Immobilization of thermotolerant intracellular enzymes on functionalized nanoporous activated carbon and application to degradation of an endocrine disruptor: kinetics, isotherm and thermodynamics studies
Saranya P,
Ranjitha S and
Sekaran G*
Environmental Technology Division, CSIR-Central Leather Research Institute (CLRI), Adyar, Chennai, India. E-mail: ganesansekaran@gmail.com; Fax: +91-44-24410232; Tel: +91-44-24452941
First published on 27th July 2015
Abstract
A bacterium, Serratia marcescens capable of degrading the endocrine disruptor, 2-nitrophloroglucinol (NPG) was isolated from tannery wastewater contaminated soil. The mixed intracellular enzymes (MICE) produced from S. marcescens were extracted and characterized. The functionalized nanoporous activated carbon matrix (FNAC) was prepared to immobilize MICE. The optimum conditions for the immobilization of MICE on FNAC were found to be time, 2.5 h; pH, 7.0; temperature, 40 °C; concentration of MICE, 4 mg; particle size of FNAC, 300 μm and mass of FNAC, 1 g. The FNAC materials before and after immobilization of MICE were characterized using scanning electron microscopy, Fourier transform-infrared spectrophotometry and an X-ray diffractometer. The thermal behaviour of the free and the immobilized MICE was studied using thermogravimetric analysis. The immobilization of MICE on FNAC obeyed the Freundlich model and the immobilization process followed a pseudo second order kinetic model. MICE-FNAC matrix was used to degrade NPG in aqueous solution. The degradation of NPG by MICE-FNAC was optimum at contact time, 3 h; pH, 7.0; temperature, 40 °C; concentration of NPG, 20 μM and agitation speed, 70 rpm. The degradation of NPG was found to be enhanced in the presence of Zn2+, Cu2+, Ca2+ and V3+ ions. The degradation of NPG by MICE-FNAC was studied using UV-visible, fluorescence and FTIR spectroscopy. The degradation of NPG by MICE-FNAC was confirmed using HPLC, NMR and GC-MS spectroscopy.
1. Introduction
Endocrine disrupting compounds are a group of environmental pollutants mimicking natural hormones, disrupting biosignalling pathways or modifying hormone receptors.1 The endocrine disrupting chemicals have received increased attention in water quality management and healthcare management due to their persistence in the environment and toxic activity even at low concentration.2Nitro phloroglucinol (NPG) has been widely used in the chemical industry as a raw material for manufacturing dyes and in the explosives industry as an ingredient for priming compositions, percussion caps and detonator formulations.3,4 NPG is a synthetic intermediate in the manufacture of benzoxazoles and benzothiazoles as selective ligands for human β-estrogen receptor (ER-β)5 and thus it has the ability to bind with the estrogen receptor. In the leather industry, the use of soluble dyes such as sulphur black 1, sulphur brown 14, sulphur red 6 and other sulphur based dyes could be the source for the presence of NPG in tannery wastewater. NPG is characterized by its density (1.771 g cm−3), boiling point (343 °C), and melting point (189–193 °C).
Nitroaromatic compounds are among the largest and most important groups of industrial chemicals in use. The ability of the nitro group to delocalize π electrons of the phenyl ring to satisfy its own charge deficiency and impart charge to the molecule. The nitro group is strongly deactivated towards electrophilic aromatic substitution of the benzene ring.6
The stability and recalcitrant nature of NPG are due to both the conjugation state and resonance properties of nitro groups attached to aromatic rings.6 The conventional biological treatment of nitro compounds has certain disadvantages such as high sludge production, high energy demand and frequent maintenance requirement7 besides large land area requirement and less flexibility in design & operation.8 Other methods such as adsorption, photolysis, chemical oxidation and electrochemical technique, are widely used for the treatment of nitro compounds containing wastewater.9 However, all these methods have their own limitations in the field level implementation due to high capital cost, large area requirement and high electrical energy demand.9
Advanced oxidation processes (AOPs), based on the in situ generation of non selective and highly reactive species such as hydroxyl radicals (˙OH), superoxide anion radicals (O2˙−), and hydrogen peroxide (H2O2) as initiators of the oxidative degradation of refractory organics, have been proposed as an alternative approach to the conventional methods in recent years.10 AOP with ozone (O3), ultraviolet (UV), hydrogen peroxide (H2O2) and/or catalyst offered a powerful water treatment solution for the reduction and/or removal of refractory organic compounds.11–13 The main disadvantages of AOP are high operation costs due to chemicals and/or energy input, formation of oxidation intermediates potentially toxic and complicated design procedures.14
The biodegradation of pollutants in wastewater mainly depends on microorganisms that attack the pollutants enzymatically and convert them into nonhazardous products.15 Bacteria that inhabit chemical waste contaminated sites possess an astonishing ability of evolving new pathways for catabolism of recalcitrant organic compounds. Such pathways are accompanied by the appearance of transcriptional regulatory circuits that adjust the level of biodegradative capacity of the bacteria. Transcriptional regulation mediated by the binding of organic effector on a regulatory protein that acts on specific protein seems to be late evolutionary development as compared to metabolic operons and enzymes.16 Biodegradation of aromatic compounds has therefore, gained an increasing attention in pollution prevention.17,18
In biodegradation of wastewater, toxic pollutants were eliminated either through intracellular accumulation or through enzymatic transformation into less/nontoxic compounds,19 applicable only to labile organic compounds. Sometimes the secondary metabolites of the pollutants are more toxic and persistent than the parent compounds.20 Hence, there has been constant research on the development of cost effective techniques for the treatment of nitro organic compounds in wastewater. The removal of aromatic nitro groups by dioxygenase was first reported by Ecker et al.21 Additional support for the reaction mechanism came from the observation recorded by Sander et al.22 that the enzyme from a Pseudomonas sp. could catalyze the elimination of the nitro group from 2,4,5-trichloronitrobenzene.
The intracellular enzymes were explored for wastewater treatment as they were highly specific and extremely efficient catalysts23 and could degrade the target pollutant. The general disadvantages observed with the application of free enzymes for the bioremediation of wastewater containing refractory organic compounds were periodic addition increased the operation cost, applied free enzyme increased COD load, and sometimes enzyme activity was lost due to the fluctuating environmental conditions. Hence, immobilization technology has been widely used for improving the activity, stability, specificity, selectivity and decreased inhibition of enzymes.24,25 The function of an intracellular enzyme depends largely on its conformation geometry. The stereo chemical configuration of an intracellular enzyme changes with harsh conditions such as high temperature, very low/high pH, high concentration of target pollutants and high ionic strength.26 The immobilized enzyme in its physical confinement to a certain region of space, retained its catalytic activity with the capacity to be used continuously.27 Few research studies have employed certain support matrices such as epoxy activated matrix, sepabeads, curdlan matrix for the enzyme immobilization.28 The immobilization of enzymes on activated carbon was observed to be nontoxic, fast, inexpensive, easy to handle and stable against denaturation by oxidants.29 However, there is no single report on the immobilization of intracellular enzymes and thereof for the degradation of NPG in wastewater. Hence, in the present investigation, mixed intracellular enzymes (MICE) were extracted from Serratia marcescens and characterized. MICE was immobilized on functionalized nanoporous activated carbon (FNAC) and used for the degradation of NPG. The kinetics and thermodynamics for the immobilization of MICE on FNAC have been evaluated.
