Bridging structure and mechanics of three-dimensional porous hydrogel with X-ray ultramicroscopy and atomic force microscopy
A. Y. Abuelfilata,
Y. Kima,
P. Millerb,
S. P. Hooc,
J. Lia,
P. Chanc and
J. Fu*a
aDepartment of Mechanical and Aerospace Engineering, Monash University, Clayton, VIC 3800, Australia. E-mail: jing.fu@monash.edu
bMonash Centre for Electron Microscopy, Clayton, VIC 3800, Australia
cDepartment of Biomedical Engineering, Swinburne University of Technology, Hawthorn, VIC 3122, Australia
First published on 15th July 2015
Abstract
The need for an in vitro 3D scaffold that can substitute specific tissue-types is becoming increasingly prevalent in tissue engineering and stem cell research. As a promising candidate for engineered complex 3D tissue scaffolds, hydrogels have emerged as synthetic or natural polymers with tissue-like stiffness, biocompatibility and high permeability for oxygen, nutrients and other water-soluble metabolites, similar to the native extracellular matrix. However, high-resolution characterization of hydrogels and their three-dimensional porous structures still remains a challenge. In this research, hydroxypropyl cellulose methacrylate (HPC-MA) hydrogels were examined for the first time through X-ray ultramicroscopy (XuM), an imaging technique based on phase contrast and with high spatial resolution, to visualise, reconstruct and analyse 3D porous structures. This Scanning Electron Microscopy (SEM) based X-ray system produced projection images of 1.67 μm pixel size, with distinguishable hydrogel membrane structures. In addition, reconstruction of the tomographic series provides the complete geometry of individual pores and their spatial distribution and interconnectivity, which play vital roles in accurate prediction of the hydrogel's porous structure prior to and during its implantation in vivo. By further incorporating Atomic Force Microscopy (AFM), the elastic modulus of the hydrogel was determined and mechanical modelling of individual pores and the bulk scaffold also proved to be feasible. The commercialised platform we utilised offers prompt visualization and specialized simulation of customized 3D scaffolds for cell growth, which will be a unique application of tissue engineering in future personalized medicine.
1. Introduction
The state of the art field of tissue engineering is currently solving major human health problems associated with loss or failure of tissues and/or organs, which is believed to cause some of the most tragic and costly problems in the health care system. In fact, this promising field has presented the option of designing patient-specific tissue engineered constructs that are tailored specifically to meet patient needs. Researchers in the field have established the ideal properties of tissue engineered scaffolds. These include a biocompatible and biodegradable three dimensional porous structure which acts as a template for initial cell attachment and subsequent tissue formation both in vitro and in vivo. In fact, the design of the biodegradable scaffold plays a crucial role in guiding the newly developed tissues while providing them with temporary mechanical support by defining and maintaining a 3D structure.1 The scaffold's highly interconnected porous structure promotes angiogenesis due to the induced tissue connectivity between the cells inside the scaffold and those from the microenvironment, mimicking the extracellular matrix (ECM)'s natural function by providing the necessary support for cells to adhere, proliferate and even differentiate.2–4 The complex interaction between cells and the ECM influences tissue morphogenesis and promotes functional tissue regeneration. Moreover, parameters including the pore diameters and their spatial distribution, as well as their connectivity at a very small scale are considered to be crucial for understanding and validating the scaffold design.5,6 Once implanted in vivo, it is believed that the correct architecture of this porous matrix will support cellular adhesion and growth and will maintain cellular differentiation by facilitating and easing the diffusion of nutrients and waste via the pores. In fact, for successful applications, the pore volumes need to be defined precisely prior to scaffold fabrication and implantation in vivo.It is also crucial to select the appropriate scaffolding material which will not only help regenerate cells but also induce their differentiation into the desired cell type and thereby restore tissue and/or organ functionality. Polymeric based hydrogel substrates supposedly have significant advantages for use as a scaffolding material, mainly since they are more flexible, offer a wide range of rigidity, can be stretched dynamically and may adopt different shapes. Furthermore, from the literature it is evident that one can tailor hydrogel properties to suit specific scaffolding design requirements, by modifying the hydrogel's chemical properties or through varying its crosslinking or polymerization conditions.7–11 In addition, hydrogels can have many other attractive material properties including biocompatibility, biodegradability and various biofunctionalities.12–14 Their hydrophilic nature and biochemical similarity to the native ECM makes them highly absorbent to water providing a hydrated matrix with tissue-like stiffness, which is an ideal microenvironment for cells to grow.15 It is noted that the stiffness of the substrate used in tissue engineering has a direct effect on stem cell differentiation, where proliferation followed by differentiation16 or differentiation along an alternative lineage,17 is increased with stiffer substrates.15 The elasticity of hydrogels over a long time scale allows for their fabrication into appropriate moulds forming 3D structures which in turn plays a crucial role in cell growth.
