Current state of the art of mass spectrometry-based metabolomics studies – a review focusing on wide coverage, high throughput and easy identification
Yang Wanga,
Shuying Liub,
Yuanjia Hua,
Peng Lia and
Jian-Bo Wan*a
aState Key Laboratory of Quality Research in Chinese Medicine, Institute of Chinese Medical Sciences, University of Macau, Taipa, Macao, China. E-mail: jbwan@umac.mo; Fax: +853-28841-358; Tel: +853-397-4873
bJilin Ginseng Academy, Changchun University of Chinese Medicine, Changchun, Jilin, China
First published on 3rd September 2015
Abstract
Metabolomics aims at the comprehensive assessment of a wide range of endogenous metabolites and attempts to identify and quantify the attractive metabolites in a given biological sample. These metabolites have diverse physicochemical properties and are presented at different concentration ranges, which makes global analysis a difficult challenge. As a commonly used analytical platform, mass spectrometry (MS) coupled with various separation techniques, such as gas chromatography (GC) and liquid chromatography (LC), has recently undergone rapid development, providing promising solutions to these problems. The ambient ionization techniques, including desorption electrospray ionization (DESI), direct analysis in real time (DART) and extractive electrospray ionization (EESI), enable rapid detection of metabolites, making it ideal for high-throughput analysis in large-scale metabolomics studies. The current applications of these approaches are described with selected illustrative examples in the present review. Furthermore, regardless of “targeted” or “non-targeted” metabolomics study, the identification of the attractive biomarkers is required to further interpret the related metabolic pathways. Therefore, in the present review, recent novel MS-based techniques that allow more robust and easier metabolite identification are summarized, and their strengths and limitations are also discussed.
1. Introduction
After genomics and proteomics, a new systems biology technique has emerged in the study of metabolism on a global level in a given biological system, termed metabolomics. This rapidly developing research field focuses on the comprehensive analysis of metabolites with molecular masses lower than 1000 Da present in biological samples, such as cells, tissues, and body fluids.1 These metabolites are the end products of cellular processes and exist in a state of dynamic equilibrium under normal condition. However, when the body is disturbed by external stimuli, the types and concentrations of these metabolites are altered accordingly. Metabolomics focuses on these changes in order to explain biological activities from a metabolic standpoint. To date, the metabolomics approach has been widely used in disease biomarker discovery for early diagnosis, disease pathogenesis and drug discovery.2–4 Additionally, the research strategy of metabolomics coincides with the integrity feature of Traditional Chinese Medicine (TCM) theory and systemic actions of herbal medicines. The emerging field of metabolomics offers a promising approach for in-depth understanding of TCM syndromes, acupuncture, as well as the therapeutic effects and toxicity of herbal medicines and their quality evaluation.5–7Due to chemical diversity and wide concentration ranges, the identification and quantification of metabolites must rely on sophisticated instrumentation. The two major platforms used in metabolomics are nuclear magnetic resonance (NMR) and MS, each of which has advantages and disadvantages. NMR does not rely on the separation of analytes, and the sample can be recovered for further analyses. Multiple types of small-molecule metabolites can be measured simultaneously, and as such, NMR is close to being a universal detector. The main advantages of NMR are high analytical reproducibility and simplicity of sample preparation. However, NMR is relatively insensitive, and much peak overlapping of the endogenous metabolites occurs when compared with MS-based techniques. In MS-based metabolomics technical platforms, the hyphenated techniques, such as gas chromatography (GC)-MS, liquid chromatography (LC)-MS and capillary electrophoresis (CE)-MS, are the most popular for determining the metabolite profile of a biological sample due to their high sensitivity and selectivity, high throughput and depth of coverage. Additionally, a number of metabolomics databases based on MS or MS/MS data are available, which offer useful information for the identification of metabolites, leading MS to attract widespread interest in this field.
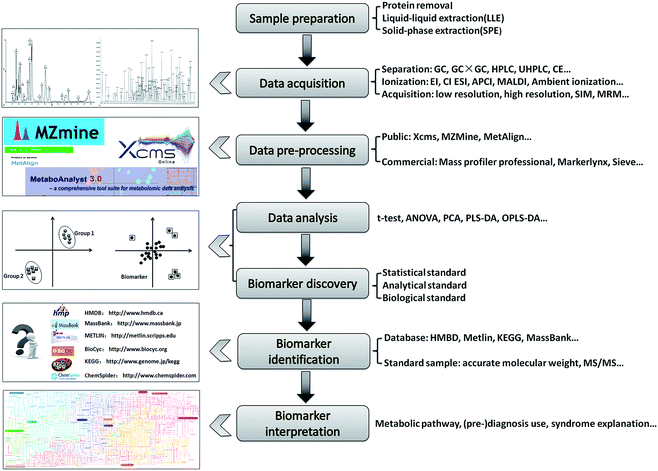
As shown in Fig. 1, sample preparation, raw data acquisition, data analysis, biomarker identification, and biological interpretation comprise the typical analytical workflow in metabolomics studies, and many recent reviews have focused on these analysis steps.8–10 However, compared with the traditional methods, several new methodologies have been developed over the past five years, including new separation columns. The use of ion-mobility MS (IMMS) in identification, as well as several newly developed ionization sources such as desorption electrospray ionization (DESI), direct analysis in real time (DART), extractive electrospray ionization (EESI), etc., have been introduced into MS-based metabolomics studies. In the present review, recent novel MS-based techniques that allow comprehensive analysis of metabolites as well as more robust and easier metabolite identification are summarized, and their strengths and limitations are also discussed.
2. Comprehensive analysis of metabolites
The known metabolites found in human serum and plasma comprise greater than 4000 chemically diverse small molecules whose concentration range spans over 9 orders of magnitude,11,12 and this number expands to over 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 when exogenous metabolites derived from foods, drugs, and microbiota are included.12,13 Similar chemical diversity also exists in plants owing to their complex secondary metabolism; it has been estimated that there are at least 200
000 when exogenous metabolites derived from foods, drugs, and microbiota are included.12,13 Similar chemical diversity also exists in plants owing to their complex secondary metabolism; it has been estimated that there are at least 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 different metabolites in the plant kingdom.14 For the comprehensive analysis of metabolites, it is essential to use strategies that offer wider coverage in terms of the type and number of metabolites present in living samples. Metabolites are chemical entities that can be analyzed using analytical chemistry tools such as NMR and MS. The resolution, sensitivity and selectivity of these technologies can be enhanced or modified by coupling them to GC or LC. However, there is currently no single approach that is amenable to comprehensive metabolite profiling because of the chemical diversity, heterogeneity and wide dynamic range of metabolites.
000 different metabolites in the plant kingdom.14 For the comprehensive analysis of metabolites, it is essential to use strategies that offer wider coverage in terms of the type and number of metabolites present in living samples. Metabolites are chemical entities that can be analyzed using analytical chemistry tools such as NMR and MS. The resolution, sensitivity and selectivity of these technologies can be enhanced or modified by coupling them to GC or LC. However, there is currently no single approach that is amenable to comprehensive metabolite profiling because of the chemical diversity, heterogeneity and wide dynamic range of metabolites.