2. Materials and methods
2.1. Material
2-Nitrophloroglucinol of high purity 98% was obtained from Alfa Aesar (India). Ethylenediamine and glutaraldehyde of purity 99.5% were purchased from Sigma-Aldrich-Fluka Chemical Co., India.2.2. Culture enrichment and isolation
The NPG degrading bacterium was isolated from the terrestrial soil acclimatized with NPG. Enrichment was carried out in M9 Minimal media (HiMedia) of volume 100 mL with samples containing 150 μM of NPG as the sole carbon source. NPG concentration in minimal media was increased from 30 to 150 μM with an incremental increase of 5 μM. A pigmented bacterium isolated from the acclimatized soil was observed to degrade NPG in the solution.2.3. Identification of the strains and phylogenic analysis
The pigmented bacterium was identified by 16S ribosomal DNA (16S rDNA) sequencing and phylogenetic analysis. The genomic DNA was isolated according to the procedure of Marmur.30 Phylogenetic analysis was performed by subjecting the deduced sequence to the 16S rDNA database to obtain the similar sequences and the phylogenetic tree was constructed using the Phylip package.312.4. Culture conditions for intracellular enzyme production
The bacterium was aseptically inoculated into Erlenmeyer flasks of volume 1000 mL containing 75 μM NPG as the substrate in 500 mL of minimal media of pH 7 for intracellular enzyme production. The culture flasks were incubated on the shaker at 150 rpm and at 35 °C for 72 h. The bacterial cells were separated from the culture flasks by centrifugation at 6000 rpm for 20 min. The bacterial biomass was washed twice with phosphate buffer (pH 7.0) and stored at −20 °C.2.5. Extraction of intracellular enzymes
The method for intracellular protein extraction was adapted from Naiem and Jai32 with some modifications. The bacterial cells were sonicated with 1 mL of 0.1 M phosphate buffer. The extracts were centrifuged at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 30 min. The cell-free supernatant was collected and dialyzed against 0.1 M phosphate buffer (pH 7.0) for overnight. The purified mixed intracellular enzymes (MICE) were lyophilized and used for further analysis and for the degradation of NPG.
000 × g for 30 min. The cell-free supernatant was collected and dialyzed against 0.1 M phosphate buffer (pH 7.0) for overnight. The purified mixed intracellular enzymes (MICE) were lyophilized and used for further analysis and for the degradation of NPG.
2.6. Assays of intracellular enzymes
The lyophilized enzyme was analyzed for various intracellular enzymes present. The enzymes involved in the degradation of NPG were determined using the respective assay procedures: catechol dioxygenase,33 dehydrogenase,34 phenol hydroxylase,35 alkaline phosphatase,36 pyruvate decarboxylase,37 nitrate reductase38 and pyruvate kinase.39 The concentration of intracellular enzymes were expressed in terms of protein content, following Lowry method,40 employing bovine serum albumin as the standard. The intracellular enzymes were quantified and used as a mixture of intracellular enzymes for the degradation of NPG.2.7. Molecular weight determination of purified intracellular enzymes
The molecular weight of enzymes was determined by using sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) according to the method followed by Laemmli,41 on a 5% stacking gel and 12% resolving gel. The protein marker ranging from 5 to 240 KDa was used as a standard marker for determination of molecular weight.2.8. Aminoacid composition of MICE by HPLC
The MICE was hydrolyzed at 100 °C for 24 h with 6 N HCl and neutralized with 1 N NaOH. The amino acid composition was analyzed using Agilent 1100 HPLC amino acid analyzer, and the data analysis was performed by using HP chem station.2.9. Preparation of functionalized nanoporous activated carbon (FNAC)
Nanoporous activated carbon (NAC) was prepared from rice husk and the functionalization of the NAC was carried out as explained by Ramani et al.42 The washed FNAC was dried at 110 °C for 6 h and it was sieved to different sizes as 100, 300, 400 and 600 μm.2.10. Immobilization of MICE on FNAC
The optimum conditions for the immobilization of MICE on FNAC were determined by varying the parameters such as time (30 to 210 min), pH (1.0–10.0), temperature (20 to 60 °C), concentration of MICE (1–5 mg), particle size of FNAC (100, 300, 400 & 600 μm) and mass of FNAC (0.5–2.5 g). The immobilization of MICE was carried out by equilibrating 1 g of FNAC with phosphate buffer of volume 15 mL at pH 7 containing a known amount of lyophilized MICE. A mg of lyophilized MICE contains 532.9 mg of protein. Immobilization capacity was measured in terms of residual protein content as shown by the mathematical expression.
 | (1) |
| qt = qe(1 − exp−k1t) | (2) |
 | (3) |
| ΔG° = ΔH° − TΔS° | (4) |
2.11. Instrumental techniques for evaluation of immobilization of MICE on FNAC
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×. The FNAC samples were coated with gold by a gold sputtering device for the clear visibility of the surface morphology.
000×. The FNAC samples were coated with gold by a gold sputtering device for the clear visibility of the surface morphology.DSC analysis was carried out by loading the required quantity (8–10 mg) of lyophilized MICE, FNAC or MICE-FNAC samples was loaded in aluminum DSC pan and thermo gravimetric analysis was carried out under reduced nitrogen atmosphere from 0 °C to 200 °C at a temperature gradient of 10°C min−1. Scans were routinely recorded as duplicates using DSC Q200 (V23.10 Build 79, TA instruments).
2.12. Kinetic studies on the degradation of NPG by MICE-FNAC
The degradation study was carried out with the immobilized MICE on FNAC in a batch reactor. The degradation experiment was conducted at different variables such as the contact time (5–210 min), pH (1–10), temperature (20° to 80 °C), agitation speed (30–150 rpm) and initial concentration of NPG (5–25 μM). The effect of contact time was determined by mixing 10 μM of NPG with 1 g of MICE-FNAC in 10 mL phosphate buffer of pH 7 and they were incubated at 35 °C under mechanical agitation of 150 rpm. The samples were withdrawn at different time intervals (5 to 210 min) and UV-visible spectra were recorded using a UV-visible spectrophotometer (Cary varion; Agilent Technologies, Middleburg, Netherlands).The optimum pH was determined by incubating the reaction mixture at various pH ranging from 1 to 10 at room temperature in the following buffers: 0.1 M KCl/HCl buffer (pH 1.0, 2.0), 0.1 M acetate buffer (pH 3.0, 4.0 and 5.0), 0.1 M phosphate buffer (pH 6.0, 7.0, and 8.0), 0.1 M Tris buffer (pH 9 and 10) at optimum time, 60 min & temperature, 35 °C under mechanical agitation at 150 rpm. The optimum temperature was determined by incubating the reaction mixture at different temperatures ranging from 20 to 80 °C at optimum time, 60 min; pH, 7 under mechanical agitation at 150 rpm. The optimum initial concentration of NPG was determined by incubating the reaction mixture with varied concentration of NPG (5–25 μM) with 1 g of MICE-FNAC at optimized contact time, pH and temperature. The agitation speed (30–150 rpm) was optimized for the degradation of NPG using MICE-FNAC at optimized time, pH, temperature and initial concentration of NPG.
