Microbial glycosylation of tanshinone IIA by Cunninghamella elegans AS 3.2028†
Wen-fei Liang,
Zi-wei Li,
Shuai Ji,
Qi Wang,
Xue Qiao,
De-an Guo and
Min Ye*
State Key Laboratory of Natural and Biomimetic Drugs, School of Pharmaceutical Sciences, Peking University, 38 Xueyuan Road, Beijing 100191, China. E-mail: yemin@bjmu.edu.cn
First published on 20th July 2015
Abstract
Tanshinone IIA (TIIA) is a natural product with significant anti-atherogenic activities. However, poor water-solubility and low bioavailability hinder its further exploitation as a drug candidate. In this study, microbial transformation of TIIA by Cunninghamella elegans AS 3.2028 was conducted to obtain two new glycosylated derivatives. Their structures were identified as hydroquinone TIIA 11-O-β-D-glucopyranoside (1) and hydroquinone TIIA 12-O-β-D-glucopyranoside (2) based on extensive NMR and MS spectral analyses. The solubility of 1 in 50% MeOH–H2O solution was approximately 50-fold that of TIIA, and 1 showed remarkably improved oral absorption in mice. Furthermore, 1 and 2 exhibited similar Nrf2 activation activity to TIIA.
Tanshinone IIA (TIIA), with a phenanthrenequinone skeleton, is a major bioactive constituent of Salvia miltiorrhiza, a well-known traditional Chinese herbal medicine, Danshen.1 Recently, Danshen has been recorded as a monograph in United States Pharmacopeia, and TIIA is one of the marker compounds.2 A big array of experimental and clinical investigations have demonstrated that TIIA can be used to treat atherosclerosis, hyperlipidemia, endangiitis, and cancer.3 However, it has very poor water solubility and fairly low bioavailability (around 3% in rats after oral administration).1 Thus, TIIA is usually prepared in sodium tanshinone IIA sulfonate injection (STS) in clinical use. However, the rapid elimination of STS led to reduced pharmacological functions,4 and serious side effects have been reported when STS was taken along with warfarin.5 In recent years, a number of groups have tried to improve solubility and pharmacokinetic properties of TIIA by structural modification, including sulfonation, bromination, and modification of the phenanthrenequinone skeleton.6 Some of these chemical reactions take place under harsh conditions, and toxic reagents are needed.
Microbial transformation is a powerful approach for structural modification of structurally complex natural products. It shows many advantages such as simple operation, mild reaction condition, and high selectivity. In some cases, the biotransformed metabolites may have enhanced biological activities.7 Furthermore, glycosylation, hydroxylation and other biocatalytic reactions could increase polarity and solubility of the substrates.8 However, the microbial transformation of TIIA has never been reported, thus far.
The Cunninghamella species have been reported to catalyze hydroxylation, glycosylation, demethylation, cyclization, and sulfation reactions.9 In this study, we report simultaneous hydrogenation and glucosylation of TIIA with >95% conversion rate by Cunninghamella elegans AS 3.2028. Two new glycosylated products (1 and 2) were obtained, and their structures were identified by extensive NMR and MS spectral analyses. Compound 1 exhibited 50-fold higher solubility in 50% MeOH–H2O solution than TIIA. Moreover, 1 showed remarkably improved oral absorption in mice. The Nrf2 activation activities of TIIA, 1 and 2 were also evaluated.
We screened 15 fungal strains derived from Aspergillus, Penicillium, Syncephalastrum, Alternaria, Cunninghamella, Mucor and Botrytis species, and found that Cunninghamella elegans AS 3.2028 was capable of metabolizing TIIA into two major products (1 and 2) according to HPLC analysis. The ratio of 1 to 2 was approximately 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 based on their peak area at 254 nm (Fig. 1), and the substrates were almost completely metabolized. The UV spectra of 1 and 2 were very similar to those of TIIA, suggesting they had a similar skeleton. In the mass spectra, 1 and 2 exhibited the same [M − H]− ion at m/z 457, 164 Da greater than TIIA, indicating that 1 and 2 should be hydrogenated (+2 Da) and glycosylated (+162 Da) derivatives of TIIA. This deduction was confirmed by the neutral loss of 162 Da (a glucosyl residue) in the MS/MS spectra (Fig. 2).
