Design of dual MMP-2/HDAC-8 inhibitors by pharmacophore mapping, molecular docking, synthesis and biological activity†
Abstract
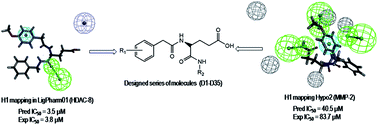
Recent analyses have highlighted the promotion of cancer migration and invasion, mediated through HDAC via MMP-2 and MMP-9. Since both class 1 HDACs and MMP-2/9 are involved in the migration and invasion of cancer, an attempt has been taken to design dual MMP-2/HDAC-8 inhibitors by pharmacophore mapping and molecular docking approaches. The designed molecules were synthesized and showed a range of inhibitory activity against different MMP subtypes. Most of these designed compounds were selective towards MMP-2 but less potent against anti-targets like MMP-8, -12, etc. The highly active MMP-2 inhibitors were also found to be active towards HDAC-8 but less potent against other class 1 HDACs (HDAC-1 and -2). Molecular dynamics simulations revealed that the designed compounds may be acting through a distinct mechanism of action in the ‘acetate ion channel’ of HDAC-8. Some potent dual MMP-2/HDAC-8 inhibitors were further explored for in vitro cellular assays against human lung carcinoma cell line A549. These analyses revealed that some of these dual inhibitors have considerable anti-migratory and anti-invasive properties. The work may help to obtain some useful dual inhibitors.

 Please wait while we load your content...
Please wait while we load your content...