2.13. Effect of metal ions on the degradation of NPG using MICE-FNAC
NPG (20 μM) was added to the solution of volume 15 mL containing 1 mM of KCl, CaCl2·2H2O, ZnCl2, MgCl2·6H2O, CuSO4·5H2O, (NH4)6Mo7O24·4H2O & VCl3 and incubated at 40 °C for 1 h to determine the stimulatory or inhibitory effects of metal ions on degradation of NPG by MICE-FNAC. The NPG with 15 mL of 0.1 M phosphate buffer (pH 7.0) solution alone served as control.2.14. Instrumental techniques for the NPG degradation by MICE-FNAC
The NPG upon degradation by MICE-FNAC was supported by the instrumental techniques including high-pressure liquid chromatography (HPLC) and fluorescence spectroscopy. The final end product of enzymatic breakdown of NPG was also confirmed using Fourier transform infrared spectroscopy (FTIR), nuclear magnetic resonance spectroscopy (NMR) and gas chromatography-mass spectroscopy (GC-MS).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 and was applied at a flow rate of 1 mL min−1.
5 and was applied at a flow rate of 1 mL min−1.3. Results and discussion
3.1. Isolation and identification of bacteria by phylogenetic analysis
The soil site selected for isolation of bacteria was acclimatized with NPG for about 45 days. The acclimatized soil sample was inoculated into minimal media (M9, HiMedia) containing NPG as the sole carbon source. After 3 days of incubation, the above sample was used as an inoculum for the fresh medium containing NPG. The process was repeated for several times in order to isolate a microorganism for degradation of NPG with high efficiency. The guanine–cytosine richness in the bacterial genome, an important factor in characterizing a species, was about 20–75%.45 The isolated microorganism was found to be highly stable with 53.46% guanine–cytosine content. The microorganism was isolated for the production of intracellular enzymes. The 16S rDNA sequencing data confirmed that the isolated organism was Serratia marcescens (Fig. 1). S. marcescens is a red pigmented bacterium that grows at pH, 7 and temperature, 35 °C.3.2. Extraction and purification of MICE
The MICE extracted from S. marcescens were purified and run on SDS-PAGE to determine their molecular weight. The SDS-PAGE analysis showed that there were several bands of enzymes present with different molecular weight. The MICE was found to contain the following enzymes: catechol dioxygenase (1.141 U mg−1), phenol hydroxylase (1.255 U mg−1), dehydrogenase (0.081 U mg−1), alkaline phosphatase (1.92 U mg−1), pyruvate decarboxylase (2.761 U mg−1), pyruvate kinase (2.296 U mg−1) and nitrate reductase (2.15 U mg−1). | ||
| Fig. 1 Rooted phylogenetic tree showing the relationship of marine isolate S. marcescens to other Serratia sp. (values shown in the parenthesis are accession number). | ||
3.3. Identification of MICE
The molecular weights of various enzymes present in the mixed intracellular enzymes were identified by SDS-PAGE (Fig. 2). The molecular weight of extracted enzymes were as follows: 1,2-catechol dioxygenase, 38 kDa; phenol hydroxylase, 28 kDa; dehydrogenase, 109 kDa; alkaline phosphatase, 70 kDa; pyruvate decarboxylase, 71 kDa; pyruvate kinase, 63 kDa; and nitrate reductase, 156 kDa. | ||
| Fig. 2 SDS-PAGE of purified MICE from S. marcescens lane 1 molecular weight of marker, lane 2 purified MICE. | ||
3.4. Aminoacid composition of MICE
The amino acid composition of MICE was determined using HPLC as shown in Table 1. It was found that the MICE contained polar amino acids by 60.0%, non polar amino acids by 27.4% and aromatic amino acids by 13%. The aminoacids such as aspartic acid, glutamic acid, serine and histidine are very important in functioning of the protein as they constitute the catalytic triad of the various intracellular enzymes.46| Amino acids | Level (nmol mL−1) |
|---|---|
| Aspartic acid | 11 |
| Glutamic acid | 91 |
| Serine | 18 |
| Histidine | 85 |
| Glycine | 248 |
| Threonine | 80 |
| Arginine | 26 |
| Alanine | 235 |
| Tyrosine | 18 |
| Methionine | 74 |
| Valine | 79 |
| Phenylalanine | 25 |
| Isoleucine | 135 |
| Leucine | 29 |
| Lysine | 150 |
| Cysteine | 97 |
3.5. Immobilization of MICE on FNAC
The effect of time on the immobilization of MICE was carried out to determine the equilibrium values. The activity of MICE and protein content were measured at different time intervals (30 to 210 min) during the process of immobilization with 2 mg of lyophilized MICE at pH 7 and at 35 °C (Fig. 3a). It was found that, the immobilization was rapid up to 90 min and then there was a slight increase in immobilization until the equilibrium (150 min) was attained. Initially, the number of adsorption sites available was higher and the driving force for the mass transfer of MICE was greater. As the immobilization time was increased, the number of bare active sites became less and the MICE molecules might become clustered inside the FNAC particles, thus impairing the diffusion of MICE. The initial protein content of MICE was found to be 1065.8 mg and the immobilization capacity of FNAC was 643 mg g−1 of the matrix.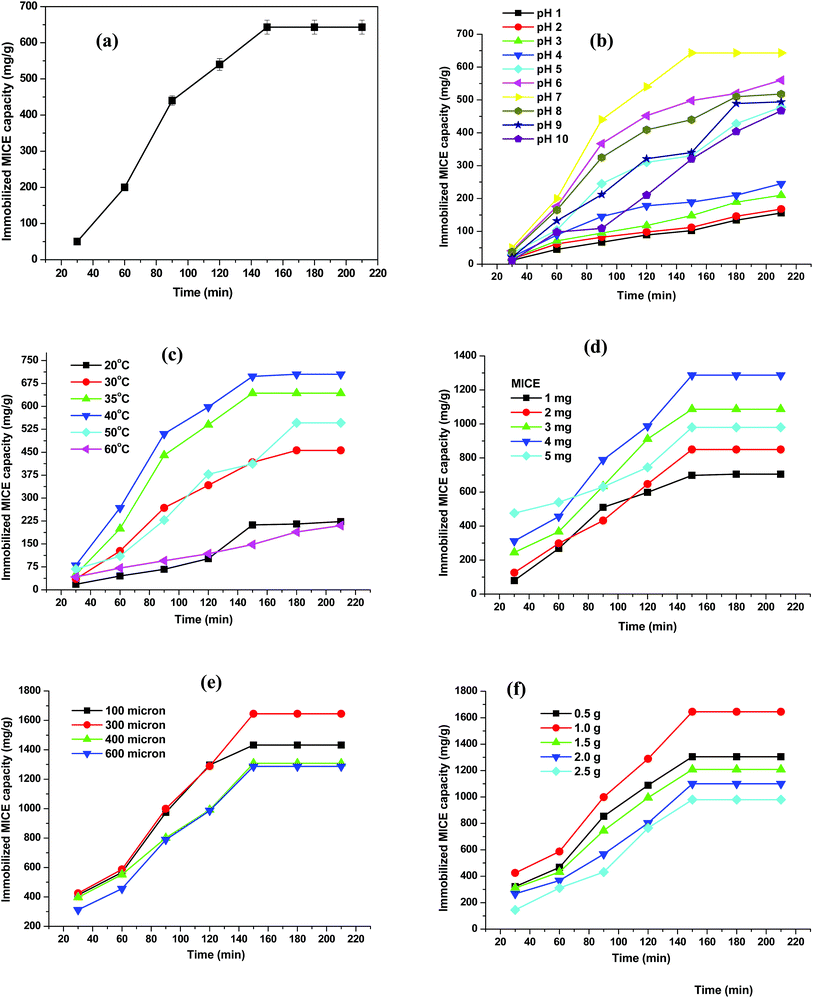 | ||
| Fig. 3 Effect of (a) time, (b) pH, (c) temperature, (d) initial concentration of MICE, (e) particle size of FNAC and (f) mass of FNAC on the immobilization of MICE on FNAC. | ||
The experiments were carried out at were carried out at a wide range of pH values between pH 1 and 10 for MICE of initial protein content of 1065.8 mg and temperature 35 °C in order to determine the optimum pH for the immobilization of the MICE onto FNAC. The immobilization of MICE on FNAC was found to be the maximum at pH 7 with immobilization capacity of 643 mg g−1 as shown in Fig. 3b. Immobilization outside this pH range was decreased, may be attributed to the factors that the enzyme may become denatured or lose its tertiary structure, and, therefore, lose its ability to function as a catalyst for the enzymatic reactions.
