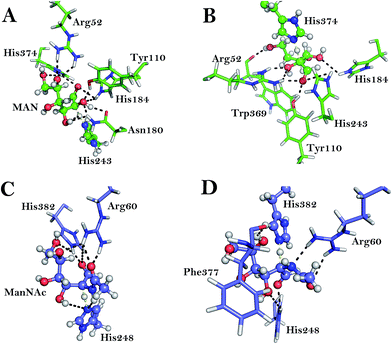
The RaCE monomer is an (α/α)6 structure, and it contains 389 residues. 10 ns MD simulation was carried out on the RaCE/MAN complex to gain the stable conformation for the further mechanism studies and the MD simulation results of RaCE/MAN was depicted in Fig. 2. The root mean square deviation (RMSD) values for the protein and ligand with respect to the crystal structure are shown in Fig. 2A. The backbone RMSD for the protein RaCE in the complex is stable at about 1.0 Å after the whole MD run. In addition, the ligand MAN reached equilibrium after the first 200 ps. Its RMSD values keep about 0.9 Å during the rest of the MD simulation. The ligand MAN has five hydroxyl groups and an aldehyde group which can be served as the donor and acceptor of hydrogen bonds. As shown in Fig. 2B, we analyzed the representative MD structure of RaCE. The ligand MAN forms a series of hydrogen bonds with many residues of RaCE, such as Arg52, Tyr110, Asn180, His184, His243 and His374. To get insights into the function of these residues, we analyzed the hydrogen bonds and salt bridge formed between MAN and the residues of RaCE (show in Fig. 2C). In the RaCE/MAN complex, each of these amino acids forms hydrogen bond with the ligand MAN in more than 90% frames in the simulation trajectories. The hydrogen bonding network and salt bridge interaction help the ligand stability in the pocket. In the active pocket of RaCE, a His184–His243–His374 triplet histidine center is regarded as the active center of the enzyme.1,2 In a similar manner, the root mean square deviation (RMSD) values for the pAGE and ligand are shown in Fig. 3A. The ligand ManNAc has four hydroxyl groups, an aldehyde group and an N-acetyl group. The representative MD structure of the pAGE/ManNAc complex was depicted in Fig. 3B. The ligand ManNAc forms a series of hydrogen bonds with many residues of pAGE, which is a set of conserved residues in pAGE family, such as Arg60, His248 and His382. The hydrogen bonds and salt bridge formed between ManNAc and the residues of pAGE were depicted in Fig. 3C. All of their side chains are in proper orientation for ideal hydrogen bonding to the substrate. In the active pocket of pAGE, a His248–His382 catalytic center is regarded as the active center of the enzyme.14,15 All the mentioned above prove that the MD simulation has been equilibrated. Other MD simulation results are in the ESI (Fig. S1 and S2†). As a result, both of the optimized structures were used as the initial model for the following QM/MM calculations.The epimerization activity of RaCE
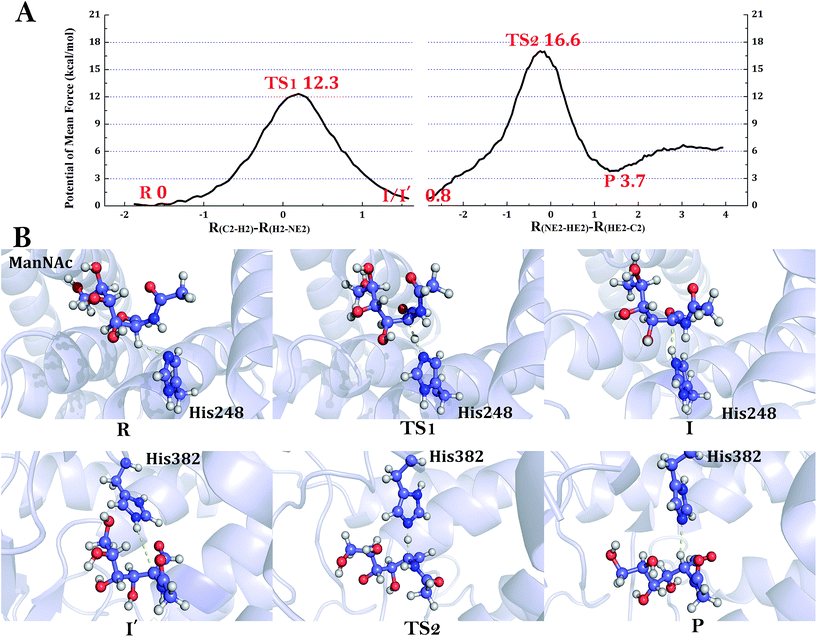