Since the regulation of cellular response and tissue integration is affected by the porous structure of hydrogels,18 clear imaging and visualisation of the three dimensional porous hydrogel has proven to be vital for the successful design of new tissue engineering scaffolds and for understanding the subsequent effect on the cellular behaviour upon interaction with the seeded cells. Lack of current methods to promptly obtain the three dimensional porous structure of hydrogels limits investigation and accurate prediction of their structure and function. Examples of current imaging techniques include Transmission Electron Microscopy (TEM),19 Scanning Electron Microscopy (SEM)20 and confocal microscopy.21,22 TEM requires thin sectioning of the sample to thickness less than hundred nanometer making 3D measurements difficult.19 SEM is restricted to the sample surface only as the detection depth is limited by the interaction volume of the electrons, typically a few microns or less. Further, confocal microscopy is limited in its ability to resolve the complete porous morphology of a typical hydrogel sample due to limited focal depth. Therefore it is not possible to fully view and measure the size, spatial distribution and interconnectivity of pores within the hydrogel structure using these three imaging techniques. In this research, we demonstrate for the first time the ability of a SEM based X-ray imaging technique named X-ray ultramicroscopy (XuM)23–26 to be used for 3D visualisation and analysis of porous hydrogel structures. The XuM is a projection X-ray microscope, a technique that has been in use for many decades27 and one that is routinely used in many X-ray imaging instruments. The projection method for X-ray microscopy is illustrated in Fig. 1a.
Interaction of the SEM's electron beam with a target generates a sub-micron X-ray source. The target positioner is mounted on the left hand side of the sample chamber providing 3 axes of movement and allowing a range of targets to be accurately positioned under the electron beam. The sample is mounted vertically on the SEM stage and X-rays from the source pass through the sample to form a projected image on the direct-detection X-ray camera mounted on the right-hand side of the sample chamber. Magnification is varied by moving the sample between the X-ray source and the camera (Y stage movement here). Magnification (M) at the camera is given by M = (R1 + R2)/R1 where R1 is the distance from the X-ray source to the sample and R2 is the distance from the sample to the camera. The field of view can be adjusted by moving the stage in the X and Z directions and a tomographic image series can be collected by rotating the sample.
A computer-controlled rotation stage is mounted on the SEM stage for tomography. This has a manually adjusted XY translation stage used to centre the sample on the rotation axis. An important consequence of the XuM experimental geometry is that almost all X-ray images will show both phase contrast and absorption contrast. Phase contrast will appear as one or more bright/dark Fresnel fringes at edges in the sample. These fringes can provide significant edge contrast even in a sample showing little absorption contrast which can be a great benefit in 2D imaging. However, these fringes will cause significant artifacts in a tomographic reconstruction. Phase retrieval algorithms can be used to extract quantitative data from XuM images, to improve image quality and to aid interpretation, and to transform the images into a form more suitable for tomographic processing (remove fringes).24 Here the transport of intensity (TIE) algorithm28 was used with the assumption that the sample is homogeneous. The Feldkamp–Davis–Kress cone-beam algorithm29 was used to reconstruct slices through the sample.