The optimum temperature for the immobilization of MICE onto FNAC was determined by varying the temperature from 20 to 60 °C at an initial MICE activity (1065.8 mg) at their optimum pH 7. The immobilization of MICE on FNAC was found to be the maximum at 40 °C. At 40 °C temperature, the maximum loading achieved was 705 mg g−1 of FNAC (Fig. 3c). The reaction rate decreased beyond the optimum temperature owing to denaturation of enzymes and thereby lost its ability to function as a catalyst in the enzymatic reaction.
The immobilization capacity of FNAC was found to increase with increase in the initial MICE concentration, however, the percentage immobilization decreased with increase in MICE concentration. The optimum MICE concentration was found to be 4 mg (2131.6 mg of protein) with the immobilization capacity of 1287 mg g−1 of FNAC at the optimum pH and temperature (Fig. 3d). The high immobilization capacity of FNAC may be due to more number of anchoring sites in the matrix.
The optimum particle size of FNAC for the immobilization of MICE was determined by varying the particle size as 100, 300, 400 & 600 μm at the optimum conditions pH, 7; temperature, 40 °C and MICE concentration, 4 mg (2131.6 mg of protein). The immobilization capacity of FNAC was increased with decrease in the particle size of FNAC. The optimum particle size of FNAC was found to be 300 μm with the immobilization capacity of 1645 mg g−1 of FNAC at the optimum conditions (Fig. 3e).
The optimum mass of FNAC for the immobilization of MICE onto FNAC was determined by varying the mass (0.5 to 2.5 g) at the optimum conditions pH, 7; temperature, 40 °C; MICE concentration, 4 mg (2131.6 mg of protein) and FNAC particle size, 300 μm. The optimum mass of FNAC was found to be 1 g with the immobilization capacity of 1645 mg g−1 of FNAC at the optimum conditions (Fig. 3f).
Thus, the optimum conditions required for the maximum immobilization of MICE on FNAC was found to be time, 150 min; pH, 7; temperature, 40 °C; MICE concentration, 4 mg (2131.6 mg of protein); FNAC particle size, 300 μm and mass of FNAC, 1 g with the immobilized MICE capacity of 1645 mg g−1.
 | (5) |
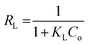 | (6) |
| Langmuir constants | Freundlich constants | ||
|---|---|---|---|
| KL (L g−1) | 3.294 | KF (mg g−1) (L mg−1) | 0.513 |
| b (L mg−1) | 5.868 | n | 3.12 |
| R2 | 0.8627 | R2 | 0.9349 |
| RL | 2.8 × 10−4 | ||
The Freundlich isotherm can be applied to non-ideal adsorption onto heterogeneous surfaces as well as multilayer sorption and is expressed by eqn (7).
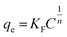 | (7) |
The adsorption isotherms indicated that immobilization of MICE on FNAC obeyed Freundlich isotherm model based on the regression coefficient (R2). The value of RL, the separation factor fell in the range of less than zero, indicating that the immobilization of MICE on FNAC was irreversible. This confirmed the immobilization of MICE, through the formation of a strong bonding with adsorption sites of FNAC.
| Temperature (°C) | qe (exp) (mg g−1) | Pseudo first order | Pseudo second order | ||||||
|---|---|---|---|---|---|---|---|---|---|
| k1(min−1) | qe (Cal) (mg g−1) | R2 | χ2 | k2 (mg−1 g min−1) | qe (Cal) (mg g−1) | R2 | χ2 | ||
| 20 | 223 | 0.0057 | 248 | 0.973 | 2.63 | 3.17 × 10−5 | 230 | 0.996 | 0.21 |
| 30 | 456 | 0.0194 | 467 | 0.943 | 0.26 | 2.42 × 10−5 | 450 | 0.969 | 0.08 |
| 35 | 643 | 0.0201 | 701 | 0.970 | 4.80 | 1.26 × 10−5 | 652 | 0.998 | 0.12 |
| 40 | 705 | 0.0203 | 789 | 0.980 | 8.94 | 1.20 × 10−5 | 692 | 0.999 | 0.24 |
| 50 | 546 | 0.0117 | 623 | 0.487 | 9.52 | 1.06 × 10−5 | 554 | 0.942 | 0.12 |
| 60 | 210 | 0.0124 | 305 | 0.852 | 29.5 | 2.46 × 10−5 | 254 | 0.983 | 7.62 |
3.6. Thermodynamic studies for immobilization of MICE on FNAC
The thermodynamic parameters were calculated using the eqn (4). The change in positive entropy value (ΔS° = 0.256 kJ mol−1 K−1) indicates that the immobilization of MICE on FNAC was accompanied with increased random distribution of it at standard temperature, 25 °C. Moreover, the negative value of enthalpy (ΔH° = −78.19 kJ mol−1) of the system indicates the immobilization of MICE on FNAC was an exothermic process at 25 °C. The negative free energy (ΔG° = −153.20 kJ mol−1) value indicates the immobilization of MICE on FNAC was spontaneous at 25 °C.3.7. Evidences for the immobilization of MICE on FNAC
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibrations of amide I. The bands at 3434 cm−1 and 1547.03 cm−1 may be attributed to N–H stretching and C–N bending. The bands at 633.59 cm−1 and 662.85 cm−1 may be attributed to C–N stretching vibrations in MICE. The bands at 2925.88 cm−1 and 2855.09 cm−1 may be due to the asymmetrical and symmetrical C–H stretching of methylene groups respectively. The band at 1162.55 cm−1 may be attributed to C–O stretching vibrations present in MICE.