RaCE has an activity of epimerization, isomerizing the D-glucose moiety at the reducing end of oligosaccharides linked by β-1,4-glycosidic linkages and generated D-mannose. In the studies of Fujiwara et al.1,2 they predicted that CE achieve reversible epimerization associated with three histidine residues (triplet histidine center). The histidine residues corresponding to RaCE-His184, RaCE-His243 (pAGE-His248) and RaCE-His374 (pAGE-His382) are completely conserved in the AGE superfamily.1,2 In this study, we simulate the reaction from the reactant open form D-mannose to the product open form D-glucose with the complex model based on the final structure of MD simulation. Fig. 2D provides the distance between the NE2 atom of His243 and H2 atom of the MAN. The catalytic residue His243 is located at alpha helix 9 from Tyr241 to Leu258. After the MD simulation, the distance between NE2 of the residue His243 and H2 is 2.8 Å, which is an appropriate distance for a reaction to take place.The results of QM/MM simulation of the catalytic reaction by RaCE are shown in Fig. 4A. There are two steps in this reaction (Fig. 1A). The first step is the deprotonation, the H2 atom from the C2 of the D-mannose to NE2 of residue His243. We scanned along the distance difference of between C2–H2 and H2–NE2 (R(C2–H2)–R(H2–NE2)) and gained an energy barrier of 10.9 kcal mol−1 (Fig. 4A). The reaction coordinate at this point (TS1) is 0.15 Å. When the H2 atom reaches the residue His243, the free energy decreases to 3.4 kcal mol−1, which is denoted as the intermediate (I) state. The detailed description is shown in Table 1. We have not only obtained the structure of the transition state (TS1) and intermediate (I) state, but also the structural information of the whole reaction process (Fig. 4B). According to the structure information, the carbonyl group attached to C2 atom will depolarize the C–H bond and help accomplishing C–H bond cleavage. The double-protonated His184 may help to lower the pKa of the hydrogen connecting the C2 of MAN and achieve the C–H activation by non-covalent interaction with the substrate MAN. That is consistent with the experiment speculation.1,2 When the H2 atom is abstracted, the substrate will form a cis-enediol intermediate, a resonance structure (Fig. 4B-I). As shown in Table 1, the C2 and O24 atom get more negative (C2: from −0.074 to −0.455 and O24: from −0.408 to −0.660) that is very beneficial to the stability of the structure because there are many positively charged amino acids around atom C2 and O24 like H184, H243, H374 and Arg52. The C1–C2 single bond will form double bond at the same time the O24 atom rotates towards the residue His184 which help to stabilize the intermediate (I) state by non-covalent interaction (Fig. 4B). That situation can also be proved by the dihedral angle results of O7, C2, C1, O24 (the dihedral angle results of the intermediate state is 0.60) (Table 1).