2.1 GC-MS-based analysis
GC-MS has long been used in metabolomics for the comprehensive analysis of metabolites due to its high separation capacity and its high selectivity and sensitivity. Furthermore, the hyphenation of GC with MS detection using electron-impact ionization (EI) with standard 70 eV ionization energy facilitates compound identification through mass spectra matching with known compounds from customized libraries or public databases. However, chemical derivatization is required for polar metabolites to improve their volatility, thermal stability, detectability, and retention index and also to minimize deleterious column adsorption. In fact, metabolome coverage by GC-MS is limited by the thermal stability of the stationary phase, as well as that of metabolites and their derivatives.15 In order to increase the coverage of detected metabolites, several novel technologies have been developed for GC-MS-based metabolomics. Ionic liquid stationary phases for GC-MS exhibit “dual nature” retention behavior; namely, they are able to separate polar molecules as if the stationary phase were polar, and nonpolar molecules as if the stationary phase were nonpolar.16 One limitation is that this type of stationary phase is unstable at high temperatures, though this can be overcome by cross-linking a new class of ionic liquid component to provide a more durable and robust stationary phase with high thermal stability (>350 °C).17 This dual-nature retention selectivity is useful for metabolite coverage in metabolomics; however, the applications of ionic liquids in metabolomics have yet to be fully explored, although the commercial availability of columns with different selectivities is expected to facilitate rapid growth in this area.18The advent of comprehensive two-dimensional (2D)-GC further enhances separation performance by coupling two columns coated with different stationary phases in series, which greatly increases peak capacity; as a result, the product of the peak capacity is in two dimensions. The investigation of volatile and derivatized metabolites in biological samples via comprehensive 2D-GC coupled with time-of-flight mass spectrometry (TOF-MS) can provide highly complex and information-rich data for comprehensive metabolomics analysis. The addition of a second separation dimension with a different chemical selectivity and fast-scanning TOF-MS offers better chemical selectivity and overall peak capacity compared with traditional one-dimensional gas chromatography (1D-GC). The use of 2D-GC-TOF-MS was recently applied to the untargeted and comprehensive analysis of the volatile composition of human urine. From the 700 total metabolites detected in the study, 294 were distributed among the chemical families of hydrocarbons, amines, amides, esters, ketones, aldehydes, alcohols, carboxylic acids, ethers, etc., providing the most complete information yet available on the volatile components of human urine.19 Optimized 2D GC-TOF-MS using a polar and nonpolar column connected in series is amenable to achieving a major increase in peak capacity to over 9000 resolved features within a short total analysis time.20
GC-MS is limited to volatile, thermally stable, and energetically stable compounds. Unfortunately, this approach is less suitable to the poor volatility of highly polar metabolites. Chromatographic analyses of these compounds usually rely on other separation techniques, such as LC and CE.
2.2 LC-MS-based analysis
LC-MS is an important tool in metabolomics because the molecular identification and quantification of polar, weakly polar, and neutral metabolites can be achieved using this technology, even at low concentrations or with high matrix effects. Soft ionization technologies such as electrospray ionization (ESI) and atmospheric pressure chemical ionization (APCI) are commonly used in LC-MS. As shown in Fig. 2, ESI is ideal for semi-polar and polar compounds, whereas APCI is more suitable for neutral or weakly polar compounds. Both of these ionization technologies can provide complementary data, which can help to detect more metabolite components. For instance, complementary ESI and APCI analyses resulted in a 20% increase in the number of detected ions in a human blood serum extract compared with ESI analysis alone.21 In addition, different polarity modes provide different information. Many MS instruments are now capable of fast polarity switching during data acquisition, and some can even obtain data in both positive and negative mode simultaneously.22 The use of both positive and negative ionization mode in LC-MS analysis provides more comprehensive metabolite coverage than the use of a single polarity mode. Because some analytes are detected only in the negative ion mode, whereas others are observed in the positive ion mode, Nordström et al.21 noted that more than 90% of the ions observed in human blood plasma in ESI or APCI positive ion mode were not detected in negative ion mode, and vice versa.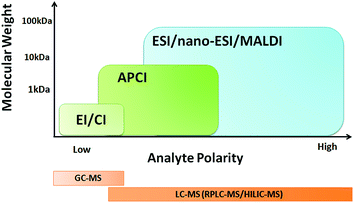 | ||
| Fig. 2 Suitability of different ionization sources for metabolomic analysis based on metabolite polarity and molecular weight. | ||
Column technology has also improved metabolite coverage and reduced analysis time. For example, instruments based on ultrahigh-pressure liquid chromatography (UHPLC) packed with 1.7 μm particle C18 columns provide higher resolution and improved sensitivity compared with common HPLC, along with a 2-fold shorter analysis time.23 The shorter analysis time also allows for two LC-MS injections, for positive and negative ion modes, in less than a single HPLC run time, thereby increasing the number and variety of small compounds that can be measured.
To date, reversed-phased (RP)-LC is the most commonly used chromatographic technique in metabolomics studies due to its good reproducibility and predictability, wide applicability to various classes of metabolites and mobile phase compatibility for coupling to ESI-MS. In addition, various reports have demonstrated the strong potential of RP-LC-MS for the global metabolic profiling of biological samples, and protocols are currently available for the analysis of urine, blood and tissues samples.24–26 Nevertheless, RP-LC systems are not suitable for all metabolites: highly polar and charged metabolites are difficult to retain and always co-elute with the void volume. Although the addition of ion-pairing agents can improve the analysis of polar compounds, this method is not suitable for RP-LC-MS due to its severe ion suppression and the possibility of ion source contamination. The advent of RP pentafluorophenylpropyl (PFPP) columns allows for metabolomics analyses in both reversed- and normal-phase retention. For instance, Yang et al. established a method using a PFPP column for metabolomics studies of bacteria, demonstrating that highly polar compounds exhibit good separation on the PFPP column and that the separation of isomers or metabolites with similar structures was improved using the PFPP column.27 Lv et al. also obtained similar results when separating a broad number of hydrophilic metabolites from biological samples with a PFPP column.28
Owing to the chemical diversity and different physicochemical properties of metabolites, single-column methods are not suitable for separating all metabolites in biological samples. Thus, orthogonal separation modes, such as the combined use of hydrophilic interaction chromatography (HILIC) and RP-LC, were applied to expand metabolome coverage of both polar and nonpolar metabolites.29 HILIC should be regarded as a complementary normal-phase LC separation mode for resolving polar metabolites that are poorly retained by RP-LC. Compared with conventional normal phase-LC, HILIC contains higher levels of organic modifiers (e.g., acetonitrile) in the mobile phase, which is more suitable for polar metabolites when using ESI-MS. Recently, a single extraction-dual LC-MS platform that combined use of RP-LC and HILIC was developed to allow the global profiling of both hydrophobic and hydrophilic metabolites.30 The value of this approach was illustrated through the metabolic profiling of bacterial cells, human cancer cells and human plasma, where the combined HILIC/RPLC-MS approach generated more than 3000 molecular features in each sample, with the highest number of unique features identified by RPLC in ESI positive mode and by HILIC in ESI negative mode. Greater coverage of human urine samples can be obtained when using HILIC-MS to complement RP-LC-MS, because urine contains a large number of polar and ionic metabolites.31 However, the major drawback of HILIC is its increased retention time drifts, as well as the rather long re-equilibration time between runs.32
Alternatively, aqueous normal phase (ANP)-LC offers minimal shift in retention time (±0.05 min) with a similar number of detectable metabolite features in human urine compared with RP-LC.33 As reported by Callahan et al., approximately 1000 compounds were reproducibly detected in human urine samples, and 400 compounds were detected in xylem fluid from soybean plants. This method was noted to greatly increase metabolite coverage over RP-only metabolite profiling in biological samples.33
2.3 CE-MS-based analysis and combination of different separation modes
CE-MS is now considered a powerful complementary analytical technique for the analysis of ionic metabolites in biological samples. Generally, CE-MS is ideal for the profiling of polar, nonpolar, and ionic classes of metabolites or degradation products from primary metabolism, as well as secondary metabolites that are weakly retained or have poor separation efficiency in RPLC or HILIC.34 Developments in and application of CE-MS for metabolomics between the years 2008 and 2014 have been well reviewed.35–37Separation techniques play a vital role in metabolomics for the comprehensive detection of metabolites in complex biological samples; different separation techniques based on chromatography and electrophoresis exhibit huge potential for different classes of metabolites. Thus, the combined use of different instrumental analytical approaches is necessary to achieve comprehensive analysis of the metabolites based on the selectivity of GC, LC, and CE, and it is beneficial to increasing the coverage of detected metabolites, which cannot be achieved by single-analysis techniques. The integrated platform also provides sensitive and reliable detection of thousands of metabolites in biological samples. t'Kindt et al. jointed the GC-MS and LC-MS platform to conduct comprehensive analysis in plant metabolomics.38 In Ibáñez's study, CE, RPLC and HILIC all coupled to TOF-MS were combined to achieve a global metabolomics investigation of the effect of dietary polyphenols on HT29 colon cancer cells.39 Due to its complementary nature, this multianalytical platform offers extensive metabolic information and coverage, which benefits the further interpretation of the biomarkers.