1 based on their peak area at 254 nm (Fig. 1), and the substrates were almost completely metabolized. The UV spectra of 1 and 2 were very similar to those of TIIA, suggesting they had a similar skeleton. In the mass spectra, 1 and 2 exhibited the same [M − H]− ion at m/z 457, 164 Da greater than TIIA, indicating that 1 and 2 should be hydrogenated (+2 Da) and glycosylated (+162 Da) derivatives of TIIA. This deduction was confirmed by the neutral loss of 162 Da (a glucosyl residue) in the MS/MS spectra (Fig. 2).
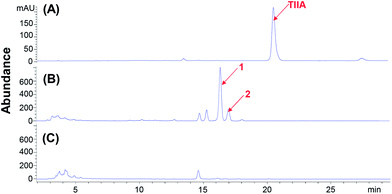 | ||
| Fig. 1 HPLC chromatograms for microbial transformation of TIIA. (A) TIIA standard. (B) Biotransformed products of TIIA by Cunninghamella elegans. (C) Blank control of C. elegans. | ||
For preparative-scale microbial transformation, a total of 32 mg of TIIA was added into C. elegans, and 8.0 mg of 1 (25.0% yield) and 1.5 mg of 2 (4.7% yield) were obtained by repeated column chromatography and semi-preparative liquid chromatography (Fig. 3). The purities were above 95% by HPLC analysis. Both 1 and 2 are new compounds.
Compound 1 had the molecular formula of C25H30O8, as established by its HRESIMS spectrum (m/z 459.2009 [M + H]+, calcd 459.2013). The 13C NMR resonances at δC 107.1, 77.0, 76.1, 74.1, 69.8 and 61.2, as well as the anomeric proton resonance at δH 4.41 (1H, d, J = 8.4 Hz), indicated the presence of a β-D-glucose residue (Table 1). In order to confirm the sugar residue, 1 was hydrolyzed by trifluoroacetic acid, and the water-soluble fraction was analyzed by ion chromatography coupled with pulsed amperometric detection (IC-PAD) as we had previously reported.10 As expected, the chromatogram showed one peak at the same retention time as D-glucopyranose (Fig. 4). The other NMR resonances were very similar to those of TIIA, except for C-11 and C-12. The two carbonyl resonances at δC 183.8 (C-11) and 175.9 (C-12) in TIIA disappeared, and two oxygenated olefinic carbon signals appeared at δC 136.1 and 142.1 in 1, indicating that a double bond formed between C-11 and C-12. In addition, a new phenolic hydroxyl group resonated at δH 9.83, and showed HMBC correlations with C-11 (136.1), C-12 (142.1) and C-13 (115.3). These evidences suggested the phenolic hydroxyl group was connected to C-12. The HMBC correlation between the anomeric proton (δH 4.41) and C-11 (136.1) indicated the β-D-glucose residue was linked to C-11. Therefore, the structure of compound 1 was identified as hydroquinone TIIA 11-O-β-D-glucopyranoside (Fig. 3).
| Position | 1 (600 MHz for 1H) | 2 (400 MHz for 1H) | ||
|---|---|---|---|---|
| δH (ppm, J in Hz) | δC type | δH (ppm, J in Hz) | δC type | |
| 1 | 3.98, m; 2.96, dt (4.2, 18.0) | 31.0, CH2 | 3.49, d (6.4) | 31.0, CH2 |
| 2 | 1.80, m; 1.50, m | 20.0, CH2 | 1.74, d (6.4) | 19.8, CH2 |
| 3 | 1.66, br s | 38.8, CH2 | 1.66, br s | 38.3, CH2 |
| 4 | 34.7, C | 34.5, C | ||
| 5 | 142.7, C | 141.9, C | ||
| 6 | 7.43, d (8.4) | 123.4, CH | 7.55, d (8.4) | 125.8, CH |
| 7 | 7.83, d (8.4) | 116.7, CH | 7.88, d (8.4) | 118.1, CH |