This method provides great potential for studies of soft materials including hydrogels, typically containing low Z elements such as C, H, O and N as shown by recent studies based on synchrotron X-ray imaging30,31 but with the access advantage of a laboratory-based technique. This imaging instrument has an ultimate spatial resolution of 100 nm or less for 2D images but under the conditions used for tomography here the resolution in the reconstructions is several microns.23,26,32–34 XuM is proven to be advantageous over other imaging techniques as it eliminates the tedious preparation and analysis of sectioned samples, while generating 2D images and reconstruction of 3D models. This enables accurate analysis of features such as size, shape, interconnectivity and spatial distribution of pores within the material.23,26,32–34
This study is the first one aiming to explore the capability of laboratory based X-ray phase contrast imaging to provide fast three dimensional visualisation of biocompatible porous hydrogels, including the dimensions and spatial distribution of pores within the hydrogel structure. In addition, nanomechanics of the same hydrogel sample were further investigated by AFM force spectroscopy,35,36 which has been a unique approach to investigate soft materials and cells.37–39 Together with the elastic modulus obtained, the reconstructed three dimensional structures allow mechanical modelling and simulation of each single pore and establish a rational approach for exploring structure–mechanics relationships.
2. Materials and methods
2.1. Hydrogel scaffold fabrication
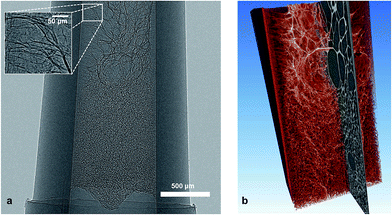
Hydroxypropyl cellulose methacrylate HPC-MA hydrogels were prepared as described in the protocol of Hoo et al.9 These hydrogel conjugates were formed through modifying hydroxypropyl cellulose (HPC) with bifunctional methacrylic anhydride (MA). After crosslinking, the crosslinked gels were washed with deionised water to remove any uncrosslinked HPC-MA conjugates and were frozen at −20 °C in a freezer, followed by lyophilisation under vacuum for 48 h using a freeze dryer (HETO PowerDry, PL6000, Thermo Scientific). Prepared hydrogel was retrieved in a micropipette tip (Fig. 1b), and was then freeze dried at −20 °C in a freezer. The samples were then transferred to the SEM chamber equipped with a XuM system for imaging (Fig. 1c).2.2. 3D imaging of porous hydrogel structure via XuM
A series of 2D X-ray images of the HPC-MA hydrogel scaffold were recorded at 0.5° steps over 180 degrees plus the fan angle. The SEM was operated at 30 keV beam energy with beam current >200 nA striking a bulk W target inclined at 45 degrees. Each image is the sum of two frames integrated over 30 s. The image magnification at the camera was 12× resulting in a voxel dimension of 1.67 μm. The inside diameter of the syringe was about 750 μm. The rotation series was processed as described above to obtain a 3D tomographic image set. A single 2D image from the image series (before phase retrieval) with an enlarged zoomed in inset showing fine details is presented in Fig. 2a. Fig. 2b shows a rendered view of the processed data set.2.3. Reconstruction of 3D models
The 3D tomographic image set was exported to software packages ImageJ (National Institutes of Health, Bethesda, MD, USA) and Avizo Fire (FEI Visualization Sciences Group, Burlington, MA, USA) for further 3D reconstruction. Pre-processing of the images involved cropping and filtering to focus on the regions of interest and to maximize the signal-to-noise ratio. Semi-automatic segmentation was performed in Avizo to identify the membrane structures of the hydrogel (Fig. 3a). After constructing the distance map, individual pores were identified and reconstructed (Fig. 3b). The volume, size, and location of each pore were exported for further analysis, and solid models of a single pore or multiple pores were reconstructed and exported as triangular meshes (stl file format) as needed.2.4. Measurement of elastic modulus with atomic force microscopy (AFM)
By probing the surface of the sample with nanoscale cantilever, force between the tip and the sample and the deflection of cantilever are constantly measured and can be further analysed to understand mechanical properties of the target sample at nanoscale level.40 In this study, Young's modulus of hydrogel was examined using an AFM instrument (JPK NanoWizard II, JPK Instruments AG, Berlin, Germany). Contact mode was used, and AFM cantilever with 0.06 N m−1 spring constant was used in order to accommodate the low modulus of hydrogel sample. Calibration of the cantilever was conducted prior to the force mapping using mica sheet, measuring the sensitivity and actual spring constant of the cantilever. Force mapping of the hydrogel sample was done by measuring 5 × 5 μm regions in multiple locations across the sample. Analysis of the force curved data was carried on using JPK data processing software (JPK Instruments AG, Berlin, Germany), which allows batch processing.2.5. Finite element analysis (FEA)