3. Rapid detection of metabolites
Hyphenated separation platforms offer improved metabolome coverage and high quality data, but relatively long separation times are needed in these technologies. Therefore, rapid analysis technologies, which provide high-throughput screening approaches, are necessary in large-scale metabolomics studies. Recently, numerous developments in MS ion sources, combined with accurate mass and/or tandem mass analyzers, have provided very dramatic approaches for metabolomics studies. Direct MS analysis, which is capable of high throughput, in conjunction with chemometrics data analysis offers a feasible method of large-scale analysis (e.g., screening in large-scale clinical trials), for which high throughput is mandatory. This section reviews a number of established and emerging technologies that allow for direct and rapid analysis in metabolomics.3.1 Infusion-related mass spectrometry
Two different strategies exist to introduce liquid samples into mass spectrometers: direct infusion mass spectrometry (DIMS) and flow infusion or flow injection mass spectrometry (FIMS) (reviewed in detail by Draper et al.40). DIMS refers to the direct analysis of complex biological samples without chromatographic separation by using a syringe pump or similar device to constantly introduce the fluid sample into the MS.41,42 Due to its rapid analysis speed and greater sensitivity relative to NMR, DIMS is very useful for large-scale metabolomics studies, with the possibility of analyzing several hundred samples per day. This technique has been successfully applied to profile low-abundance species in small samples on high-resolution mass spectrometers.43,44 The utilization of nanoelectrospray emitters also reduces the ionization suppression effects caused by competitive ionization with other components in the matrix due to the increased ionization efficiency of nano-MS.45FIMS introduces samples into an LC-MS running solvent, which then flows to the electrospray ionization interface at a rate of 100–1000 μl min−1. Because the peak width typically ranges between 5 and 15 s, close to 300 samples can be analyzed in an hour, and 1000–2000 samples can be achieved per day with a standard LC-MS device.46 Sévin et al. detected 8961 ions by using FIMS in Escherichia coli, of which 535 could be annotated based on the features of a theoretical m/z within a 0.001 Da mass tolerance.47 Madalinski et al. reported that a few thousand m/z signals were observed in yeast extracts using FIMS with an acquisition of 3 min, 400 of which were found to be analytically relevant (e.g., the intensity was 3-fold higher than that of the background noise, and the signals comprised at least 60% of the acquisition profiles under the same experimental conditions).48 Dense metabolite coverage and high sensitivity can be achieved in a short analysis time using FIMS; however, as with DIMS, chemical isomers cannot be distinguished by this rapid screening technique, and ion suppression is a concern in ESI-MS. A compromise between rapid analysis speed and the separation of isomers is required when using these methods. The separation of isobaric compounds needs other separation methods. Furthermore, matrix effects can be reduced by proper sample pretreatment, for instance, sample desalting via solid phase extraction (SPE);49 more than 5000 injections per day are possible using online high-throughput SPE/MS.50
3.2 Ambient ionization techniques
Ambient ionization MS allows the rapid analysis of samples or objects in the open environment with little or no sample preparation and promises to enable in situ investigation of diverse analytes. The development of ambient ionization was initiated with the introduction of DESI by Cooks in 2004.51 Since then, a variety of approaches based on different desorption and ionization methods have been developed, and almost 30 ambient ionization approaches have been used in MS analysis, as reviewed by Monge et al.52 Among these, two recent techniques in ambient ionization sources, named DESI51 and DART,53 are the most popular and have been widely used for rapid analyses across a diverse range of applications.54,55 DESI is a novel MS ionization technique that was pioneered by the Cooks group in 2004. In the DESI process, a spray of charged micro-droplets (e.g., acidic methanol–water) from a pneumatically assisted electrospray needle is directed towards the analyte of interest under atmospheric conditions, picking up the sample through interactions with the surface and then generating charged ions of the analyte in the gas phase that can be directed into a mass spectrometer. By this means, mass spectra similar to those obtained from ESI can be acquired rapidly and with little or no sample preparation. DESI-MS has proven its usefulness in the high-throughput differential metabolomics of biological samples without sample preparation.56 Jackson et al. monitored the presence of targeted central carbon metabolites in the cell extracts and quench supernatants of Escherichia coli by using DESI-MS.57DART is another ambient ionization technology, which was developed by Cody and colleagues in 2005.53 This solvent-free method relies on the desorption of analytes using a stream of hot gas (usually helium gas or nitrogen gas); the gas carries active species derived from a plasma discharge flowing from the ion source onto a sample surface and is responsible for the desorption and ionization of analyte molecules from the sample surface. DART ionization is followed by APCI processes and thus results in APCI-like spectra. Although developed later than DESI-MS, DART-MS has shown tremendous potential for high-throughput and real-time direct analyses of small molecules from biological matrices in metabolomics. Zhou et al.58 firstly reported a rapid approach by using DART-TOF-MS for the metabolomics fingerprinting of human serum, and each DART run requires only 1.2 min. Since the first report by Zhou et al., DART applications in the field of metabolomics have grown substantially. Over 2010–2014, DART-MS has been used in many aspects of metabolomics: DART-MS combined with multivariate statistical analysis was employed as a tool for beer origin recognition,59 for the authentication of tomatoes and peppers from organic and conventional farming,60 and for discrimination analysis of the geographical origin of Angelica gigas roots collected from Korea and China;61 a high-throughput DART-MS approach also enabled the examination of chicken muscle and feed by recording the metabolomic fingerprints of ionizable compounds;62 breast cancer detection has been performed by combining DART and NMR metabolomics data;63 and DART has been used for other applications, including skin metabolome changes64 and fish metabolomics.65
Other ambient ionization techniques that display potential for high-throughput analysis have also been used in metabolomics studies. EESI, an extension of DESI, uses two separate sprayers, one to nebulize a sample solution and another to produce charged droplets from a spray solution. Analyte molecules undergo interactions and collisions with the charged droplets and become ionized for mass spectral analysis.66 EESI-MS provides a means to analyze metabolites directly from biological fluids (e.g., urine) without sample preparation.67,68 A paper spray (PS) ionization approach was recently developed for the rapid, high-throughput and direct analysis of complex mixtures on a paper substrate in open-environment conditions.69,70 The sample is preloaded onto a triangular piece of paper using wetting solution or is transferred from the surface by using the paper as a wipe. A high voltage (3–5 kV) is applied to the paper, and ions of the analyte are generated for MS analysis. PS-MS allows for the rapid and direct detection of different types of samples, such as hormones, lipids, and therapeutic drugs, with minimal sample preparation and short analysis times (less than 1 min) using a small volume of tissue sample (typically 1 mm3 or less).71 Hamid and coworkers72 used PS-MS combined with multivariate statistical analysis for the rapid discrimination of bacteria, and no sample preparation or lipid extraction steps were required prior to analysis. Laser ablation-electrospray ionization (LAESI), another ambient ionization technique, uses the native water content of a sample as the matrix and exhibits potential for high-throughput metabolomics. Nemes et al. reported that this technique is capable of detecting a variety of compounds and size ranges (up to 66 kDa) without any sample preparation.73 Recently, Sripadi et al. extended this technique to the in vitro analysis of animal tissue metabolites from an untreated Torpedo californica electric organ.74
