Blocking the heat shock response and depleting HSF-1 levels through heat shock protein 90 (hsp90) inhibition: a significant advance on current hsp90 chemotherapies†
Yen Chin Koay,
Jeanette R. McConnell,
Yao Wang and
Shelli R. McAlpine*
School of Chemistry, University of New South Wales, Sydney, NSW 2052, Australia. E-mail: s.mcalpine@unsw.edu.au
First published on 30th June 2015
Abstract
Clinical inhibitors of heat shock protein 90 (hsp90) modulate the N-terminus of the protein, which elicits a cell rescue cascade known as the heat shock response. This cytoprotective mechanism counteracts the impact of hsp90 chemotherapeutic agents. Inhibiting hsp90's activity via the C-terminus does not produce a heat shock response. Herein we report an extensive structure–activity relationship on 41 molecules that are based on the SM class of cyclic pentapeptides. This class of compounds control hsp90's C-terminus function, which induces rapid cell death without activating the heat shock response. We show that modifying single and dual side-chains was one route for producing active molecules. Moving the N-methyl residue around the ring also impact the biological activity of the molecule. Two of the most potent analogues were evaluated for hsp90 inhibitory activity and for their ability to reduce the heat shock response while simultaneously killing cancer cells. In addition, analysis of the most effective molecules in pharmacokinetic studies are described highlighting the compound's potential as a therapeutic drug.
Introduction
Heat shock protein 90 (hsp90) has been a clinical oncogenic target since 2001.1 Recent clinical outcomes show that molecules that modulate hsp90 via binding to its ATP binding site at the N-terminal domain have several drawbacks.2 One of the most prominent issues faced by these “classical” inhibitors is the induction of a rescue cascade, which produces high levels of cytoprotective heat shock proteins (hsps), termed a heat shock response (HSR), a process governed by the transcription factor heat shock factor 1 (HSF-1). The classical hsp90 inhibitors that control hsp90's function by blocking ATP produce high levels of hsp70.1,3–6 These hsps facilitate cancer cell survival by maintaining numerous oncogenic pathways, and promoting resistance mechanisms.7 In contrast to these classical inhibitors, modulating hsp90's C-terminus controls its oncogenic function without inducing a HSR.3–6 Thus, recent efforts have focused on developing new compounds that control access to hsp90's C-terminus.Over the past several years, McAlpine and co-workers have published mechanistic data on four macrocycles, SM122,3 SM145, SM249 and SM253,5 all of which modulate hsp90's C-terminus. These compounds block co-chaperone access to hsp90's C-terminus, inhibit numerous oncogenic pathways regulated by hsp90, and control hsp90's protein folding function, all without inducing the HSR. Indeed, this class of compound causes a significant reduction in hsp levels.3–5
Herein we describe an extensive structure–activity relationship on derivatives that are based on McAlpine's most recent compounds SM249 and SM253 (Fig. 1). Synthesis and anticancer activity of 41 analogues in HCT116 colon cancer, and MiaPaCa-2 pancreatic cancer cell lines are described, with 35 of these compounds being reported here for the first time. The most effective analogues were then mechanistically evaluated for their ability to: (a) rapidly induce apoptosis, (b) cause phase-specific cell cycle arrest, (c) stop direct binding events between hsp90 and C-terminal co-chaperones (d) inhibit hsp90's protein folding function, and (e) reduce hsp levels associated with the HSR.
Results and discussion
Design
Our initial studies started with designing molecules based on SM249, which had a GI50 = 7.7 μM in HCT116, and 12.8 μM in MiaPaCa-2 cell lines. SM249 has a C![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 9.0 (Fig. 1). Thus, the molecule was reasonably potent as a lead, but was highly insoluble. The synthesis and biological activity of series I molecules (Fig. 1), where R1 was moiety a–f (i.e. compounds 1–6) were recently published (Fig. 1).5 That study produced the lead compound SM253, (4), where a thiazole moiety was placed at R1. SM253 had a 3 fold greater GI50 over SM249, as well as a significantly lower C
P = 9.0 (Fig. 1). Thus, the molecule was reasonably potent as a lead, but was highly insoluble. The synthesis and biological activity of series I molecules (Fig. 1), where R1 was moiety a–f (i.e. compounds 1–6) were recently published (Fig. 1).5 That study produced the lead compound SM253, (4), where a thiazole moiety was placed at R1. SM253 had a 3 fold greater GI50 over SM249, as well as a significantly lower C![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P (7.4). Thus, the incorporation of alternative moieties at R1 was investigated. The histidine derivatives (g and h), e.g. compounds 7 and 8, mimic the thiazole, while the glutamic acid derivatives (i–k) e.g. compounds 9–11, increase the polarity of the molecule, thus improving the solubility.