After reconstruction, 3D models in triangular mesh were exported first, which represented the actual porous structures obtained. These meshes were first validated by Solidworks software (Dassault Systèmes, Solidworks Corp., Waltham, Massachusetts, USA) to avoid compatibility issues and ensure the correct unit. The final models were then imported to FEA simulation software ANSYS (ANSYS, Inc., Canonsburg, Pennsylvania, USA), which recognised the models as single solid bodies. Prior to simulation, hydrogel properties were setup based on the average elastic modulus obtained with AFM whereby average densities were measured. Boundary conditions were applied, and finally stress, strain and deformation of the input structure were calculated and analysed after converging of the simulation.3. Results and discussion
3.1. Imaging results of porous hydrogels
The results of a single phase contrast image by XuM is presented in Fig. 2a demonstrating the fine resolution with pixel size at 1.67 μm. For the enlarged region in the inset, hydrogel membrane structures down to ∼5 μm in thickness were distinguishable from the background. The large pores in the top sections were also clearly shown, and densely packed smaller pores were revealed in the lower half of the sample. A rendered visualization of the tomographic series is presented in Fig. 2b, and the overall 3D porous structures are clearly shown. Compared to previous visualization of hydrogel with synchrotron based X-ray phase contrast,30,31 additional details were revealed, in particular the interconnectivity of pores. This is possibly due to the fact that hydrated samples were imaged in previous reports, and contrast and stage stability may have been significantly affected. In the current study, the vacuum chamber of SEM and the internal rotating stage provided excellent isolation of noise and vibration, and that allowed high precision in tomographic operation and imaging. Although dehydrated samples were imaged in this study, freeze drying or phase separation is a typical step in porous hydrogel fabrication,9,41 and the imaging results obtained in this study are representative of the actual structures under physiological conditions.The initial visualization (Fig. 2), revealed an interesting characteristic in which that pore size significantly decreased towards the bottom of the sample (increasing diameter of the micropipette tip). This phenomenon was barely visible by inspecting the internal walls of the micropipette tip with an optical microscope, and only evident from the XuM applied in this study. The top region was in a polar surface shape due to capillary effects, and the results from this study imply that lower density of the hydrogel prior to phase separation will finally result in larger pores.
3.2. Pore size, volume and distribution
A typical segmentation of pores and hydrogel materials from the horizontal plane is illustrated in Fig. 3a. Segmentation was done based on intensity thresholding of voxels representing regions of different densities to distinguish the pores (voids) from the actual hydrogel materials, with the final surface reconstructed (Fig. 3b). Fig. 3c presents a cross-section which demonstrates the reconstruction of all the individual pores in the imaged hydrogel scaffold based on segmentations. Quantitative analysis was then performed to obtain pore volumes and distribution throughout the imaged hydrogel structure, whereby statistically significant features were revealed indicating the increase in the pore size in the top region of the tip.An overview of the geometric dimensions of reconstructed individual pores in the hydrogel sample is presented in Table 1. Both of the top half (tip) and bottom half (bottom) contains 450 horizontal images converted from the tomographic series with voxel size of 1.67 μm. From the numerical values, it is evident that the larger pores are located in the top half, as the median pore volume and pore area are doubled compared to those in the bottom half. Histograms of the pore volume and pore area presented in Fig. 4a and b show that all the data of pore volume and area follow lognormal distributions, with all the calculated goodness-of-fit levels >0.99. Comparison of the pore size and pore volume distributions in the top and bottom halves resulted in p values <0.01 based on z test, which provide statistically relevant confirmation for the initial hypothesis of pore variance.