In summary, the ambient ionization techniques described above provide ionization in open air under atmospheric pressure. Although most of these techniques are still in the early stages, progress in their development will meaningfully impact the analysis of biological samples. These techniques hold great promise for rapid, in situ metabolome analysis with high throughput. But these techniques are not suitable for all metabolites; for instance, DART is suitable for volatile compounds (volatile oil, small molecule coumarin), nitrogen-containing compounds (alkaloids) and some carbonyl-containing compounds (flavonoids and anthraquinone), but it is not suitable for glucosides without derivatization.75 Thus, we should consider the analyte's chemical characteristics to choose the appropriate technique in metabolomics study.
4. Easy identification of metabolites
The identification of metabolites in MS-based metabolomics studies largely relies on accurate mass, isotopic distribution, tandem mass spectral fragmentation patterns and other information provided by MS, as well as the reference standards. For GC-MS, retention time and mass spectra can be compared with the NIST76 or Fiehn metabolomics databases77 for identification of metabolites due to the reproducible fragmentation patterns produced by EI. For LC-MS, a high mass accuracy can be obtained by using a high-resolution mass spectrometer (e.g., TOF, Orbitrap and FT-ICR), which can greatly reduce the number of potential molecular formulas related to one mass peak. When combined with the fragmentation patterns, numerous isomers corresponding to a single formula can be reduced to a narrow set of structures or even one structure. In practice, the development of metabolomics databases has also provided useful information on metabolite identification. Several commonly used public databases for metabolite identification or pathway analyses are listed in Table 1. Additionally, there are also commercially accessible databases, such as the Wiley MS libraries and NIST. Many of these databases support spectral searches and spectral matching assessment (e.g., NIST and MassBank). In addition to these spectral matching resources, several databases also allow the use of high-accuracy mass weight to help with metabolite identification. They support mass searching and mass deconvolution (including adducts, multiply charged and neutral loss fragments) such as HMDB and METLIN. Several commercial chemical calculators are available to generate reasonable chemical formulas from accurate mass data and isotopic distribution. Once a formula is determined, the compound can be tentatively identified by searching databases (Table 1). Although there are many different databases related to the metabolomics, the identification of metabolites is still very tough work because of the isomers and the lack of metabolite standards.| Database | No. Records | Note | Website | References |
|---|---|---|---|---|
| NIST Chemistry WebBook | N/A | MS | http://webbook.nist.gov/chemistry/ | 86 |
| Golm Metabolome Database (GMD) | N/A | MS | http://gmd.mpimp-golm.mpg.de/ | 87 |
| ChemSpider | ∼25 million unique chemical compounds | N/A | http://www.chemspider.com/ | 88 |
| Human Metabolome Database (HMDB) | ∼41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 818 metabolites 818 metabolites |
MS, MS/MS | http://www.hmdb.ca | 13 |
| Metabolite and Tandem MS Database (METLIN) | >64![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 metabolites 000 metabolites |
MS, MS/MS, | http://metlin.scripps.edu/index.php | 89 |
| MassBank | ∼40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 889 spectra 889 spectra |
MS, MSn | http://www.massbank.jp/ | 90 |
| PubChem | >700![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 compounds 000 compounds |
N/A | https://pubchem.ncbi.nlm.nih.gov/ | 91 |
| Madison Metabolomics Consortium Database (MMCD) | ∼19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 metabolites 700 metabolites |
MS | http://mmcd.nmrfam.wisc.edu/ | 92 |
| Manchester Metabolomics Database (MMD) | 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 687 endogenous and exogenous metabolite species 687 endogenous and exogenous metabolite species |
N/A | http://dbkgroup.org/MMD/ | 93 |
| KNApSAcK | 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 899 metabolites and 109 899 metabolites and 109![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 820 metabolite species entries 820 metabolite species entries |
N/A | http://kanaya.naist.jp/KNApSAcK/ | 94 |
| MyCompound ID | 8021 known human endogenous metabolites and their predicted metabolic products | MS/MS fragmentation search | http://www.mycompoundid.org/mycompoundid_IsoMS/index.jsp | 95 and 96 |
| Platform for Riken Metabolomics (PRIMe) | >1 million entries of untargeted MS/MS data of plant metabolites | MS, MS/MS | http://prime.psc.riken.jp | 97 and 98 |
| Spectral Data Base (SDBS) | ∼34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 compounds 000 compounds |
MS | http://sdbs.db.aist.go.jp/sdbs/cgi-bin/direct_frame_top.cgi | |
| Chemical Entities of Biological Interest (ChEBI) | >12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 molecular entities, groups and classes 000 molecular entities, groups and classes |
N/A | http://www.ebi.ac.uk/chebi/ | 99 |
| Lipid Bank | ∼7009 lipid class entries | Lipid metabolomics | http://lipidbank.jp/index.html | 100 |
| Lipid Maps | ∼37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 566 lipids 566 lipids |
Lipid metabolomics | http://www.lipidmaps.org/ | 101 and 102 |
| Kyoto Encyclopedia of Genes and Genomes (KEGG) | ∼17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 348 metabolites and other small molecules, ∼343 348 metabolites and other small molecules, ∼343![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 127 pathway maps 127 pathway maps |
Pathway analysis | http://www.genome.jp/kegg/ | 103 |
| MassTRIX | Mass translator into pathways | http://masstrix.org | 104 | |
| MetaCyc | ∼2260 pathways from 2600 different organisms | Pathway database (organism) | http://metacyc.org/ | 105 |
| BioCyc | ∼5500 pathways | Pathway database | http://biocyc.org | 105 |
| EcoCyc | N/A | Pathway database (bacterium Escherichia coli K-12) | http://ecocyc.org | 106 |
| HumanCyc | N/A | Pathway database (human) | http://humancyc.org | 107 |
| Plant Metabolic Network (PMN) | ∼350 plant species metabolic pathways | Pathway databases (plant) | http://www.plantcyc.org | 105 |
| Reactome | ∼7088 human proteins participating in 6744 reactions | Pathway database | http://www.reactome.org/ | 108 |
Recently, several new identification methodologies based on traditional methods have been rapidly developed. In GC-MS-based metabolomics studies, the identification of unknown peaks and subsequent structural elucidation remain necessary due to the chemical diversity of metabolites, and true unknown metabolites or novel derivatives of known components cannot be identified through database searches. Peterson et al. combined GC with Q-Orbitrap MS, which can provide high-resolution, mass-accurate MS detection to archive a strict filter for candidate chemical formulas.78,79 13C- and 15N-isotope labeling coupled with molecular ion-directed acquisition (MIDA) was then applied to obtain information-rich MS/MS spectra and also to count the number of carbon and nitrogen atoms present in the precursor and product ion species, which provides a novel way to identify the metabolites. In order to reduce complexity and simplify spectral interpretation of unknown metabolites, a new approach termed ratio analysis of mass spectrometry (RAMSY), which facilitates improved compound identification in complex MS spectra, was presented by Gu et al.80 This approach is based on the principle that under the same experimental conditions, the abundance/intensity ratios between mass fragment patterns from the same compound are relatively steady. This method has been used with both GC/MS and LC/MS. RAMSY has typically been shown to perform better than correlation methods and also to improve unknown metabolite identification for MS users in metabolomics or other fields.