| 8 | 114.2, C | 118.6, C | ||
| 9 | 126.2, C | 121.9, C | ||
| 10 | 132.4, C | 133.7, C | ||
| 11 | 136.1, C | 142.9, C | ||
| 12 | 142.7, C | 135.0, C | ||
| 13 | 115.3, C | 116.3, C | ||
| 14 | 148.6, C | 144.6, C | ||
| 15 | 116.3, C | 117.2, C | ||
| 16 | 7.73, d (1.2) | 141.1, CH | 7.69, s | 141.4, CH |
| 17 | 2.38, d (1.2) | 9.6, CH3 | 2.39, s | 9.6, CH3 |
| 18 | 1.30, s | 31.5,CH3 | 1.32, s | 32.0, CH3 |
| 19 | 1.29, s | 32.5, CH3 | 1.30, s | 31.9, CH3 |
| glc-1 | 4.41, d (8.4) | 107.6, CH | 4.57, d (8.0) | 106.7, CH |
| glc-2 | 3.43, m | 74.1, CH | 3.40, br s | 74.0, CH |
| glc-3 | 3.25, m | 76.1, CH | 3.30, br s | 76.2, CH |
| glc-4 | 3.21, m | 69.8, CH | 3.17, br s | 69.8, CH |
| glc-5 | 3.04, m | 77.0, CH | 3.17, br s | 77.5, CH |
| glc-6 | 3.57, d (10.8); 3.44, m | 61.2, CH2 | 3.68, m; 3.49, m | 61.0, CH2 |
| OH | 9.83, s | 9.21, s | ||
Compound 2 had the same molecular formula as 1, as deduced from the HRESIMS spectrum (m/z 459.2008 [M + H]+, calcd 459.2013). Its 1H and 13C NMR spectra were very similar to those of 1 (Table 1). The carbon resonances at δC 106.7, 77.5, 76.2, 74.0, 69.8 and 61.0, as well as the anomeric proton resonance at δH 4.57 (1H, d, J = 8.0 Hz), indicated the presence of a β-D-glucose residue. This was confirmed by IC-PAD analysis with a reference standard (Fig. 4). By careful analysis of the 2D NMR spectrum, we found the phenolic hydroxyl group at δH 9.21 showed HMBC correlations with C-9 (δC 121.9), C-11 (δC 142.9) and C-12 (δC 135.0) in 2, suggesting it was connected to C-11. In addition, the HMBC correlation between the anomeric proton (δH 4.57) and C-12 (135.0) indicated the β-D-glucose residue was linked to C-12. Based on the above evidences, the structure of compound 2 was established as hydroquinone TIIA 12-O-β-D-glucopyranoside.
Hydroquinone TIIA 11-O-β-D-glucopyranoside (1): brick-red amorphous powder; [α]D25 + 0.087 (c 0.1, MeOH); UV (MeOH) λmax (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 260 (3.34) nm; IR (KBr) νmax 3433, 2926, 1636, 1065 cm−1; HRESIMS m/z 459.2009 for [M + H]+ (calcd 459.2013), corresponding to C25H30O8; 1H NMR (600 MHz, DMSO-d6) and 13C NMR (150 MHz, DMSO-d6) data, see Table 1.
ε) 260 (3.34) nm; IR (KBr) νmax 3433, 2926, 1636, 1065 cm−1; HRESIMS m/z 459.2009 for [M + H]+ (calcd 459.2013), corresponding to C25H30O8; 1H NMR (600 MHz, DMSO-d6) and 13C NMR (150 MHz, DMSO-d6) data, see Table 1.
Hydroquinone TIIA 12-O-β-D-glucopyranoside (2): brick-red amorphous powder; [α]D25 + 0.019 (c 0.1, MeOH); UV (MeOH) λmax (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 260 (3.34) nm; IR (KBr) νmax 3433, 2927, 1674, 1636, 1068 cm−1. HRESIMS m/z 459.2008 for [M + H]+ (calcd 459.2013), corresponding to C25H30O8; 1H NMR (400 MHz, DMSO-d6) and 13C NMR (100 MHz, DMSO-d6) data, see Table 1.
ε) 260 (3.34) nm; IR (KBr) νmax 3433, 2927, 1674, 1636, 1068 cm−1. HRESIMS m/z 459.2008 for [M + H]+ (calcd 459.2013), corresponding to C25H30O8; 1H NMR (400 MHz, DMSO-d6) and 13C NMR (100 MHz, DMSO-d6) data, see Table 1.