| Tip | Bottom | |
|---|---|---|
| Number of slices | 450 | |
| Voxel size (μm) | 1.67 | |
| Number of pores reconstructed | 1117 | 3573 |
| Mean pore volume (μm3) | 280![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 248 248 |
95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 916 916 |
| Median pore volume (μm3) | 81![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 748 748 |
43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 585 585 |
| Mean pore area (μm2) | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 627 627 |
12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 012 012 |
| Median pore area (μm2) | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 643 643 |
7612 |
 | ||
| Fig. 4 Comparison of the individual hydrogel pores at the tip and bottom of a micropipette tip, with regard to (a) pore volume and (b) pore area. | ||
3.3. Exploring structure–mechanics: from interconnectivity to FEA
Another notable achievement based on XuM imaging is the feasibility of exploring the interconnectivity of hydrogel pores, which has not been demonstrated based on phase contrast. Porous bone has been a popular target sample for conventional X-ray microtomography, and the feasibility has been proved for deriving the interconnectivity from the 3D model reconstructed.42,43 Medial surface or “skeleton” was first constructed from the pores, and evolved into a 3D graph(s) representing the connectivity of individual pores.44 With the fine details captured by XuM in this study, the corresponding skeleton and the final graphs revealing interconnectivity could be constructed and demonstrated in Fig. 5a. The nodes representing the individual pores are shown in red, while the white segments represent the mutual access of two individual pores. The final result consists of 6 separate graphs, while one single graph contains 99% of the pores implying the pores are effectively interconnected with each other.This assessment of interconnectivity is expected to be crucial for the successful design of new tissue engineering scaffolds, due to its subsequent effect on permeability and proliferation of cells. One recent nanotomography approach was to iteratively image the porous structure with SEM after thin layers of hydrogel were removed with Focused Ion Beam (FIB).45 Considering that the thickness of removed layers can be tens of nanometers and SEM resolution approaches single digit nanometer, this approach allowed the reconstruction of a 3D volume of porous hydrogel with unambiguous interconnectivity evidence. Comparison of the porosity measurements from different approaches also suggested that the results from mercury porosimetry might be lacking significant information compared to those from imaging. Acquisition rate, however, is the major concern for the FIB-SEM approach due to slow material removal through FIB, while the proposed XuM approach is capable of imaging a much larger volume with sufficient resolution.
As the resolution of current phase contrast X-ray approaches submicron levels, it is now feasible to model the mechanics of individual pores as well as their collective performance as a scaffold. The elastic modulus of the target sample could first be measured by AFM force spectroscopy as demonstrated in Fig. 5b, and a lognormal distribution was fitted to the dataset. The average value of 18.17 MPa was supplemented as the elastic modulus of “solid” hydrogel, and with the 3D structure from XuM, the bulk porous scaffold could be modelled. One subvolume example is presented in Fig. 5c showing FEA simulation of a reconstructed porous structure, and the geometric deformation and stresses could then be investigated in this virtual environment under various loading conditions. By further estimating an internal pressure due to containing water, the simulated average elasticity of “bulk” porous hydrated hydrogel is approximately 700 kPa. An additional AFM force spectroscopy measurement on the same porous hydrogel in the bulk hydrated form confirmed that the stiffness is significantly lower with average 200 kPa compared to the material modulus of approximately 20 MPa (Fig. 5d), while the range is consistent with the simulation outputs. All these results demonstrate the feasibility of linking the material properties of a hydrogel with its porous structures as facilitated by phase contrast X-ray imaging, and modelling a complete porous hydrogel sample is only constrained by the current computational capability.