For the purpose of obtaining MS/MS information for most detected metabolites, a procedure named gas-phase fractionation (GPF) was used for serum analysis by Calderón-Santiago et al., which provided useful MS/MS information for at least 80% of entities detected in the MS scan, compared with 48–57% from the traditional auto MS/MS data acquisition mode.81 The MS/MS information obtained using the GPF method will be helpful in metabolite identification. In target MS/MS data acquisition mode, narrow isolation windows should always be used to minimize contaminating MS fragments; however, even narrow windows can reduce the selectivity and sensitivity. In addition, a considerable portion of compounds are not purely isolated in the collision cell for MS fragmentation due to the chemical complexity of most biological samples. To address this issue, a novel metabolomics workflow was introduced to obtain high-quality MS/MS data for metabolite structure identification. This approach involves the experimental deconvolution of metabolomics MS/MS data acquired by using wide MS isolation windows (e.g., 9 Da) shifted over the precursor m/z region of interest. An R package implementing the algorithms named decoMS2 used in this workflow provided an unbiased mechanism for producing high-quality MS/MS spectra for structure identification.82
Metabolites with modifications such as phosphorylation, acetylation, methylation, glucuronidation, and sulfation are commonly assigned as unknowns, which causes a significant loss of valuable information in metabolomics studies. In order to gain some insight into these unknown metabolites, an untargeted, modification-specific metabolomics (NT-MSM) strategy was developed.83 The modification information obtained using this method greatly reduces the number of matches for metabolite identification during database searching and thus significantly cuts down the time required in this process. Similarly, Mitchell et al. developed an algorithm for the detection of functional groups within metabolite databases to increase metabolite identification rates in untargeted metabolomics studies.84
IMMS is an emerging separation technique in metabolomics, especially during the metabolite identification step. As we all know, the variation among samples caused by different matrixes or sample loading methods can cause shifts in retention times, which complicates the use of retention time for the purpose of metabolite identification. In IMMS, ions are separated according to their mobility in a carrier buffer gas based on their size-to-charge ratio and their interactions with the buffer gas, which provides a complement to traditional MS analysis. The collision cross-section (CCS) measured by IMMS is a unique physicochemical molecular property that can be used as an orthogonal molecule descriptor in addition to retention time and m/z. The use of CCS offers an opportunity to improve metabolite identification and also makes identification more robust and reproducible. The CCS approach remains largely unexplored in metabolomics studies; however, many scientists have applied this method in metabolite identification. Paglia et al. measured CCS values for 125 metabolites using IMMS across three independent laboratories, indicating high intra- and inter-laboratory reproducibility.85 They also generated a CCS database of common cellular metabolites to confidently identify metabolite changes related to the epithelial–mesenchymal transition. This group also investigated the use of CCS as a highly specific molecular characteristic for identifying lipids in biological samples. However, the CCS database is still in the early stage; further studies should be encouraged to extend and populate the present metabolite database with CCS values for metabolomics and other small-molecule applications.
All these methods make the identification of metabolites easier, but some of them are very complicated for users because they require programming knowledge. Thus, approaches that are easy to operate should be explored in the future.
5. Summary and perspective
In the field of metabolomics, scientists seek the comprehensive analysis, rapid detection and easy identification of metabolites. As a commonly used analytical platform, MS plays an crucial role in these studies due to its high sensitivity, selectivity, and easy combination with other chromatographic techniques. The development of novel MS and chromatographic techniques has provided approaches to detect a wide range of metabolites. Furthermore, the emergence of ambient ionization sources allows the rapid detection of metabolites. The established metabolome databases and the evolving identification methodologies simplify the process of metabolite identification. However, so far there is no single approach that can simultaneously provide high throughput and comprehensive metabolite coverage; thus, a compromise between these two aspects is required in metabolomics studies. In addition, some false-positive and false-negative structural information still arise during metabolite identification. Compared with genomics and proteomics, the analytical platform for metabolomics is not yet mature. There is still a considerable need to develop new technologies to enable expanded metabolite coverage, reduced detection time, and simplified metabolite identification.Conflict of interest
The authors declare no conflicts of interest.Acknowledgements
The present study was supported by grants from the Research Committee of the University of Macau (MYRG123-ICMS12 and MYRG111-ICMS13 to JB Wan) and from the Macao Science and Technology Development Fund (010/2013/A1 to JB Wan).References
- S. G. Villas-Bôas, S. Mas, M. Åkesson, J. Smedsgaard and J. Nielsen, Mass Spectrom. Rev., 2005, 24, 613–646 CrossRef PubMed
.
- R. Kaddurah-Daouk, B. S. Kristal and R. M. Weinshilboum, Annu. Rev. Pharmacol. Toxicol., 2008, 48, 653–683 CrossRef CAS PubMed
.
- D. Beyoğlu and J. R. Idle, Biochem. Pharmacol., 2013, 85, 12–20 CrossRef PubMed
.
- J. Stewart and H. Bolt, Arch. Toxicol., 2011, 85, 3–4 CrossRef CAS PubMed
.