| R | TS1 | I | |
|---|---|---|---|
| Distance | |||
| R(C2–H2) | 1.14 ± 0.03 | 1.38 ± 0.03 | 2.37 ± 0.02 |
| R(H2–NE2) | 2.75 ± 0.01 | 1.24 ± 0.02 | 0.98 ± 0.02 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Mulliken charge | |||
| C2 | −0.074 | −0.346 | −0.455 |
| O24 | −0.408 | −0.406 | −0.660 |
| O7 | −0.394 | −0.420 | −0.346 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Dihedral | |||
| O7–C2–C1–O24 | −45.6 ± 1.4 | −5.1 ± 0.8 | 0.6 ± 0.1 |
The second step of epimerization reaction is reprotonation reaction. The residue His374 provide a proton to the C2 atom of the cis-enediol intermediate in the opposite direction across to the C1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2 double bond. The distance difference between NE2–HE2 and HE2–C2 (R(NE2–HE2)–R(HE2–C2)) was scanned, and the results of QM/MM simulation of the catalytic reaction by pAGE are shown in Fig. 4A. The structural information of the reprotonation reaction is shown in Fig. 4B. It is noteworthy that I and I′ are the same conformation only in different perspectives (I in the perspective of His243, I′ in the perspective of His374). The reaction coordinate corresponds to the highest energy point (TS2) of the second step is about −0.2 Å. The detailed description is shown in Table 2. After that, the HE2 atom of residue His374 is dragged toward C2 of the ligand and form a covalent bond. The second step decreases the system free energy to 0.7 kcal mol−1. Consequently, the reprotoned reaction has a barrier of 12.5 kcal mol−1, and the energy of the product relative to the reactant is about 2.7 kcal mol−1. The catalytic residue His374 is located at alpha helix 13 from Tyr373 to Arg385. After the MD simulation, the distance between HE2 of the residue His374 and C2 is 4.1 Å, which is a bit far for a reaction to take place. But the distance decrease to 3.4 Å after the deprotonation activity. In the reprotonation step, the C2–C3 bond will rotate about 70° to make the orientation of C1
C2 double bond. The distance difference between NE2–HE2 and HE2–C2 (R(NE2–HE2)–R(HE2–C2)) was scanned, and the results of QM/MM simulation of the catalytic reaction by pAGE are shown in Fig. 4A. The structural information of the reprotonation reaction is shown in Fig. 4B. It is noteworthy that I and I′ are the same conformation only in different perspectives (I in the perspective of His243, I′ in the perspective of His374). The reaction coordinate corresponds to the highest energy point (TS2) of the second step is about −0.2 Å. The detailed description is shown in Table 2. After that, the HE2 atom of residue His374 is dragged toward C2 of the ligand and form a covalent bond. The second step decreases the system free energy to 0.7 kcal mol−1. Consequently, the reprotoned reaction has a barrier of 12.5 kcal mol−1, and the energy of the product relative to the reactant is about 2.7 kcal mol−1. The catalytic residue His374 is located at alpha helix 13 from Tyr373 to Arg385. After the MD simulation, the distance between HE2 of the residue His374 and C2 is 4.1 Å, which is a bit far for a reaction to take place. But the distance decrease to 3.4 Å after the deprotonation activity. In the reprotonation step, the C2–C3 bond will rotate about 70° to make the orientation of C1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2 double bond to His374 in order to C2 reprotoned by HE2 of the residue His374 and then rotate back about 20° (Table 2). That situation can also be proved by the dihedral angle results of O8, C3, C2, C1 (Table 2). After reprotonation reaction, the charge of the C2, O7 and O24 atom returned to the original level of the reactant state (Table 2). This is consistent with the Fujiwara et al.'s conclusion2 that the C2–C3 bond will rotate for helping the reprotoned from the His374.
C2 double bond to His374 in order to C2 reprotoned by HE2 of the residue His374 and then rotate back about 20° (Table 2). That situation can also be proved by the dihedral angle results of O8, C3, C2, C1 (Table 2). After reprotonation reaction, the charge of the C2, O7 and O24 atom returned to the original level of the reactant state (Table 2). This is consistent with the Fujiwara et al.'s conclusion2 that the C2–C3 bond will rotate for helping the reprotoned from the His374.
