Graphene oxide nanoparticle attachment and its toxicity on living lung epithelial cells†
Abstract
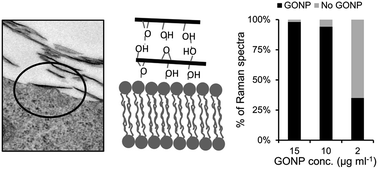
Since its discovery, graphene and its oxidized form, graphene oxide (GO), have attracted interest in a wide range of technical applications. Concerns about their potential toxicity calls for scrutinized studies, but hitherto conflicting results have been reported which partly may be due to variations of synthesis and exposure procedures. Here we report on the attachment and toxicity of contamination-free graphene oxide nanoparticles (GONP) in living lung epithelial cells. The synthesis of chemically pure GONP was made by an improvement of the Hummer's method based on graphene exfoliated from graphite using high-intensity ultrasonication, resulting in two dimensional sheets with a lateral dimension in the range 200 nm to 3 μm and thickness of 0.9 nm. Confocal Raman spectroscopy combined with multivariate analysis was used to study the interaction of GONP and living cells. It is shown that overlapping Raman bands due to GONPs and biomolecules in the cells can clearly be separated with this approach. Orthogonal partial least squares discriminant analysis was used to compare spectral data collected from cells exposed to GONP with spectral data collected from non-exposed control cells, and spectral data from cells exposed to a surfactant known to induce apoptosis. Our analyses show that GONP readily attach to the cells, forming sheets which cover a large fraction of the cell surfaces, and induce small chemical changes. In particular, chemical modifications of proteins and lipids in lung epithelial cells are inferred. GONPs do not, however, decrease cell viability. In contrast, enhanced cell proliferation is observed. Our results shed new light on the interactions of GO, and in contrast to some previous reports, suggest that GO is not toxic. The hyperspectral Raman spectroscopy analysis employed here should be applicable for other fields in nanomedicine as a label-free non-perturbing analytical method.

 Please wait while we load your content...
Please wait while we load your content...