- M. Wang, R. J. A. Lamers, H. A. Korthout, J. H. van Nesselrooij, R. F. Witkamp, R. van der Heijden, P. J. Voshol, L. M. Havekes, R. Verpoorte and J. van der Greef, Phytother. Res., 2005, 19, 173–182 CrossRef CAS PubMed
.
- A. Buriani, M. L. Garcia-Bermejo, E. Bosisio, Q. Xu, H. Li, X. Dong, M. S. Simmonds, M. Carrara, N. Tejedor and J. Lucio-Cazana, J. Ethnopharmacol., 2012, 140, 535–544 CrossRef PubMed
.
- C. Hu and G. Xu, TrAC, Trends Anal. Chem., 2014, 61, 207–214 CrossRef CAS PubMed
.
- R.-J. Raterink, P. W. Lindenburg, R. J. Vreeken, R. Ramautar and T. Hankemeier, TrAC, Trends Anal. Chem., 2014, 61, 157–167 CrossRef CAS PubMed
.
- M. M. Koek, R. H. Jellema, J. van der Greef, A. C. Tas and T. Hankemeier, Metabolomics, 2011, 7, 307–328 CrossRef CAS PubMed
.
- D. S. Wishart, Bioanalysis, 2011, 3, 1769–1782 CrossRef CAS PubMed
.
- D. S. Wishart, C. Knox, A. C. Guo, R. Eisner, N. Young, B. Gautam, D. D. Hau, N. Psychogios, E. Dong and S. Bouatra, Nucleic Acids Res., 2009, 37, D603–D610 CrossRef CAS PubMed
.
- N. Psychogios, D. D. Hau, J. Peng, A. C. Guo, R. Mandal, S. Bouatra, I. Sinelnikov, R. Krishnamurthy, R. Eisner and B. Gautam, PLoS One, 2011, 6, e16957 CAS
.
- D. S. Wishart, T. Jewison, A. C. Guo, M. Wilson, C. Knox, Y. Liu, Y. Djoumbou, R. Mandal, F. Aziat and E. Dong, Nucleic Acids Res., 2012, gks1065 Search PubMed
.
- O. Fiehn, Plant Mol. Biol., 2002, 48, 155–171 CrossRef CAS
.
- E. Kaal and H.-G. Janssen, J. Chromatogr. A, 2008, 1184, 43–60 CrossRef CAS PubMed
.
- J. L. Anderson, D. W. Armstrong and G.-T. Wei, Anal. Chem., 2006, 78, 2892–2902 CrossRef
.
- J. L. Anderson and D. W. Armstrong, Anal. Chem., 2005, 77, 6453–6462 CrossRef CAS PubMed
.
- C. F. Poole and S. K. Poole, J. Sep. Sci., 2011, 34, 888–900 CrossRef CAS PubMed
.
- S. M. Rocha, M. Caldeira, J. Carrola, M. Santos, N. Cruz and I. F. Duarte, J. Chromatogr. A, 2012, 1252, 155–163 CrossRef CAS PubMed
.
- M. M. Koek, B. Muilwijk, L. L. van Stee and T. Hankemeier, J. Chromatogr. A, 2008, 1186, 420–429 CrossRef CAS PubMed
.
- A. Nordström, E. Want, T. Northen, J. Lehtiö and G. Siuzdak, Anal. Chem., 2008, 80, 421–429 CrossRef PubMed
.
- M. Yuan, S. B. Breitkopf, X. Yang and J. M. Asara, Nat. Protoc., 2012, 7, 872–881 CrossRef CAS PubMed
.
- A. M. Evans, C. D. DeHaven, T. Barrett, M. Mitchell and E. Milgram, Anal. Chem., 2009, 81, 6656–6667 CrossRef CAS PubMed
.
- W. B. Dunn, D. Broadhurst, P. Begley, E. Zelena, S. Francis-McIntyre, N. Anderson, M. Brown, J. D. Knowles, A. Halsall and J. N. Haselden, Nat. Protoc., 2011, 6, 1060–1083 CrossRef CAS PubMed
.
- E. J. Want, P. Masson, F. Michopoulos, I. D. Wilson, G. Theodoridis, R. S. Plumb, J. Shockcor, N. Loftus, E. Holmes and J. K. Nicholson, Nat. Protoc., 2013, 8, 17–32 CrossRef CAS PubMed
.
- E. J. Want, I. D. Wilson, H. Gika, G. Theodoridis, R. S. Plumb, J. Shockcor, E. Holmes and J. K. Nicholson, Nat. Protoc., 2010, 5, 1005–1018 CrossRef CAS PubMed
.
- S. Yang, M. Sadilek and M. E. Lidstrom, J. Chromatogr. A, 2010, 1217, 7401–7410 CrossRef CAS PubMed
.
- H. Lv, G. Palacios, K. Hartil and I. J. Kurland, J. Proteome Res., 2011, 10, 2104–2112 CrossRef CAS PubMed
.
- P. Yin, D. Wan, C. Zhao, J. Chen, X. Zhao, W. Wang, X. Lu, S. Yang, J. Gu and G. Xu, Mol. BioSyst., 2009, 5, 868–876 RSC
.
- J. Ivanisevic, Z.-J. Zhu, L. Plate, R. Tautenhahn, S. Chen, P. J. O'Brien, C. H. Johnson, M. A. Marletta, G. J. Patti and G. Siuzdak, Anal. Chem., 2013, 85, 6876–6884 CrossRef CAS PubMed
.
- T. Zhang, D. J. Creek, M. P. Barrett, G. Blackburn and D. G. Watson, Anal. Chem., 2012, 84, 1994–2001 CrossRef CAS PubMed
.
- R. Ramautar and G. J. de Jong, Bioanalysis, 2014, 6, 1011–1026 CrossRef CAS PubMed
.
- D. L. Callahan, D. D. Souza, A. Bacic and U. Roessner, J. Sep. Sci., 2009, 32, 2273–2280 CrossRef CAS PubMed
.
- N. L. Kuehnbaum and P. Britz-McKibbin, Chem. Rev., 2013, 113, 2437–2468 CrossRef CAS PubMed
.
- R. Ramautar, O. A. Mayboroda, G. W. Somsen and G. J. de Jong, Electrophoresis, 2011, 32, 52–65 CrossRef CAS PubMed
.
- R. Ramautar, G. W. Somsen and G. J. de Jong, Electrophoresis, 2013, 34, 86–98 CrossRef CAS PubMed
.
- R. Ramautar, G. W. Somsen and G. J. de Jong, Electrophoresis, 2015, 36, 212–224 CrossRef CAS PubMed
.
- R. t'Kindt, K. Morreel, D. Deforce, W. Boerjan and J. van Bocxlaer, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2009, 877, 3572–3580 CrossRef PubMed
.
- C. Ibáñez, C. Simó, V. García-Cañas, Á. Gómez-Martínez, J. A. Ferragut and A. Cifuentes, Electrophoresis, 2012, 33, 2328–2336 CrossRef PubMed
.
- J. Draper, A. J. Lloyd, R. Goodacre and M. Beckmann, Metabolomics, 2013, 9, 4–29 CrossRef CAS
.
- R. Goodacre, S. Vaidyanathan, G. Bianchi and D. B. Kell, Analyst, 2002, 127, 1457–1462 RSC
.