| I | TS2 | P | |
|---|---|---|---|
| Distance | |||
| R(NE2–HE2) | 1.09 ± 0.03 | 1.26 ± 0.03 | 1.90 ± 0.03 |
| R(HE2–C2) | 3.39 ± 0.03 | 1.45 ± 0.02 | 1.07 ± 0.02 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Mulliken charge | |||
| C2 | −0.455 | −0.443 | −0.053 |
| O24 | −0.660 | −0.403 | −0.362 |
| O7 | −0.346 | −0.356 | −0.357 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Dihedral | |||
| O8–C3–C2–C1 | 148.9 ± 1.2 | 72.4 ± 2.6 | 95.1 ± 2.8 |
The epimerization activity of pAGE
While CE catalyzes epimerization of unmodified sugars, AGE reacts with modified sugars. Both the two types of enzymes have a similar epimerization reaction, but their detailed mechanisms have some differences. Thus, the reaction from the reactant open form ManNAc to the product open form GluNAc with the complex model was simulated. Fig. 3D provides the distance between the NE2 atom of His248 and H2 atom of the ManNAc. The catalytic residue His248 is located at alpha helix 12 from Pro246 to Arg262. After the MD simulation, the distance between NE2 of the residue His248 and H2 is 3.0 Å which is an appropriate distance for a reaction to take place.The result of QM/MM calculation of the catalytic reaction by pAGE is illustrated in Fig. 5A. In the deprotonation step, at the reactant (R) state of the epimerization reaction, this distance is shortened to about 2.8 Å (Fig. 5A). We defined the R state as the zero point of the potential of mean force and scanned along the distance difference of between C2–H2 and H2–NE2 (R(C2–H2)–R(H2–NE2)). When the reaction coordinate changes from −1.9 Å to 1.6 Å, the system reaches the transition state of the first step (TS1). The reaction coordinate at this point (TS1) is 0.20 Å. The energy barrier of the first step is 12.3 kcal mol−1. Then the free energy decreases to 0.8 kcal mol−1 at the intermediate (I) state of this step. In this step, the residue His248 gets the proton from ManNAc as shown in Fig. 5B. The Mulliken charge results clearly reveal that the O24 atom and the C2 atom of substrate ManNAc is getting negative (Table 3). In a similar manner, when the H2 atom is abstracted, the C1–C2 single bond will form double bond and the substrate will form a cis-enediol intermediate, a resonance structure. It is noteworthy that the O24 atom rotated in an opposite direction corresponds to the O24 atom of RaCE. That situation can also be proved by the dihedral angle results of N7, C2, C1, O24 (Table 3). When the H2 proton reaches the residue His248, the free energy decreases to 0.8 kcal mol−1, which is denoted as the intermediate (I) state, much lower than the intermediate of RaCE. After the H2 leaving, the N-acetyl group bound to C2 will form the resonance structures, the negative charge from C2 to O not N7, and help stabilize the structure of intermediate (I) state (Table 3). The detailed description of the charge results of the C2, O24, N13, O atoms is shown in Table 3.
| R | TS1 | I | |
|---|---|---|---|
| Distance | |||
| R(C2–H2) | 1.10 ± 0.01 | 1.45 ± 0.02 | 2.59 ± 0.02 |
| R(H2–NE2) | 2.80 ± 0.02 | 1.25 ± 0.02 | 1.00 ± 0.02 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Mulliken charge | |||
| C2 | −0.101 | −0.368 | −0.487 |
| O24 | −0.392 | −0.444 | −0.645 |
| N13 | −0.306 | −0.278 | −0.277 |
| O | −0.572 | −0.541 | −0.603 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Dihedral | |||
| N7–C2–C1–O24 | −147.3 ± 2.1 | −144.2 ± 1.6 | −175.3 ± 1.9 |
The second step of epimerization reaction is reprotonation reaction. The distance difference between NE2–HE2 and HE2–C2 (R(NE2–HE2)–R(HE2–C2)) was scanned, and the result of QM/MM calculation of the catalytic reaction by pAGE is illustrated in Fig. 5A and B. Similar with the reprotonation step of RaCE, the C2–C3 bond of substrate in the pAGE system will rotate about 60° to make the orientation of C1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2 double bond to His382 in order to C2 reprotoned by HE2 of the residue His382 and then rotate back about 20°. That situation can also be proved by the dihedral angle results of O8, C3, C2, C1 (Table 4). That may explain why the reprotonation of pAGE has high energy barrier, it is just because of the functional group is too big difficult to rotate C2–C3 bond. The reaction coordinate corresponds to the highest energy point (TS2) of the second step is about −0.25 Å. The detailed description is shown in Table 4. After that, the HE2 atom of residue His382 is dragged toward C2 of the ligand and form a covalent bond. The proton transfer process can also be proved by the Mulliken charge results of C2, O24, N7, O (Table 4). Consequently, the reprotoned reaction has a barrier of 16.6 kcal mol−1, and the energy of the product relative to the reactant is about 3.7 kcal mol−1. The structural information of the whole reaction process is shown in Fig. 5B.