4. Conclusion
In this study we have utilized a high spatial resolution X-ray ultramicroscopy imaging technique based on phase contrast, to visualise three dimensional hydrogel structure, which plays a crucial role as a scaffolding biomaterial in numerous biomedical applications including tissue engineering and drug discovery systems. Specific sample preparation protocols have been developed and demonstrated for the first time the capability of this tomographic imaging technique to capture the complete structure of porous hydrogel structures at a resolution of a few microns. This SEM based X-ray approach avoids the use of synchrotron radiation, and proves to be excellent for scaffold imaging in term of contrast, resolution and stability. The following analysis presented in this study also demonstrates the capabilities of the proposed approach to not only acquire the pore dimensions, but to also quantitatively determine the spatial distribution and connections of pores, which play a vital role in accurate prediction of hydrogel porous structure prior to and during its implantation in vivo. By incorporating nanomechanics testing with AFM, mechanical modelling of individual pores and the bulk scaffold also becomes feasible for the first time. Although dehydrated samples were demonstrated in the current study, hydrated samples can also be imaged by incorporating integrated sample cells26 or other membrane bound liquid cells which allow sufficient penetration of X-ray.46 We expect the developed platform will offer prompt visualization and modelling for customized hydrogel and 3D scaffold development for cell growth, which will be unique for future personalized medicine.Acknowledgements
The authors would like to acknowledge the use of facilities within the Monash Centre for Electron Microscopy (MCEM) and support of Monash Interdisciplinary Research (IDR) Seed Fund and Monash Engineering International Postgraduate Research Scholarship (FEIPRS). This research also used equipment funded by Australian Research Council grant LE0882821.References
- W. Yeong, C. Chua, K. Leong and M. Chandrasekaran, Rapid prototyping in tissue engineering: challenges and potential, Trends Biotechnol., 2004, 22, 643 CrossRef CAS PubMed
.
- F. Rosso, G. Marino, A. Giordano, M. Barbarisi, D. Parmeggiani and A. Barbarisi, Smart materials as scaffolds for tissue engineering, J. Cell. Physiol., 2005, 203, 465 CrossRef CAS PubMed
.
- M. E. Furth, A. Atala and M. E. van Dyke, Smart biomaterials design for tissue engineering and regenerative medicine, Biomaterials, 2007, 28, 5068 CrossRef CAS PubMed
.
- D. W. Hutmacher, Scaffold design and fabrication technologies for engineering tissues—state of the art and future perspectives, J. Biomater. Sci., Polym. Ed., 2001, 12, 107 CrossRef CAS PubMed
.
- S.-M. Lien, L.-Y. Ko and T.-J. Huang, Effect of pore size on ECM secretion and cell growth in gelatin scaffold for articular cartilage tissue engineering, Acta Biomater., 2009, 5, 670 CrossRef CAS PubMed
.
- F. J. O'Brien, B. A. Harley, I. V. Yannas and L. J. Gibson, The effect of pore size on cell adhesion in collagen-GAG scaffolds, Biomaterials, 2005, 26, 433 CrossRef PubMed
.
- A. Al-Abboodi, J. Fu, P. M. Doran, T. T. Y. Tan and P. P. Y. Chan, Injectable 3D Hydrogel Scaffold with Tailorable Porosity Post-Implantation, Adv. Healthcare Mater., 2014, 3, 725 CrossRef CAS PubMed
.
- S. J. Bryant, J. L. Cuy, K. D. Hauch and B. D. Ratner, Photo-patterning of porous hydrogels for tissue engineering, Biomaterials, 2007, 28, 2978 CrossRef CAS PubMed
.
- S. P. Hoo, Q. L. Loh, Z. Yue, J. Fu, T. T. Tan, C. Choong and P. P. Chan, Preparation of a soft and interconnected macroporous hydroxypropyl cellulose methacrylate scaffold for adipose tissue engineering, J. Mater. Chem. B, 2013, 1, 3107 RSC
.
- Q. Liu, E. L. Hedberg, Z. Liu, R. Bahulekar, R. K. Meszlenyi and A. G. Mikos, Preparation of macroporous poly(2-hydroxyethyl methacrylate) hydrogels by enhanced phase separation, Biomaterials, 2163, 21, 2000 Search PubMed
.
- Y. Kim, A. Abuelfilat, S. Hoo, A. Al-Abbood, B. Liu, T. W. Ng, P. P. Y. Chan and J. Fu, Tuning the surface properties of hydrogel at nanoscale with focused ion irradiation, Soft Matter, 2014, 10, 8448 RSC
.
- L. J. Lee, Polymer nanoengineering for biomedical applications, Ann. Biomed. Eng., 2006, 34, 75 CrossRef PubMed
.
- A. S. Hoffman, Hydrogels for biomedical applications, Adv. Drug Delivery Rev., 2002, 54, 3 CrossRef CAS
.