O stretching vibrations of amide I. The bands at 3434 cm−1 and 1547.03 cm−1 may be attributed to N–H stretching and C–N bending. The bands at 633.59 cm−1 and 662.85 cm−1 may be attributed to C–N stretching vibrations in MICE. The bands at 2925.88 cm−1 and 2855.09 cm−1 may be due to the asymmetrical and symmetrical C–H stretching of methylene groups respectively. The band at 1162.55 cm−1 may be attributed to C–O stretching vibrations present in MICE.
The FT-IR spectrum of FNAC shows the peak corresponding to the N–H stretching vibration of a secondary amine at 3416.26 cm−1. The significant increase in the intensity of the band at 1715.57 cm−1 in FNAC corresponds to C–O stretching vibrations of carbonyl or aldehyde groups. The bands at 2923.82 cm−1 and 1365.59 cm−1 may be attributed to methylene stretching and bending vibrations respectively. This may be due to the addition of ethylenediamine and glutaraldehyde during the preparation of FNAC. This confirms the condensation of ethylenediamine and glutaraldehyde onto nanoporous activated carbon.42
The FT-IR spectrum of MICE-FNAC (Fig. 6c) showed that the frequency at 1652.34 cm−1 may be attributed to amide I of immobilized MICE. The peak corresponding to C–O stretching vibration (1715.57 cm−1) in MICE-FNAC of ketone and aldehyde groups were masked because of the strong binding of NH2 group of the MICE with the aldehyde group of FNAC. The wide peak observed at 3414.43 cm−1 in MICE-FNAC is due to the N–H stretching vibration of secondary amine.
The TGA of FNAC (Fig. 8b) showed 3.17% weight loss at 101.34 °C due to the removal of moisture. A major weight loss occurred from 194.24 to 533.58 °C and 62.05% of the sample remained as fixed residue at 800 °C, indicating the thermal resistance of the constituents of FNAC.
The thermogram of MICE-FNAC (Fig. 8c) showed 8.05% weight loss at 90.31 °C due to the removal of moisture. A weight loss of 9.64% was observed at 190.39 °C which could be due to decomposition of smaller particles in MICE. A major weight loss occurred from 215.12 °C to 376.89 °C is due to the decomposition of the major components of the MICE-FNAC. At the end of the scan (800 °C), 62.60% of the sample remained as fixed residue, indicating the higher thermal resistance of the constituents of MICE-FNAC than MICE.
The DSC of MICE (Fig. 8d) shows that there was an endothermic transition at 75.97 °C, 208.05 °C with the enthalpy of transition 144.7 J g−1 and 187.2 J g−1 respectively. The DSC of FNAC (Fig. 8e) shows there was a thermal transition at 118.09 °C with the enthalpy of transition 203.9 J g−1. The DSC of MICE-FNAC (Fig. 8f) showed a sharp endothermic thermal transition at 111.38 °C and small bumps at 171.34 °C & 233.33 °C with the enthalpy of transition 178.9 J g−1.
These results suggest that the thermal stability of the MICE was enhanced due to the stabilized bonding between MICE and functional groups of FNAC. And also the results suggested that there was not much change in their enthalpy of transition, indicating that the nature of MICE was not being altered upon immobilization.
3.8. Degradation kinetics of NPG using MICE-FNAC
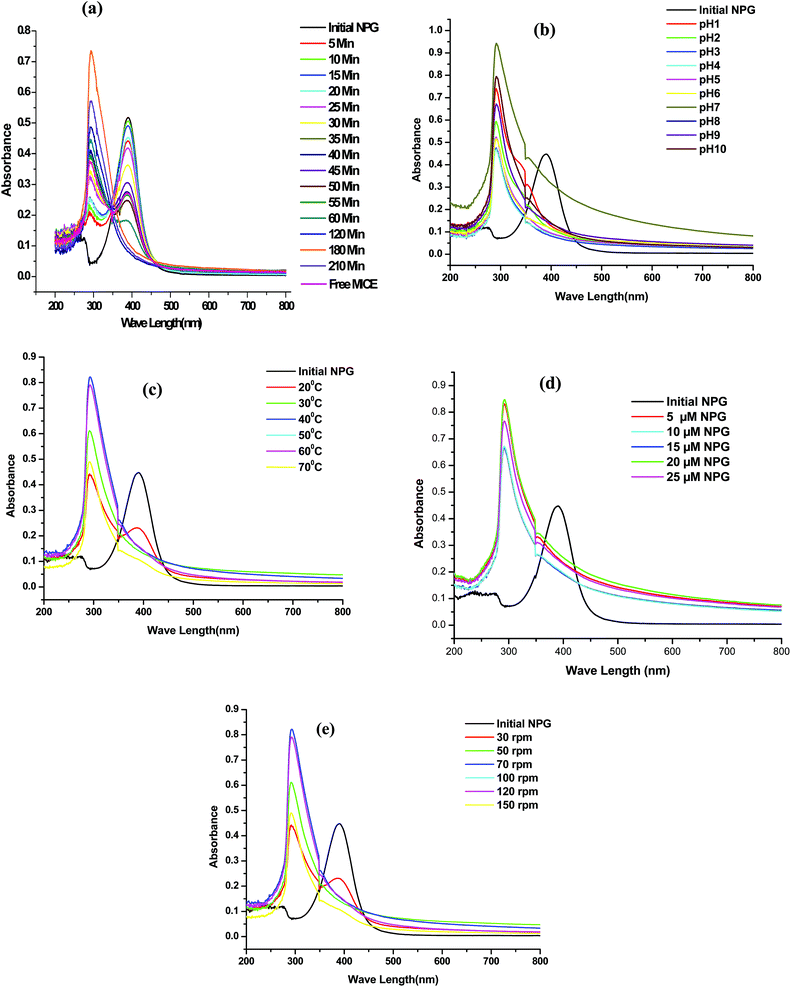
The effect of various factors such as contact time (5–210 min), pH (1–10), temperature (20° to 80 °C), agitation speed (30–150 rpm), initial concentration of NPG (5–25 μM) was studied on degradation of NPG using 1 g of immobilized MICE (1645 mg g−1 of FNAC). The degradation of NPG as a function of time (Fig. 9a) shows the disappearance of characteristic peak of NPG (λ390 nm) with increase in time and increase in the intensity of the formation of new compound at λ290 nm using MICE-FNAC. The degradation of NPG using free MICE also showed the disappearance of characteristic peak of NPG (λ390 nm) and the formation of new compound at λ290 nm (Fig. 9a). The maximum degradation was attained in 3 h at pH, 7; temperature, 30 °C; initial NPG concentration, 10 μM with agitation speed of 70 rpm. The degradation of NPG using FNAC was used as the control; where there is no formation of new compound at λ290 nm suggesting MICE mediates the degradation of NPG.The effect of pH on the degradation of NPG using MICE-FNAC was studied. It was observed that MICE-FNAC was able to degrade NPG at different pH conditions from 1–10 (Fig. 9b) and the maximum degradation of NPG was observed at pH 7.0 using MICE-FNAC. This also suggests that the carrier matrix, FNAC provides MICE the ability to retain the geometry from denaturation despite the wide fluctuations in pH. The chemical resistance of MICE in its immobilized state may be correlated with the strong binding established with FNAC.