- A. Koulman, B. A. Tapper, K. Fraser, M. Cao, G. A. Lane and S. Rasmussen, Rapid Commun. Mass Spectrom., 2007, 21, 421–428 CrossRef CAS PubMed
.
- P. Chumnanpuen, M. A. E. Hansen, J. Smedsgaard and J. Nielsen, Int. J. Genomics, 2014, 2014, 894296 Search PubMed
.
- C. Junot, G. Madalinski, J.-C. Tabet and E. Ezan, Analyst, 2010, 135, 2203–2219 RSC
.
- A. D. Southam, T. G. Payne, H. J. Cooper, T. N. Arvanitis and M. R. Viant, Anal. Chem., 2007, 79, 4595–4602 CrossRef CAS PubMed
.
- T. Fuhrer, D. Heer, B. Begemann and N. Zamboni, Anal. Chem., 2011, 83, 7074–7080 CrossRef CAS PubMed
.
- D. C. Sévin and U. Sauer, Nat. Chem. Biol., 2014, 10, 266–272 CrossRef PubMed
.
- G. Madalinski, E. Godat, S. Alves, D. Lesage, E. Genin, P. Levi, J. Labarre, J.-C. Tabet, E. Ezan and C. Junot, Anal. Chem., 2008, 80, 3291–3303 CrossRef CAS PubMed
.
- M. Rogeberg, H. Malerod, H. Roberg-Larsen, C. Aass and S. R. Wilson, J. Pharm. Biomed. Anal., 2014, 87, 120–129 CrossRef CAS PubMed
.
- W. Jian, M. V. Romm, R. W. Edom, V. P. Miller, W. A. LaMarr and N. Weng, Anal. Chem., 2011, 83, 8259–8266 CrossRef CAS PubMed
.
- Z. Takats, J. M. Wiseman, B. Gologan and R. G. Cooks, Science, 2004, 306, 471–473 CrossRef CAS PubMed
.
- M. E. Monge, G. A. Harris, P. Dwivedi and F. M. Fernández, Chem. Rev., 2013, 113, 2269–2308 CrossRef CAS PubMed
.
- R. B. Cody, J. A. Laramée and H. D. Durst, Anal. Chem., 2005, 77, 2297–2302 CrossRef CAS PubMed
.
- J. H. Gross, Anal. Bioanal. Chem., 2014, 406, 63–80 CrossRef CAS PubMed
.
- D. R. Ifa, C. Wu, Z. Ouyang and R. G. Cooks, Analyst, 2010, 135, 669–681 RSC
.
- H. Chen, Z. Pan, N. Talaty, D. Raftery and R. G. Cooks, Rapid Commun. Mass Spectrom., 2006, 20, 1577–1584 CrossRef CAS PubMed
.
- A. U. Jackson, S. R. Werner, N. Talaty, Y. Song, K. Campbell, R. G. Cooks and J. A. Morgan, Anal. Biochem., 2008, 375, 272–281 CrossRef CAS PubMed
.
- M. Zhou, J. F. McDonald and F. M. Fernández, J. Am. Soc. Mass Spectrom., 2010, 21, 68–75 CrossRef CAS PubMed
.
- T. Cajka, K. Riddellova, M. Tomaniova and J. Hajslova, Metabolomics, 2011, 7, 500–508 CrossRef CAS
.
- H. Novotná, O. Kmiecik, M. Gałązka, V. Krtková, A. Hurajová, V. Schulzová, E. Hallmann, E. Rembiałkowska and J. Hajšlová, Food Addit. Contam., Part A, 2012, 29, 1335–1346 CrossRef PubMed
.
- H. J. Kim, Y. T. Seo, S.-I. Park, S. H. Jeong, M. K. Kim and Y. P. Jang, Metabolomics, 2015, 11, 64–70 CrossRef CAS
.
- T. Cajka, H. Danhelova, M. Zachariasova, K. Riddellova and J. Hajslova, Metabolomics, 2013, 9, 545–557 CrossRef CAS
.
- H. Gu, Z. Pan, B. Xi, V. Asiago, B. Musselman and D. Raftery, Anal. Chim. Acta, 2011, 686, 57–63 CrossRef CAS PubMed
.
- H. M. Park, H. J. Kim, Y. P. Jang and S. Y. Kim, Biomol. Ther., 2013, 21, 470 CrossRef PubMed
.
- T. Cajka, H. Danhelova, A. Vavrecka, K. Riddellova, V. Kocourek, F. Vacha and J. Hajslova, Talanta, 2013, 115, 263–270 CrossRef CAS PubMed
.
- H. Chen, A. Venter and R. G. Cooks, Chem. Commun., 2006, 2042–2044 RSC
.
- Z. Zhou, M. Jin, J. Ding, Y. Zhou, J. Zheng and H. Chen, Metabolomics, 2007, 3, 101–104 CrossRef CAS
.
- H. Gu, H. Chen, Z. Pan, A. U. Jackson, N. Talaty, B. Xi, C. Kissinger, C. Duda, D. Mann and D. Raftery, Anal. Chem., 2007, 79, 89–97 CrossRef CAS PubMed
.
- J. Liu, H. Wang, N. E. Manicke, J.-M. Lin, R. G. Cooks and Z. Ouyang, Anal. Chem., 2010, 82, 2463–2471 CrossRef CAS PubMed
.
- L. Shen, J. Zhang, Q. Yang, N. E. Manicke and Z. Ouyang, Clin. Chim. Acta, 2013, 420, 28–33 CrossRef CAS PubMed
.
- H. Wang, N. E. Manicke, Q. Yang, L. Zheng, R. Shi, R. G. Cooks and Z. Ouyang, Anal. Chem., 2011, 83, 1197–1201 CrossRef CAS PubMed
.
- A. M. Hamid, A. K. Jarmusch, V. Pirro, D. H. Pincus, B. G. Clay, G. Gervasi and R. G. Cooks, Anal. Chem., 2014, 86, 7500–7507 CrossRef CAS PubMed
.
- P. Nemes and A. Vertes, Anal. Chem., 2007, 79, 8098–8106 CrossRef CAS PubMed
.
- P. Sripadi, J. Nazarian, Y. Hathout, E. P. Hoffman and A. Vertes, Metabolomics, 2009, 5, 263–276 CrossRef CAS
.
- Y. Wang, C. Li, L. Huang, L. Liu, Y. Guo, L. Ma and S. Liu, Anal. Chim. Acta, 2014, 845, 70–76 CrossRef CAS PubMed
.
- H. Wu, R. Xue, C. Lu, C. Deng, T. Liu, H. Zeng, Q. Wang and X. Shen, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2009, 877, 3111–3117 CrossRef CAS PubMed
.
- T. Kind, G. Wohlgemuth, D. Y. Lee, Y. Lu, M. Palazoglu, S. Shahbaz and O. Fiehn, Anal. Chem., 2009, 81, 10038–10048 CrossRef CAS PubMed
.
- A. C. Peterson, J. P. Hauschild, S. T. Quarmby, D. Krumwiede, O. Lange, R. A. Lemke, F. Grosse-Coosmann, S. Horning, T. J. Donohue, M. S. Westphall, J. J. Coon and J. Griep-Raming, Anal. Chem., 2014, 86, 10036–10043 CrossRef CAS PubMed
.