- J. A. Hubbell, Materials as morphogenetic guides in tissue engineering, Curr. Opin. Biotechnol., 2003, 14, 551 CrossRef CAS PubMed
.
- J. da Silva, et al., The cavity-to-cavity migration of leukaemic cells through 3D honey-combed hydrogels with adjustable internal dimension and stiffness, Biomaterials, 2010, 31, 2201 CrossRef CAS PubMed
.
- H. J. Kong, T. R. Polte, E. Alsberg and D. J. Mooney, FRET measurements of cell-traction forces and nano-scale clustering of adhesion ligands varied by substrate stiffness, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 4300 CrossRef CAS PubMed
.
- A. J. Engler, S. Sen, H. L. Sweeney and D. E. Discher, Matrix elasticity directs stem cell lineage specification, Cell, 2006, 126, 677 CrossRef CAS PubMed
.
- Y.-C. Chiu, S. Kocagöz, J. C. Larson and E. M. Brey, Evaluation of Physical and Mechanical Properties of Porous Poly(Ethylene Glycol)-co-(L-Lactic Acid) Hydrogels During Degradation, PLoS One, 2013, 8, e60728 CAS
.
- A. Leal-Egana, U.-D. Braumann, A. Diaz-Cuenca, M. Nowicki and A. Bader, Determination of pore size distribution at the cell-hydrogel interface, J. Nanobiotechnol., 2011, 9, 24 CrossRef CAS PubMed
.
- S. H. Kim and C. C. Chu, Pore structure analysis of swollen dextran–methacrylate hydrogels by SEM and mercury intrusion porosimetry, J. Biomed. Mater. Res., 2000, 53, 258 CrossRef CAS
.
- S. M. Paterson, Y. S. Casadio, D. H. Brown, J. A. Shaw, T. V. Chirila and M. V. Baker, Laser scanning confocal microscopy versus scanning electron microscopy for characterization of polymer morphology: sample preparation drastically distorts morphologies of poly(2-hydroxyethyl methacrylate)-based hydrogels, J. Appl. Polym. Sci., 2013, 127, 4296 CrossRef CAS PubMed
.
- M. A. Kotlarchyk, E. L. Botvinick and A. J. Putnam, Characterization of hydrogel microstructure using laser tweezers particle tracking and confocal reflection imaging, J. Phys.: Condens. Matter, 2010, 22, 194121 CrossRef CAS PubMed
.
- S. Mayo, P. Miller, S. Wilkins, T. Davis, D. Gao, T. Gureyev, D. Paganin, D. Parry, A. Pogany and A. Stevenson, Quantitative X-ray projection microscopy: phase-contrast and multi-spectral imaging, J. Microsc., 2002, 207, 79 CrossRef CAS
.
- D. Paganin, S. Mayo, T. E. Gureyev, P. R. Miller and S. W. Wilkins, Simultaneous phase and amplitude extraction from a single defocused image of a homogeneous object, J. Microsc., 2002, 206, 33 CrossRef CAS
.
- S. Mayo, T. Davis, T. Gureyev, P. Miller, D. Paganin, A. Pogany, A. Stevenson and S. Wilkins, X-ray phase-contrast microscopy and microtomography, Opt. Express, 2003, 11, 2289 CrossRef CAS
.
- D. Gao, S. W. Wilkins, D. J. Parry, T. E. Gureyev, P. R. Miller and E. Hanssen, X-ray ultramicroscopy using integrated sample cells, Opt. Express, 2006, 14, 7889 CrossRef
.
- C. V. E. A. W. C. Nixon, The X-ray shadow microscope, J. Appl. Phys., 1953, 24, 616 CrossRef PubMed
.
- M. Reed Teague, Deterministic phase retrieval: a Green's function solution, J. Opt. Soc. Am., 1983, 73, 1434 CrossRef
.
- L. Feldkamp, L. Davis and J. Kress, Practical cone-beam algorithm, J. Opt. Soc. Am. A, 1984, 1, 612 CrossRef
.