C2 double bond to His382 in order to C2 reprotoned by HE2 of the residue His382 and then rotate back about 20°. That situation can also be proved by the dihedral angle results of O8, C3, C2, C1 (Table 4). That may explain why the reprotonation of pAGE has high energy barrier, it is just because of the functional group is too big difficult to rotate C2–C3 bond. The reaction coordinate corresponds to the highest energy point (TS2) of the second step is about −0.25 Å. The detailed description is shown in Table 4. After that, the HE2 atom of residue His382 is dragged toward C2 of the ligand and form a covalent bond. The proton transfer process can also be proved by the Mulliken charge results of C2, O24, N7, O (Table 4). Consequently, the reprotoned reaction has a barrier of 16.6 kcal mol−1, and the energy of the product relative to the reactant is about 3.7 kcal mol−1. The structural information of the whole reaction process is shown in Fig. 5B.
| I | TS2 | P | |
|---|---|---|---|
| Distance | |||
| R(NE2–HE2) | 1.03 ± 0.02 | 1.22 ± 0.01 | 2.55 ± 0.03 |
| R(HE2–C2) | 3.84 ± 0.01 | 1.44 ± 0.02 | 1.09 ± 0.03 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Mulliken charge | |||
| C2 | −0.487 | −0.426 | −0.090 |
| O24 | −0.645 | −0.460 | −0.372 |
| N7 | −0.277 | −0.251 | −0.327 |
| O | −0.603 | −0.565 | −0.515 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Dihedral | |||
| O8–C3–C2–C1 | 124.6 ± 1.9 | 66.5 ± 3.0 | 80.4 ± 1.2 |
Overall, we reconstruct the whole potential of mean force of the epimerization reaction of RaCE and pAGE and obtained the structural information of the whole reaction process. Through the above analysis, the rate-limiting steps of the two reactions both are the second step, the reprotonation activity. This step of RaCE needs to overcome an energy barrier of 12.5 kcal mol−1 (16.6 kcal mol−1 in pAGE), and reaches the product state at a low energy level (2.7 kcal mol−1 in RaCE, 3.7 kcal mol−1 in pAGE). The Ito's group3 and Lee's group15 reported the kinetics assay data of the RaCE and the pAGE, respectively. The rate constant (kcat) of the RaCE is 63.8 s−1 (pH 7.5, 30 °C), the pAGE is 23.5 s−1 (pH 8.0, 37 °C). According to the transition state theory (TST),35,36 the estimated energy barrier is about 15.7 kcal mol−1 for the epimerization of RaCE, 16.3 kcal mol−1 for the pAGE. As a result, our QM/MM simulation methods are reasonable, and the results of umbrella sampling method are close to the experimental data. In addition, the C–H bond activation in RaCE and pAGE have adopted a different strategy, that His184 help to depolarize 2-OH described above by non-covalent interaction in RaCE while the π-electron delocalization caused by the resonance structures of the N-acetyl group help to activate C–H bond at the position of C2 in pAGE.
Other residues help stabilize the transition state
The function of an enzyme is to lower activation energy by tight binding to the unstable transition state structure. Therefore, the transition state structure is central to understanding catalysis. In the both complexes, there are many amino acids around the substrate help stabilize the transition state by non-covalent interactions. Among them, the hydrogen bonding network and salt bridge interaction is a major contribution (Fig. 6). During the epimerization activities of RaCE, the residues Arg52, Tyr110, Asn180, His184, His243 and His374 have hydrogen bonds with the transition state, helping to stabilize both of TS1 and TS2 (Fig. 6A and B). The key distances (with deviation) between MAN and the residues of RaCE of TS1, intermediates and TS2 are shown in Table S1 in the ESI.† In a similar vein, the hydrogen bonding network of TS1 and TS2 of pAGE are shown in Fig. 6C and D. The key distances (with deviation) between ManNAc and the residues of pAGE of TS1, intermediates and TS2 are shown in Table S2 in the ESI.† These residues are crucial for the epimerization activities of RaCE and pAGE.Conclusions