P (7.4). Thus, the incorporation of alternative moieties at R1 was investigated. The histidine derivatives (g and h), e.g. compounds 7 and 8, mimic the thiazole, while the glutamic acid derivatives (i–k) e.g. compounds 9–11, increase the polarity of the molecule, thus improving the solubility.
Series II (Fig. 1) involved synthesizing a complete set of analogues where R2 = a–k, (compounds 12–22) thus providing the opportunity to compare activity when the same moieties were placed at different a position around the ring (Fig. 1).
Series III was only generated as a single analog, the thiazole at R3 (Fig. 1). Finally, synthesis of molecules with moieties a–d were placed at position R4 generating structures 24–27. The biphenyl was well established to be essential to the activity of the molecule and was therefore not modified.
Synthesis
The synthesis of 35 new analogues was accomplished via a combination of solid phase and solution phase chemistry (Scheme 1). Analogues where positions other than R2 were made via the strategy shown below, but by modifying R1, R3, or R4's substituent respectively. Using a pre-loaded 2-chlorotrityl-leucine resin and then sequentially coupling fluorenylmethyloxy carbonyl (Fmoc) protected amino acids using three equivalents of coupling reagent of 1-hydroxybenzotriazole (HOBt) or 1-hydroxy-7-azabenzotriazole (HOAt) and six equivalents N,N′-diisopropylcarbodiimide (DIC) followed by deprotection of the amine led to the desired resin bound linear pentapeptide. The peptide was cleaved from the resin with 50% trifluoroethanol in dichloromethane, which generated the double de-protected linear pentapeptide, which was then cyclized using three coupling reagents 4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholin-4-ium chloride (DMTMM), 2-(1H-7-azabenzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HATU), and O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium tetrafluoroborate (TBTU) at 0.8 equivalent each, together with 8.0 equivalents of N,N-diisopropylethylamine (DIPEA) under dilute conditions.N-Methylation of the individual amino acid was performed on solid supports during the linear peptide elongation. The synthesis begins with the coupling of the first amino acid on resin and, followed by the subsequent removal of the Fmoc group and the introduction of the nosyl group (2-nitrobenzenesulfonyl group), which then N-methylated using Mitsunobu reaction. Removal of the nosyl group is completed by nucleophilic addition reaction using 1,8-diazabicycloundec-7-ene (DBU) and 2-mercaptoethanol (Scheme 2). However, on-resin methylation on 3,3-diphenyl-D-alanine of 40 repeatedly failed, possibly because of the steric hindrance from the biphenyl ring. Thus, 3,3-diphenyl-D-alanine was methylated through nucleophilic attack of a methylation reagent, iodomethane (MeI) in the presence of a reducing agent, sodium hydride (60% dispersion in mineral oil) in anhydrous THF (0.30 M) in solution.
Structure–activity relationship study
The SAR analysis involved testing the new analogues for their cytotoxicity against HCT116 and MiaPaCa-2 cells (Table 1). The potency of these molecules was compared to the lead compound SM253 (4, Table 1). This parent molecule has an GI50 of approximately 5 μM.5 Of the new changes made to R1, only the inclusion of the tert-butyl protected glutamic acid (9) improved cytotoxicity, with an GI50 of approximately 3 μM and 5 μM in the two cell lines respectively (Table 1). Incorporating a thiazole (15), methoxyphenyl (16), tryptophan (17), or a methylated glutamic acid (22) at R2, improved cytotoxicity compared to SM253, while all the other modifications did not (Table 1).| Cmpd # | Rx | GI50 (μM) HCT-116 | GI50 (μM) MiaPaCa-2 | C![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P P |
|
|---|---|---|---|---|---|
| a Indicates that structure has been reported previously. | |||||
| 1 | R1 | aa | 18.1 ± 0.4 | 23.6 ± 2.5 | 7.60 |