- A. C. Peterson, A. J. Balloon, M. S. Westphall and J. J. Coon, Anal. Chem., 2014, 86, 10044–10051 CrossRef CAS PubMed
.
- H. Gu, G. N. Gowda, F. C. Neto, M. R. Opp and D. Raftery, Anal. Chem., 2013, 85, 10771–10779 CrossRef CAS PubMed
.
- M. Calderón-Santiago, F. Priego-Capote and M. D. Luque de Castro, Anal. Chem., 2014, 86, 7558–7565 CrossRef PubMed
.
- I. Nikolskiy, N. G. Mahieu, Y.-J. Chen, R. Tautenhahn and G. J. Patti, Anal. Chem., 2013, 85, 7713–7719 CrossRef CAS PubMed
.
- W. Dai, P. Yin, Z. Zeng, H. Kong, H. Tong, Z. Xu, X. Lu, R. Lehmann and G. Xu, Anal. Chem., 2014, 86, 9146–9153 CrossRef CAS PubMed
.
- J. M. Mitchell, T. W.-M. Fan, A. N. Lane and H. N. Moseley, Front. Genet., 2014, 5, 237 Search PubMed
.
- G. Paglia, J. P. Williams, L. Menikarachchi, J. W. Thompson, R. Tyldesley-Worster, S. Halldorsson, O. Rolfsson, A. Moseley, D. Grant, J. Langridge, B. O. Palsson and G. Astarita, Anal. Chem., 2014, 86, 3985–3993 CrossRef CAS PubMed
.
- S. Stein, R. Brown, P. Linstrom and W. Mallard, NIST Chemistry WebBook, NIST Standard Reference Database Number 69, National Institute of Standards and Technology (NIST), 2011 Search PubMed
.
- J. Hummel, N. Strehmel, J. Selbig, D. Walther and J. Kopka, Metabolomics, 2010, 6, 322–333 CrossRef CAS PubMed
.
- H. E. Pence and A. Williams, J. Chem. Educ., 2010, 87, 1123–1124 CrossRef CAS
.
- R. Tautenhahn, K. Cho, W. Uritboonthai, Z. Zhu, G. J. Patti and G. Siuzdak, Nat. Biotechnol., 2012, 30, 826–828 CrossRef CAS PubMed
.
- H. Horai, M. Arita, S. Kanaya, Y. Nihei, T. Ikeda, K. Suwa, Y. Ojima, K. Tanaka, S. Tanaka and K. Aoshima, J. Mass Spectrom., 2010, 45, 703–714 CrossRef CAS PubMed
.
- Y. Wang, J. Xiao, T. O. Suzek, J. Zhang, J. Wang and S. H. Bryant, Nucleic Acids Res., 2009, 37, W623–W633 CrossRef CAS PubMed
.
- Q. Cui, I. A. Lewis, A. D. Hegeman, M. E. Anderson, J. Li, C. F. Schulte, W. M. Westler, H. R. Eghbalnia, M. R. Sussman and J. L. Markley, Nat. Biotechnol., 2008, 26, 162–164 CrossRef CAS PubMed
.
- M. Brown, W. B. Dunn, P. Dobson, Y. Patel, C. Winder, S. Francis-McIntyre, P. Begley, K. Carroll, D. Broadhurst and A. Tseng, Analyst, 2009, 134, 1322–1332 RSC
.
- Y. Shinbo, Y. Nakamura, M. Altaf-Ul-Amin, H. Asahi, K. Kurokawa, M. Arita, K. Saito, D. Ohta, D. Shibata and S. Kanaya, in Plant Metabolomics, ed. K. Saito, R. A. Dixon and L. Willmitzer, Springer, 2006, pp. 165–181 Search PubMed
.
- L. Li, R. Li, J. Zhou, A. Zuniga, A. E. Stanislaus, Y. Wu, T. Huan, J. Zheng, Y. Shi and D. S. Wishart, Anal. Chem., 2013, 85, 3401–3408 CrossRef CAS PubMed
.
- Y. Tang, R. Li, G. Lin and L. Li, Anal. Chem., 2014, 86, 3568–3574 CrossRef CAS PubMed
.
- K. Akiyama, E. Chikayama, H. Yuasa, Y. Shimada, T. Tohge, K. Shinozaki, M. Y. Hirai, T. Sakurai, J. Kikuchi and K. Saito, In Silico Biol., 2008, 8, 339–345 CAS
.
- T. Sakurai, Y. Yamada, Y. Sawada, F. Matsuda, K. Akiyama, K. Shinozaki, M. Y. Hirai and K. Saito, Plant Cell Physiol., 2013, 54, e5 CrossRef CAS PubMed
.
- K. Degtyarenko, P. de Matos, M. Ennis, J. Hastings, M. Zbinden, A. Mcnaught, R. Alcántara, M. Darsow, M. Guedj and M. Ashburner, Nucleic Acids Res., 2008, 36, D344–D350 CrossRef CAS PubMed
.
- K. Watanabe, E. Yasugi and M. Oshima, Trends Glycosci. Glycotechnol., 2000, 12, 175–184 CrossRef CAS
.
- E. Fahy, M. Sud, D. Cotter and S. Subramaniam, Nucleic Acids Res., 2007, 35, W606–W612 CrossRef PubMed
.
- M. Sud, E. Fahy, D. Cotter, A. Brown, E. A. Dennis, C. K. Glass, A. H. Merrill, R. C. Murphy, C. R. Raetz and D. W. Russell, Nucleic Acids Res., 2007, 35, D527–D532 CrossRef CAS PubMed
.
- M. Kanehisa, S. Goto, Y. Sato, M. Kawashima, M. Furumichi and M. Tanabe, Nucleic Acids Res., 2014, 42, D199–D205 CrossRef CAS PubMed
.
- K. Suhre and P. Schmitt-Kopplin, Nucleic Acids Res., 2008, 36, W481–W484 CrossRef CAS PubMed
.
- R. Caspi, T. Altman, R. Billington, K. Dreher, H. Foerster, C. A. Fulcher, T. A. Holland, I. M. Keseler, A. Kothari and A. Kubo, Nucleic Acids Res., 2014, 42, D459–D471 CrossRef CAS PubMed
.
- I. M. Keseler, A. Mackie, M. Peralta-Gil, A. Santos-Zavaleta, S. Gama-Castro, C. Bonavides-Martínez, C. Fulcher, A. M. Huerta, A. Kothari and M. Krummenacker, Nucleic Acids Res., 2013, 41, D605–D612 CrossRef CAS PubMed
.
- P. Romero, J. Wagg, M. L. Green, D. Kaiser, M. Krummenacker and P. D. Karp, Genome Biol., 2004, 6, R2 CrossRef PubMed
.
- D. Croft, A. F. Mundo, R. Haw, M. Milacic, J. Weiser, G. Wu, M. Caudy, P. Garapati, M. Gillespie and M. R. Kamdar, Nucleic Acids Res., 2014, 42, D472–D477 CrossRef CAS PubMed
.
| This journal is © The Royal Society of Chemistry 2015 |