- A. A. Appel, J. C. Larson, S. Somo, Z. Zhong, P. P. Spicer, F. K. Kasper, A. B. Garson III, A. M. Zysk, A. G. Mikos and M. A. Anastasio, Imaging of poly(α-hydroxy-ester) scaffolds with x-ray phase-contrast microcomputed tomography, Tissue Eng., Part C, 2012, 18, 859 CrossRef CAS PubMed
.
- E. M. Brey, A. Appel, Y.-C. Chiu, Z. Zhong, M.-H. Cheng, H. Engel and M. A. Anastasio, X-ray imaging of poly(ethylene glycol) hydrogels without contrast agents, Tissue Eng., Part C, 2010, 16, 1597 CrossRef CAS PubMed
.
- DigitalMicrograph®, G.a. X-ray ultra Microscope (XuM) Model 502, Product Brochure.
- D. Wu, D. Gao, S. C. Mayo, J. Gotama and C. Way, X-ray ultramicroscopy: a new method for observation and measurement of filler dispersion in thermoplastic composites, Compos. Sci. Technol., 2008, 68, 178 CrossRef CAS PubMed
.
- J. S.-P. XuM, Image below the surface and add another dimension to microscopy using the SEM.
- M. Radmacher, Measuring the elastic properties of biological samples with the AFM, IEEE Eng. Med. Biol. Mag., 1997, 16, 47 CrossRef CAS
.
- M. E. Dokukin and I. Sokolov, Quantitative mapping of the elastic modulus of soft materials with HarmoniX and PeakForce QNM AFM modes, Langmuir, 2012, 28, 16060 CrossRef CAS PubMed
.
- Y. Shan and H. Wang, The structure and function of cell membranes examined by atomic force microscopy and single-molecule force spectroscopy, Chem. Soc. Rev., 2015, 44, 3617 RSC
.
- F. W. Stetter, S. Kienle, S. Krysiak and T. Hugel, Investigating Single Molecule Adhesion by Atomic Force Spectroscopy, J. Visualized Exp., 2015, e52456 Search PubMed
.
- B. Liu, M. H. Uddin, T. W. Ng, D. L. Paterson, T. Velkov, J. Li and J. Fu, In situ probing the interior of single bacterial cells at nanometer scale, Nanotechnology, 2014, 25, 415101 CrossRef PubMed
.
- J. Zlatanova, S. M. Lindsay and S. H. Leuba, Single molecule force spectroscopy in biology using the atomic force microscope, Prog. Biophys. Mol. Biol., 2000, 74, 37 CrossRef CAS
.
- H.-W. Kang, Y. Tabata and Y. Ikada, Fabrication of porous gelatin scaffolds for tissue engineering, Biomaterials, 1999, 20, 1339 CrossRef CAS
.
- A. C. Jones, C. H. Arns, D. W. Hutmacher, B. K. Milthorpe, A. P. Sheppard and M. A. Knackstedt, The correlation of pore morphology, interconnectivity and physical properties of 3D ceramic scaffolds with bone ingrowth, Biomaterials, 2009, 30, 1440 CrossRef CAS PubMed
.
- J. R. Jones, G. Poologasundarampillai, R. C. Atwood, D. Bernard and P. D. Lee, Non-destructive quantitative 3D analysis for the optimisation of tissue scaffolds, Biomaterials, 2007, 28, 1404 CrossRef CAS PubMed
.
- T.-C. Lee, R. L. Kashyap and C.-N. Chu, Building skeleton models via 3-D medial surface axis thinning algorithms, CVGIP: Graphical Models and Image Processing, 1994, 56, 462 CrossRef
.
- A. Al-Abboodi, J. Fu, P. M. Doran and P. P. Y. Chan, Three-dimensional nanocharacterization of porous hydrogel with ion and electron beams, Biotechnol. Bioeng., 2013, 110, 318 CrossRef CAS PubMed
.
- N. D. Jonge, D. B. Peckys, G. J. Kremers and D. W. Piston, Electron microscopy of whole cells in liquid with nanometer resolution, Proc. Natl. Acad. Sci. U. S. A., 2159, 106, 2009 Search PubMed
.
| This journal is © The Royal Society of Chemistry 2015 |