The effect of temperature on the degradation of NPG using MICE-FNAC was studied. The NPG degradation increases with increase in temperature. It was observed that the MICE-FNAC could withstand wide range of temperatures. The intensity of formation of new compound varied but the degradation of NPG was achieved. It was observed that the characteristic peak of NPG at λ390 nm was reduced in its intensity and a new peak appeared at λ290 nm indicating partial degradation of NPG at 20 °C (Fig. 9c). The peak at λ390 nm was completely disappeared at 30 °C and the maximum amount of new compound at λ290 nm was found at 40 °C. As the temperature increased, the intensity of characteristic peak at λ390 nm decreased progressively and the intensity of new peak at λ290 nm increased steadily.
The initial concentration of NPG (Fig. 9d) was varied and it was found that the optimum concentration of NPG was about 20 μM to obtain the maximum concentration of new compound. The effect of agitation speed on degradation of NPG using MICE-FNAC was studied. It was observed that there was a small peak at λ390 nm and a new peak at λ290 nm indicating partial degradation of NPG at agitation speed of 30 rpm (Fig. 9d). As the agitation speed increased, the peak at λ390 nm was completely eliminated but a new compound of high intensity appeared at 70 rpm.
The optimum conditions for the degradation of NPG using MICE-FNAC was found to be time, 3 h; pH, 7; temperature, 40 °C, initial concentration of NPG, 20 μM and agitation speed, 70 rpm.
3.9. Effect of metal ions
Most enzymes require the presence of metal ion activators to express their catalytic activity. Metal ions act as cofactors for many enzymes to increase their catalytic activity at lower concentrations while the catalytic activity was reduced due to the toxic effect of metal ions at high concentrations.47 The effect of various metal ions such as K+, Zn2+, Cu2+, Ca2+, Mg2+, Mo6+ and V3+ on the degradation of NPG using MICE-FNAC was studied in the present investigation. Zn2+, Cu2+, Ca2+ and V3+ enhanced the catalytic effect on degradation of NPG using MICE-FNAC (Fig. 10). All other metal ions inhibited the degradation of NPG as evidenced from the absence of new compound formation at λ290 nm. The metal ions such as Zn2+, Cu2+, Ca2+ and V3+ had stimulatory effect on the degradation of NPG were used in combination (Fig. 10). The intensity of new compound formed at λ290 nm was in the decreasing order as V3+ > all metal ions > Cu2+ > Ca2+ > Zn2+ ions.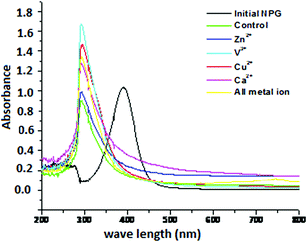 | ||
| Fig. 10 UV-visible spectra of degradation of NPG by MICE-FNAC in the presence of metal ion enhancers. | ||
The MICE from S. marcescens such as dehydrogenase and alkaline phosphatase could be enhanced by the presence of zinc ion.46 Copper ion enhanced the activity of dioxygenase,48 while the kinase was activated by calcium and reductase by vanadium ion.49 In protein metal-binding sites, the metal ion is coordinated by different combinations of protein side chains, including the nitrogen of histidine, the oxygen of aspartate or glutamate and the sulfur of cysteine; among these, histidine is the most commonly observed, followed by cysteine.50
The Zn2+ activates alkaline phosphatase by binding to the ligands three histidine and two water molecules, while Zn2+ activates dehydrogenase enzyme by binding with two cysteine, one histidine and a water molecule. Copper ion activates dioxygenase by forming square planar coordination with two histidine, one cysteine and a water molecule while calcium ion forms a octahedral coordination with two aspartate residues (or one aspartate and one asparagine) bound in a monodentate manner, one glutamate residue bound in a bidentate manner, one main-chain carbonyl group and one water molecule. The sixth ligand is variable and may be water, serine, asparagine or aspartate.51 Vanadium ion activates reductase by binding with the carboxylate groups in aspartate, glutamate and cysteine molecules. The metal ion in the active site of intracellular enzymes causes polarisation of hydrogen–oxygen bond, making the oxygen slightly more negative, thereby weakening the peptide enhancing the enzyme activity.52
3.10. Instrumentation evidences for the degradation of NPG
NPG was treated with MICE-FNAC at the optimized conditions and the degradation was studied using the following instrumentation techniques.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C in aromatic ring. The bands at 1549.43 cm−1 and 1588.17 cm−1 are due to the asymmetric stretching vibrations of N
C in aromatic ring. The bands at 1549.43 cm−1 and 1588.17 cm−1 are due to the asymmetric stretching vibrations of N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O in nitro group. The band at 1312.91 cm−1 is attributed to symmetric N
O in nitro group. The band at 1312.91 cm−1 is attributed to symmetric N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration in nitro group. The stretching of aromatic C–H is observed at 3096.1 cm−1. The C–N stretching in NPG is confirmed at 836.67 cm−1 and 699.02 cm−1. The bands at 770.25 cm−1, 520.92 cm−1, 470.13 cm−1 are due to the out of plane bending vibrations of C–H in aromatic ring. The inplane aromatic C–H bending vibrations were confirmed at 1081.8 cm−1 and 1015.6 cm−1. The band at 3456.25 cm−1 is attributed to the strong bonded O–H vibrations.
O stretching vibration in nitro group. The stretching of aromatic C–H is observed at 3096.1 cm−1. The C–N stretching in NPG is confirmed at 836.67 cm−1 and 699.02 cm−1. The bands at 770.25 cm−1, 520.92 cm−1, 470.13 cm−1 are due to the out of plane bending vibrations of C–H in aromatic ring. The inplane aromatic C–H bending vibrations were confirmed at 1081.8 cm−1 and 1015.6 cm−1. The band at 3456.25 cm−1 is attributed to the strong bonded O–H vibrations.
The degradation of NPG by MICE-FNAC was confirmed from the FT-IR spectrum (Fig. 12b). The band at 1642.17 cm−1 is attributed to asymmetric stretching of carboxylate ion in degraded sample. The peak at 1450.3 cm−1 is attributed to asymmetric bending of CH3 group. The band at 3436.68 cm−1 is due to the stretching of O–H group. The peak at 1084.12 cm−1 may be attributed to stretching of C–O group. The FT-IR spectrum confirmed the degradation of NPG and the presence of a keto group concludes the degraded end product could be pyruvic acid.
The 13C NMR spectrum of NPG was scanned between δ 0.0 and δ 200 ppm. The chemical shift around δ 130–140 ppm indicates the evidence for phenyl carbon in the aromatic ring (Fig. 15a). The occurrence of chemical shifts at δ 118.45, δ 118.22 and δ 115.41 ppm confirm the presence of carbon atoms attached to hydroxyl groups. The chemical shift at δ 78.93 ppm can be assigned to the carbon attached to nitro groups. The 13C-NMR spectrum of MICE-FNAC degraded NPG sample shows that the peaks around δ 110–140 ppm were disappeared. The chemical shift at δ 208 ppm indicates the evidence for keto carbon. The chemical shift at δ 18.71 ppm indicates the evidence for methyl carbon (Fig. 15b). The absence of carboxylic peak δ 180 ppm may be due to the masking effect of the solvent. The absence of peaks at δ 78.93, 118.45, δ 118.22 and δ 115.41 ppm confirms the degradation of NPG by MICE-FNAC.