A time-course study showed TIIA could be almost completely metabolized within 10 h of co-incubation with C. elegans (Fig. 5). Compound 1 was the major product and its relative amount increased to 80% at 10 h. Interestingly, the relative amount of 2 maintained at around 20% during 24 h. Similarly, previous reports had revealed that only hydroquinone TIIA 11-O-β-D-glucuronic acid, but not hydroquinone TIIA 12-O-β-D-glucuronic acid, was detected as a major metabolite of TIIA in rats after oral administration.11
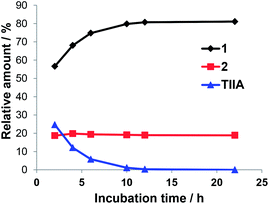 | ||
| Fig. 5 Time-course for the biotransformation of TIIA by Cunninghamella elegans (the relative amounts were calculated on the basis of peak areas in the HPLC chromatogram at 254 nm). | ||
Glycosylation is a feasible way to prepare natural product derivatives with improved solubility and bioactivities.12 Although a big number of TIIA derivatives have been synthesized by chemical means, this is the first time the glycosides of TIIA were prepared. Moreover, the biocatalytic reaction could take place efficiently under mild reaction conditions.
We tested the solubility of TIIA and 1 (2 was not tested due to its limited amount). Since both compounds were almost insoluble in H2O, their solubility in 50% MeOH–H2O solution was determined. As expected, glucosylation could significantly enhance the solubility of TIIA. TIIA was only slightly soluble in 50% MeOH–H2O (1.7 μg mL−1). In contrast, the solubility of 1 increased by almost 50-fold to 84.6 μg mL−1 (Table S2†).
In order to compare the bioavailability of TIIA and 1, they were orally administered to mice at the same mole dose (0.17 mmol kg−1). The plasma samples were collected at 1 h and 2 h after administration, because the time of occurrence for maximum drug concentration (Tmax) for TIIA in rats was around 1 h.1 The plasma samples were combined and analyzed by HPLC/DAD/ESI-MSn. The results indicated both TIIA and 1 could be absorbed into circulation within 2 h, and only the prototypes were detected (Fig. S2†). According to LC/SRM-MS quantitative analysis, the concentrations of TIIA and 1 in the plasma samples were 0.10 nM and 1.39 nM, respectively, indicating that glycosylation could remarkably increase the oral absorption of TIIA (Fig. S3†).
The antioxidant activity of TIIA could contribute to its cardiovascular therapeutic effects.13 In this study, the Nrf2 transcription activation activities of TIIA, 1 and 2 were evaluated by using a luciferase reporter assay in HepG2 human hepatocellular carcinoma cells.14 The glycosylated products 1 and 2 had similar antioxidant activities to TIIA, and all of them could significantly activate Nrf2 transcription in a concentration-dependent manner (around 4-fold of the control at 20 μM) (Fig. 6).
Conclusions
In summary, we obtained two new glycosylated products of TIIA (1 and 2) by microbial transformation of Cunninghamella elegans AS 3.2028. Their structures were established based on extensive NMR and MS spectroscopic analyses. The solubility of 1 in 50% MeOH–H2O solution was approximately 50 times higher than that of TIIA, and the oral absorption in mice also increased remarkably when TIIA was glycosylated. Furthermore, 1 and 2 exhibited similar antioxidant activities to TIIA. This is the first report on the glycosylation of TIIA by microbial transformation. The glycosylated products show noticeable druggability, and warrant to be further evaluated in future work.Acknowledgements
This work was supported by National Natural Science Foundation of China (No. 81222054), and the Program for New Century Excellent Talents in University from Chinese Ministry of Education (No. NCET-11-0019). We wish to thank Dr Zhenbang Jiang (Thermo-Dionex Inc.) for his technical help with IC-PAD analysis.Notes and references
- S. Gao, Z. P. Liu, H. Li, P. J. Little, P. Q. Liu and S. W. Xu, Atherosclerosis, 2012, 220, 3–10 CrossRef CAS PubMed.
- United States Pharmacopoeia <Salvia 1-3> Dietary Supplements, USP 37/NF, 32 edn, U. S. Pharmacopoeia Convention, Rockville, MD, 2014.
- (a) Y. Zhang, P. X. Jiang, M. Ye, S. H. Kim, C. Jiang and J. X. Lü, Int. J. Mol. Sci., 2012, 13, 13621–13666 CrossRef CAS PubMed; (b) F. T. Tang, X. Q. Wu, T. Q. Wang, P. Wang, R. F. Li, H. J. Zhang, J. Gao, S. R. Chen, L. P. Bao, H. Q. Huang and P. Q. Liu, Vasc. Pharmacol., 2007, 46, 427–438 CrossRef CAS PubMed; (c) W. Y. Chen, F. T. Tang, S. R. Chen and P. Q. Liu, China Pharm., 2008, 19, 884–887 CAS; (d) B. Wei, W. W. Li, J. Ji, Q. H. Hu and H. Ji, Atherosclerosis, 2014, 235, 318–327 CrossRef CAS PubMed; (e) Y. Zhang, S. H. Won, C. Jiang, H. J. Lee, S. J. Jeong, E. O. Lee, J. H. Zhang, M. Ye, S. H. Kim and J. X. Lü, Pharm. Res., 2012, 29, 1595–1608 CrossRef CAS PubMed.