Ruminococcus albus cellobiose 2-epimerase (RaCE) and N-acetyl-D-glucosamine 2-epimerase from porcine kidney (pAGE) both belong to AGE superfamily. Both of them main activity is to catalyze the epimerization at the C2 position. But RaCE reacts with unmodified oligosaccharides and AGE reacts with modified monosaccharides. In our study, we started from the complex of the RaCE/MAN and pAGE/ManNAc to investigate both the epimerization of the unmodified sugar and the modified sugar by employing the QM/MM umbrella sampling simulations. The results show that the deprotonation step is accomplished by removing the hydrogen from the C2 atom of the MAN to the NE2 atom of the His243 in RaCE (from the C2 atom of the ManNAc to the NE2 atom of the His248 in pAGE). The C1–C2 single bond will form double bond and the substrate will form a cis-enediol intermediate. The difference of the two reactions is that the orientation of the atoms O24 is opposite. In the reprotonation step, the C2–C3 bond will rotate to help C2 reprotoned by HE2 of the residue RaCE-His374 (pAGE-His382). In the process of the reaction, His184 in RaCE system and the functional group N-acetyl in pAGE system play a similar role in both the C–H bond activation and the transition state stability. The estimated energy barriers of deprotonation and reprotonation activities are 10.9 and 12.5 kcal mol−1 in RaCE (12.3 and 16.6 kcal mol−1 in pAGE), which are in agreement with the previous experimental data.3,15 The energy barrier of the deprotonation reaction is much lower than the reprotonation reaction, which proves that the reprotonation activity is the rate-limiting step of epimerization at the position C2. Furthermore, we obtained the structural information of the transition states of deprotonation and reprotonation and the stable intermediate states and analyzed the enzymes function of lowering activation energy. These data suggest that these reactions are completed with the help of some important residues in the active site, such as Arg52, Tyr110, Asn180 and His184.Acknowledgements
This work is supported by Natural Science Foundation of China (Grant No. 21273095).References
- T. Fujiwara, W. Saburi, S. Inoue, H. Mori, H. Matsui, I. Tanaka and M. Yao, FEBS Lett., 2013, 587, 840–846 CrossRef CAS PubMed.
- T. Fujiwara, W. Saburi, H. Matsui, H. Mori and M. Yao, J. Biol. Chem., 2014, 289, 3405–3415 CrossRef CAS PubMed.
- S. Ito, S. Hamada, H. Ito, H. Matsui, T. Ozawa, H. Taguchi and S. Ito, Biotechnol. Lett., 2009, 31, 1065–1071 CrossRef CAS PubMed.
- T. Tyler and J. Leatherwood, Arch. Biochem. Biophys., 1967, 119, 363–367 CrossRef CAS PubMed.
- S. Ito, S. Hamada, K. Yamaguchi, S. Umene, H. Ito, H. Matsui, T. Ozawa, H. Taguchi, J. Watanabe, J. Wasaki and S. Ito, Biochem. Biophys. Res. Commun., 2007, 360, 640–645 CrossRef CAS PubMed.
- T. Senoura, H. Taguchi, S. Ito, S. Hamada, H. Matsui, S. Fukiya, A. Yokota, J. Watanabe, J. Wasaki and S. Ito, Biosci., Biotechnol., Biochem., 2009, 73, 400–406 CrossRef CAS PubMed.
- H. Taguchi, T. Senoura, S. Hamada, H. Matsui, Y. Kobayashi, J. Watanabe, J. Wasaki and S. Ito, FEMS Microbiol. Lett., 2008, 287, 34–40 CrossRef CAS PubMed.
- M. Krewinkel, M. Gosch, E. Rentschler and L. Fischer, J. Dairy Sci., 2014, 97, 155–161 CrossRef CAS PubMed.
- J. Watanabe, M. Nishimukai, H. Taguchi, T. Senoura, S. Hamada, H. Matsui, T. Yamamoto, J. Wasaki, H. Hara and S. Ito, J. Dairy Sci., 2008, 91, 4518–4526 CrossRef CAS PubMed.
- M. Nishimukai, J. Watanabe, H. Taguchi, T. Senoura, S. Hamada, H. Matsui, T. Yamamoto, J. Wasaki, H. Hara and S. Ito, J. Agric. Food Chem., 2008, 56, 10340–10345 CrossRef CAS PubMed.
- W. Saburi, T. Yamamoto, H. Taguchi, S. Hamada and H. Matsui, Biosci., Biotechnol., Biochem., 2010, 74, 1736–1737 CrossRef CAS PubMed.
- H. Sato, W. Saburi, T. Ojima, H. Taguchi, H. Mori and H. Matsui, Biosci., Biotechnol., Biochem., 2012, 76, 1584–1587 CrossRef CAS PubMed.