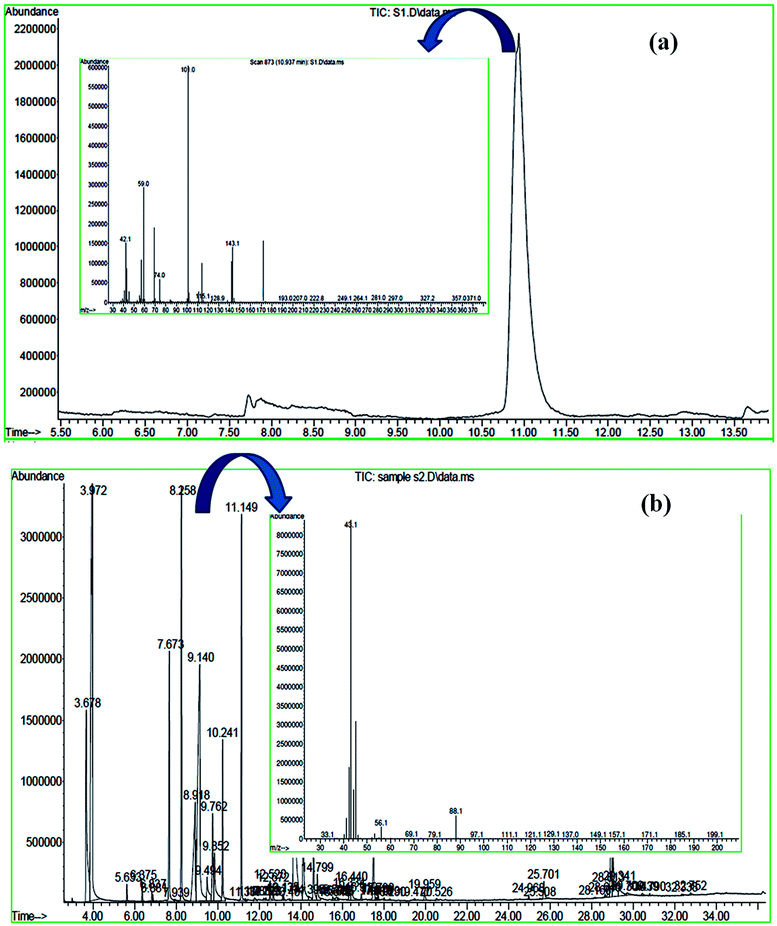
The MICE-FNAC degraded NPG sample showed the presence of pyruvic acid, corresponding to a molecular ion peak at m/z = 88 with the retention time of 8.258 min. Pyruvic acid, on fragmentation of the COOH group, yielded CH3CO+, corresponding to a peak at m/z = 43 and on elimination of the CH3 group, a peak corresponding to m/z = 33 (Fig. 16b). The fragments with molecular ions confirm the presence of pyruvic acid as the product of degradation of NPG by MICE-FNAC.
The plausible pathway for the degradation of NPG by MICE-FNAC involves the reduction of the nitro group in NPG by reductase to form 2-amino phloroglucinol which is further converted into 2-hydroxy phloroglucinol by the elimination of ammonia catalysed by monooxygenase. The next step is ring cleavage by dioxygenase to form 3, 5, dihydro muconate. This intermediate was further catalysed by kinase, dehydrogenase, phosphatase and pyruvate decarboxylase to form non-toxic, pyruvic acid (Fig. 17). The pyruvic acid was confirmed by functional groups present in the NPG degraded sample through FT-IR spectroscopy and mass fragmentation evidence from GC-MS spectroscopy.
4. Conclusions
The MICE from S. marcescens, capable of degrading NPG were extracted and characterized. The mixture of intracellular enzyme was purified and the molecular weight of enzymes was determined using SDS-PAGE. The optimum conditions for the immobilization of MICE on FNAC was found to be time, 2.5 h; pH, 7.0; temperature, 40 °C; concentration of MICE, 4 mg; particle size of FNAC, 300 μm and mass of FNAC, 1 g. The SEM images showed that the MICE was immobilized onto the carrier matrices. The immobilization of MICE was confirmed using FT-IR spectroscopy and XRD pattern analysis. The thermal behaviour of the free and the immobilized MICE was studied using thermogravimetric analysis. The immobilization of MICE on FNAC followed pseudo second order kinetic model and adsorption process obeyed Freundlich model. The degradation of NPG by MICE-FNAC was optimum at contact time, 3 h; pH, 7.0; temperature, 40 °C; concentration of NPG, 20 μM and agitation speed, 70 rpm. The degradation efficiency of NPG was enhanced with higher rate of conversion into pyruvic acid in the presence of metal ions (Zn2+, Cu2+, Ca2+, V3+ and AM). The confirmation of the NPG degradation by MICE-FNAC was justified using UV-visible, fluorescence, FTIR spectroscopy, HPLC, NMR and GC-MS spectroscopy.Acknowledgements
P. Saranya is grateful to the Council of Scientific and Industrial Research (CSIR), India. The financial assistance under the STRAIT (CSC0201) programme is also gratefully acknowledged.References
- A. M. Soto, H. Justicia, J. W. Wray and C. P. Sonnenschein, Environ. Health Perspect., 1991, 92, 167–173 CrossRef CAS.
- T. J. Kubiak, H. J. Harris, L. M. Smith, T. R. Schwartz, D. L. Stalling, J. A. Trick, L. Sileo, D. E. Doucherty and T. C. Erdman, Arch. Environ. Contam. Toxicol., 1989, 18, 706–727 CrossRef CAS.
- H. Rathsburg and B. Pat, Angew. Chem., 1928, 41, 1285 Search PubMed.
- W. Fredrich, Z. Ges and U. Schiess, Spengstoffw, 1933, 80, 113 Search PubMed.
- PCT Int. Appl., WO 2002051821 A1 20020704, 2002.
- K. Ju and R. E. Parales, Microbiol. Mol. Biol. Rev., 2010, 74, 250–272 CrossRef CAS PubMed.
- S. K. Liehr, A. R. Rubin and B. Tonning, Water Environ. Res., 2003, 76, 1191–1237 CrossRef.
- T. Robinson, G. McMullan, R. Marchant and P. Nigam, Bioresour. Technol., 2001, 77, 247–255 CrossRef CAS.
- P. C. Vandevivere, R. Bianchi and W. Verstraete, J. Chem. Technol. Biotechnol., 1988, 72, 289 CrossRef.
- J. C. Sin, S. M. Lam, A. R. Mohamed and K. T. Lee, Int. J. Photoenergy, 2012, 185159 Search PubMed.
- S. Sharma, J. P. Ruparelia and M. L. Patel, International Conference on Current Trends in Technology, Ahmedabad, 2011 Search PubMed.
- G. Tchobanoglous, F. Burton and H. Stensel, Wastewater Engineering: Treatment and Reuse, McGraw Hill, New York, NY, 4th edn, 2003 Search PubMed.