- W. Z. Chen, Y. L. Dong, C. G. Wang and G. S. Ding, Acta Pharmacol. Sin., 1979, 14, 277–282 CAS.
- (a) J. Liu, X. R. Wang, Z. W. Cai and F. S. C. Lee, J. Am. Soc. Mass Spectrom., 2008, 19, 1568–1575 CrossRef CAS PubMed; (b) J. Liu and Z. P. Chen, Nurs. Prac. Res., 2009, 6, 63 Search PubMed.
- (a) W. G. Huang, J. Y. Li, Y. Luo, J. Li and W. Lu, Chin. Chem. Lett., 2009, 20, 1461–1464 CrossRef CAS PubMed; (b) Y. F. Bi, H. W. Xu, X. Q. Liu, X. J. Zhang, Z. J. Wang and H. M. Liu, Bioorg. Med. Chem. Lett., 2010, 20, 4892–4894 CrossRef CAS PubMed; (c) C. J. Sun and D. L. Bai, Acta Pharmacol. Sin., 1985, 20, 39–43 CAS; (d) X. Z. Bu, Z. S. Huang, M. Zhang, L. Ma, G. W. Xiao, Z. P. Zhong and L. Q. Gu, Chin. J. Org. Chem., 2001, 3, 223–230 Search PubMed.
- (a) V. Křen, J. Kubisch, P. Sedmera, P. Halada, V. Přikrylová, A. Jegorov, L. Cvak, R. Gebhardt, J. Ulrichová and V. Šimánek, J. Chem. Soc., Perkin Trans. 1, 1997, 2467–2474 RSC; (b) H. X. Ge, J. Zhang, L. Chen, J. P. Kou and B. Y. Yu, Bioorg. Med. Chem., 2013, 21, 62–69 CrossRef CAS PubMed; (c) X. B. Yang, J. Hou, D. Liu, S. Deng, Z. B. Wang, H. X. Kuang, C. Y. Wang, J. H. Yao, K. X. Liu and X. C. Ma, J. Mol. Catal. B: Enzym., 2013, 88, 1–6 CrossRef CAS PubMed; (d) K. Shimoda and H. Hamada, Nutrients, 2010, 2, 171–180 CrossRef CAS PubMed.
- J. R. Jiang, S. Yuan, J. F. Ding, S. C. Zhu, H. D. Xu, X. T. Chen, X. D. Cong, W. P. Xu, H. Y. Yi and Y. J. Dai, Appl. Microbiol. Biotechnol., 2008, 81, 647–657 CrossRef CAS PubMed.
- H. Cao, X. Q. Chen, A. R. Jassbi and J. B. Xiao, Biotechnol. Adv., 2015, 33, 214–223 CrossRef CAS PubMed.
- W. Song, L. Si, S. Ji, H. Wang, X. M. Fang, L. Y. Yu, R. Y. Li, L. N. Liang, D. Zhou and M. Ye, J. Nat. Prod., 2014, 77, 1632–1643 CrossRef CAS PubMed.
- P. Li, G. Wang, J. Li, H. Hao and C. Zheng, J. Mass Spectrom., 2006, 41, 670–684 CrossRef CAS PubMed.
- V. K. Kapoor and A. Kaur, Mini-Rev. Med. Chem., 2013, 13, 584–596 CrossRef CAS.
- H. S. Zhang and S. W. Wang, Biochem. Pharmacol., 2007, 73, 1358–1366 CrossRef CAS PubMed.
- (a) T. W. Kensler and N. Wakabayashi, Annu. Rev. Pharmacol. Toxicol., 2007, 47, 89–116 CrossRef CAS PubMed; (b) J. D. Hayes and A. T. Dinkova-Kostova, Trends Biochem. Sci., 2014, 39, 199–218 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: NMR and HRESIMS spectra for 1 and 2, and experimental details. See DOI: 10.1039/c5ra09745b |
| This journal is © The Royal Society of Chemistry 2015 |




