Performance of wastewater biological phosphorus removal under long-term exposure to CuNPs: adapting toxicity via microbial community structure adjustment†
Abstract
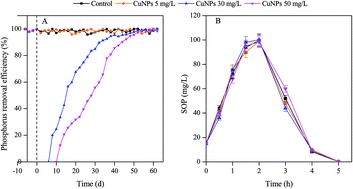
Copper nanoparticles (CuNPs) have been used in a wide range of applications, and the released CuNPs entering wastewater treatment plants (WWTP) might pose potential risks to the wastewater biological treatment process, such as phosphorus removal. Here we present the possible long-term effect of CuNPs on biological phosphorus removal, and simultaneously compare this with their acute impact. It was found that the terribly deteriorated phosphorus removal process under shock load of CuNPs returned to a normal level after long-term exposure to 50 mg L−1 of CuNPs; also, the inhibited transformations of intracellular metabolites, such as polyhydroxyalkanoates (PHA) and glycogen were gradually recovered. However, long-term exposure to 50 mg L−1 of CuNPs made both the bacterial diversity and the abundance of functional bacteria (polyphosphate accumulation organisms, PAO) in the EBPR system decrease, indicating that bacteria sensitive to CuNPs were washed out, and bacteria left via microbial community structure adjustment could undertake the task of phosphorus removal. Further mechanistic investigation revealed that the enhanced reactive oxygen species (ROS) and the decreased enzyme activity under the shock load of CuNPs returned to normal as well after long-term exposure to CuNPs.

 Please wait while we load your content...
Please wait while we load your content...