- T. Oppenländer, Photochemical purification of water and air, Advanced Oxidation Processes (AOPs): Principles, reaction mechanisms, reactor concepts, Wiley-VCH, Winheim, Germany, 2003 Search PubMed.
- S. Kommineni, J. Zoeckler, A. Stocking, S. Liang, A. Flores and M. Kavanaugh, Advanced oxidation processes, in National water Research Institute, URL, 2008 Search PubMed.
- C. S. Karigar and S. S. Rao, Decontamination, Wiley-VCH, Weinhein, 2011, pp. 1–11 Search PubMed.
- V. de Lorenz, R. Silva rocha, G. Carborosa, T. C. Galvao and I. Case, Sensing xenobiotic compounds: Lessons from bacteria that face pollutant in environment, Sensory Mechanisms in Bacteria: Molecular Aspects of Signal Recognition, ed. S. Spiro and R. Dixon, 2010, pp. 81–92 Search PubMed.
- M. Alexander, Biodegradation and Bioremediation, Academic Press, San Diego, USA, 2nd edn, 1999, pp. 325–327 Search PubMed.
- W. Fritsche and M. Hofrichter, Aerobic degradation by microorganisms, in Environmental processes II—soil decontamination, Biotechnology, ed. J. Klein, Wiley-VCH, Weinheim, Germany, 2nd edn, 2000, vol. 11b, pp. 146–155 Search PubMed.
- S. K. Brar, M. Verma, R. Y. Surampalli, K. Misra, R. D. Tyagi, N. Meunier and J. F. Blais, Pract. Period. Hazard., Toxic, Radioact. Waste Manage., 2006, 10, 59–72 CrossRef CAS.
- D. V. V. Gnanasalomi, G. R. Jebapriya and J. J. Gnanadoss, International Journal of computing algorithm, 2013, 2, 273–278 Search PubMed.
- S. Ecker, T. Widmann, H. Lenke, O. Dickel, P. Fischer, C. Bruhn and H. J. Knackmuss, Arch. Microbiol., 1992, 158, 149–154 CrossRef CAS.
- P. Sander, R. M. Wittalch, P. Fortnagcl, H. Wilkes and W. Francke, Appl. Environ. Microbiol., 1991, 57, 1430–1440 CAS.
- C. Nelson and M. Cox, Principles of Biochemistry, W. H. Freeman, New York, 4th edn, 2004, pp. 47–50 Search PubMed.
- R. Fernandez-Lafuente, J. M. Guisan, S. Ali and D. Cowan, Enzyme Microb. Technol., 2000, 26, 568–573 CrossRef CAS.
- P. V. Iyer and L. Ananthanarayan, Process Biochem., 2008, 43, 1019–1032 CrossRef CAS PubMed.
- J. Karam and J. A. Nicell, J. Chem. Technol. Biotechnol., 1997, 69, 141–153 CrossRef CAS.
- M. B. Pescod, FAO Irrigation and Drainage, Food and Agriculture Organization of United nations, Rome, Italy, 1992 Search PubMed.
- C. Mateo, J. M. Palomo, G. Fernandez-Lorente, J. M. Guisan and R. Fernandez-Lafuente, Enzyme Microb. Technol., 2007, 40, 1451–1463 CrossRef CAS PubMed.
- E. Kalogeris, Y. Sanakis, D. Mamma, P. Christakopoulos, D. Kekos and H. Stamatis, Enzyme Microb. Technol., 2006, 39, 1113–1121 CrossRef CAS PubMed.
- J. A. Marmur, J. Mol. Biol., 1961, 3, 208–218 CrossRef CAS.
- J. Felsenstein, PHYLIP (Phylogeny Inference Package) version 3.5c, Department of Genetics, University of Washington, Seattle, 1993 Search PubMed.
- H. N. Naiem and S. G. Jai, Res. J. Environ. Earth Sci., 2011, 3, 608–613 Search PubMed.
- G. Cenci and G. Caldini, Appl. Microbiol. Biotechnol., 1997, 47, 306–308 CrossRef CAS.
- K. M. Mayer and F. H. Arnold, J. Biomol. Screening, 2002, 7, 135–140 CrossRef CAS PubMed.
- H. Y. Neujahr and A. Gaal, Eur. J. Biochem., 1973, 35, 386–390 CrossRef CAS PubMed.
- K. N. Fernley, The Enzymes,ed. P. D. Boger, Academic press, New York, 1971, vol. 4, pp. 417–447 Search PubMed.
- M. T. Flikweertt, L. van der Zanden, W. M. Janssen, H. Y. Steensma, J. P. van Dijken and J. T. Pronk, Yeast, 1996, 12, 247–257 CrossRef.
- H. Ridley, C. A. Watts, D. J. Richardson and C. S. Butler, Appl. Environ. Microbiol., 2006, 72, 5173–5180 CrossRef CAS PubMed.
- K. Abbe and T. Yamada, J. Bacteriol., 1982, 149, 299–305 CAS.
- O. H. Lowry, N. J. Rosebrough, A. L. Farr and J. Randal, J. Biol. Chem., 1951, 193, 265–275 CAS.
- U. K. Laemmli, Nature, 1970, 227, 680–685 CrossRef CAS PubMed.
- K. Ramani, S. Karthikeyan, R. Boopathy, L. John Kennedy, A. B. Mandal and G. Sekaran, Process Biochem., 2012, 47, 435–445 CrossRef CAS PubMed.
- S. Lagergren, K. Sven. Vetenskapsakad. Handl., 1898, 24, 1–39 Search PubMed.
- Y. S. Ho and G. Mckay, Trans. Inst. Chem. Eng., 1998, 76(B), 183–191 Search PubMed.
- F. Hildebrand, A. Meyer and A. Eyre-Walker, PLoS Genet., 2010, 6, 1–9 Search PubMed.
- B. L. Vallee and D. S. Auld, Biochemistry, 1990, 87, 220–224 CAS.
- H. Rodriguez, B. de las Rivas, C. Gomez-Cordoves and R. Munoz, Int. J. Food Microbiol., 2008, 121, 92–98 CrossRef CAS PubMed.
- F. Fusetti, K. H. Schröter, R. A. Steiner, P. I. van Noort, T. Pijning, H. J. Rozeboom, K. H. Kalk, M. R. Egmond and B. W. Dijkstra, Structure, 2002, 10, 259–268 CrossRef CAS.
- M. Zizic, M. Zivic, I. Spasojevic, J. B. Pristov, M. Stanic, T. Cvetic-Antic and J. Zakrzewska, Res. Microbiol., 2013, 164, 61–69 CrossRef CAS PubMed.
- D. S. Gregory, A. C. R. Martin, J. C. Cheetham and A. R. Rees, Protein Eng., 1993, 6, 29–35 CrossRef CAS PubMed.
- J. P. Glusker, A. K. Katz and C. W. Bock, Rigaku J., 2000, 17, 8–16 Search PubMed.
- P. Saranya, R. Muneeswari and G. Sekaran, J. Chem. Technol. Biotechnol., 2014 DOI:10.1002/jctb.4558.
| This journal is © The Royal Society of Chemistry 2015 |