- T. Itoh, B. Mikami, W. Hashimoto and K. Murata, J. Mol. Biol., 2008, 377, 1443–1459 CrossRef CAS PubMed.
- T. Itoh, B. Mikami, I. Maru, Y. Ohta, W. Hashimoto and K. Murata, J. Mol. Biol., 2000, 303, 733–744 CrossRef CAS PubMed.
- Y.-C. Lee, H.-M. Wu, Y.-N. Chang, W.-C. Wang and W.-H. Hsu, J. Mol. Biol., 2007, 367, 895–908 CrossRef CAS PubMed.
- S. Ito, H. Taguchi, S. Hamada, S. Kawauchi, H. Ito, T. Senoura, J. Watanabe, M. Nishimukai, S. Ito and H. Matsui, Appl. Microbiol. Biotechnol., 2008, 79, 433–441 CrossRef CAS PubMed.
- R. A. Friesner and V. Guallar, Annu. Rev. Phys. Chem., 2005, 56, 389–427 CrossRef CAS PubMed.
- J. Gao, S. Ma, D. T. Major, K. Nam, J. Pu and D. G. Truhlar, Chem. Rev., 2006, 106, 3188–3209 CrossRef CAS PubMed.
- Y. Luo, Y. Luo, J. Qu and Z. Hou, Organometallics, 2011, 30, 2908–2919 CrossRef CAS.
- C. R. Søndergaard, M. H. Olsson, M. Rostkowski and J. H. Jensen, J. Chem. Theory Comput., 2011, 7, 2284–2295 CrossRef.
- D. Case, T. Darden, T. E. Cheatham III, C. Simmerling, J. Wang, R. Duke, R. Luo, R. Walker, W. Zhang and K. Merz, AMBER 12, University of California, San Francisco, 2012, p. 142 Search PubMed.
- J. W. Ponder and D. A. Case, Adv. Protein Chem., 2003, 66, 27–85 CrossRef CAS PubMed.
- J. Wang, R. M. Wolf, J. W. Caldwell, P. A. Kollman and D. A. Case, J. Comput. Chem., 2004, 25, 1157–1174 CrossRef CAS PubMed.
- D. A. Pearlman, D. A. Case, J. W. Caldwell, W. S. Ross, T. E. Cheatham, S. DeBolt, D. Ferguson, G. Seibel and P. Kollman, Comput. Phys. Commun., 1995, 91, 1–41 CrossRef CAS.
- V. Kräutler, W. F. van Gunsteren and P. H. Hünenberger, J. Comput. Chem., 2001, 22, 501–508 CrossRef.
- T. Darden, D. York and L. Pedersen, J. Chem. Phys., 1993, 98, 10089–10092 CrossRef CAS.
- W. L. DeLano, The PyMOL molecular graphics system, DeLano Scientific LLC, San Carlos, CA, 2002 Search PubMed.
- W.-T. Chu, Q.-C. Zheng and H.-X. Zhang, Phys. Chem. Chem. Phys., 2014, 16, 3946–3954 RSC.
- J. Kästner, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2011, 1, 932–942 CrossRef.
- S. Kumar, J. M. Rosenberg, D. Bouzida, R. H. Swendsen and P. A. Kollman, J. Chem. Phys., 1992, 13, 1011–1021 CAS.
- M. Souaille and B. Roux, Comput. Phys. Commun., 2001, 135, 40–57 CrossRef CAS.
- G. M. Torrie and J. P. Valleau, J. Comput. Phys., 1977, 23, 187–199 CrossRef.
- A. Monari, J.-L. Rivail and X. Assfeld, Acc. Chem. Res., 2012, 46, 596–603 CrossRef PubMed.
- M. J. Dewar, E. G. Zoebisch, E. F. Healy and J. J. Stewart, J. Am. Chem. Soc., 1985, 107, 3902–3909 CrossRef CAS.
- E. V. Anslyn and D. A. Dougherty, Modern physical organic chemistry, University Science Books, 2006 Search PubMed.
- K. J. Laidler and M. C. King, J. Phys. Chem., 1983, 87, 2657–2664 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: The MD results of the RaCE/MAN complex and the pAGE/ManNAc complex, RMSF and total potential energy; other active site residues help stabilize the transition state. See DOI: 10.1039/c5ra14091a |
| This journal is © The Royal Society of Chemistry 2015 |