| 2 | ba | 20.8 ± 2.5 | 23.0 ± 2.8 | 7.60 | |
| 3 | ca | 15.7 ± 2.9 | 18.5 ± 5.3 | 7.60 | |
| 4 (253) | da | 5.0 ± 0.5 | 5.5 ± 0.2 | 7.44 | |
| 5 | ea | 3.9 ± 0.2 | 4.5 ± 0.9 | 9.02 | |
| 6 | fa | 5.1 ± 0.6 | 4.4 ± 0.7 | 9.09 | |
| 7 | g | ≥30 | ≥30 | 7.91 | |
| 8 | h | ≥30 | ≥30 | 6.70 | |
| 9 | i | 3.0 ± 1.4 | 5.0 ± 1.8 | 8.40 | |
| 10 | j | ≥30 | ≥30 | 6.79 | |
| 11 | k | 9.6 ± 1.5 | 13.3 ± 2.1 | 7.17 | |
| 12 | R2 | a | ≥30 | ≥30 | 7.56 |
| 13 | b | 8.8 ± 1.0 | 10.3 ± 0.2 | 7.56 | |
| 14 | c | ≥30 | ≥30 | 7.56 | |
| 15 | d | 3.0 ± 1.2 | 3.0 ± 0.8 | 7.40 | |
| 16 | e | 5.3 ± 0.9 | 11.5 ± 0.5 | 8.98 | |
| 17 | f | 6.0 ± 0.2 | 4.4 ± 0.9 | 9.05 | |
| 18 | g | ≥30 | ≥30 | 7.87 | |
| 19 | h | ≥30 | ≥30 | 6.66 | |
| 20 | i | ≥30 | ≥30 | 8.37 | |
| 21 | j | ≥30 | ≥30 | 6.75 | |
| 22 | k | 5.9 ± 0.3 | 4.8 ± 0.6 | 7.13 | |
| 23 | R3 | d | 6.2 ± 0.5 | 6.4 ± 0.6 | 7.93 |
| 24 | R4 | a | 6.3 ± 0.1 | 5.4 ± 1.0 | 7.56 |
| 25 | b | 15.4 ± 0.8 | 12.4 ± 2.0 | 7.56 | |
| 26 | c | 22.2 ± 3.6 | 17.5 ± 1.8 | 7.56 | |
| 27 | d | ≥30 | ≥30 | 7.40 | |
Since the more hydrophobic nature of t-butyl protected glutamic acid, tryptophan, and methoxyphenyl substituents, resulting in high C![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values (8.4–9.0) and had comparable cytotoxicity to the thiazole moiety, which has a much lower C
P values (8.4–9.0) and had comparable cytotoxicity to the thiazole moiety, which has a much lower C![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, we only evaluated the thiazole at position R3, 23. Compound 23 had a similar GI50 value (∼6 μM) to SM253 (Table 1). Moreover, to investigate the influence of different substituents at R4, heterocyclic moieties a–d introduced at R4 produced surprising results; the compound with a thiazole side-chain at this position (27) was not active, while the most effective substitution at R4 was the ortho-pyridal alanine, which had poor activity when placed at R2 and R3. Thus, placing a thiazole at positions R1, R2, and R3 gave effective inhibitors, while a thiazole at position R4 produced an inactive molecule.
P, we only evaluated the thiazole at position R3, 23. Compound 23 had a similar GI50 value (∼6 μM) to SM253 (Table 1). Moreover, to investigate the influence of different substituents at R4, heterocyclic moieties a–d introduced at R4 produced surprising results; the compound with a thiazole side-chain at this position (27) was not active, while the most effective substitution at R4 was the ortho-pyridal alanine, which had poor activity when placed at R2 and R3. Thus, placing a thiazole at positions R1, R2, and R3 gave effective inhibitors, while a thiazole at position R4 produced an inactive molecule.
From these series, six potent molecules were synthesized, however, these all maintained moderate cytotoxicity, with GI50 values of 3–6 μM. Completion of the SAR analysis using a single substitution at each position, allowed us to select the best side chains for synthesizing dual modified analogues (Table 2 and Fig. 2). Given the advantages of a thiazole moiety as a side chain, we maintained the thiazole (d) at R1 and modified R2 or R4. Similarly, maintaining the thiazole (d) at R2 we modified R1 or R4. The most intriguing aspect of the dual modified molecules is that all 9 compounds were less active than the lead structure, or the single modification. Thus no further analogues based on this premise were synthesized.
| Cmpd # | Rx | GI50 (μM) HCT-116 | GI50 (μM) MiaPaCa-2 | C![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P P |
|||
|---|---|---|---|---|---|---|---|
| 28 | R1 | d | R2 | d | ≥30 | 20.2 ± 4.3 | 5.75 |
| 29 | d | e | 8.4 ± 0.5 | 8.8 ± 0.4 | 7.32 | ||
| 30 | d | i | 12.4 ± 2.2 | 9.2 ± 1.0 | 6.71 | ||
| 31 | d | j | ≥30 | ≥30 | 5.10 | ||
| 32 | d | k | ≥30 | ≥30 | 5.47 | ||
| 33 | e | e | 5.2 ± 2.2 | 8.7 ± 1.0 | 8.90 | ||
| 34 | e | d | 10.4 ± 2.8 | 7.5 ± 0.9 | 7.32 | ||
| 35 | R1 | d | R4 | b | ≥30 | ≥30 | 5.91 |
| 36 | R2 | d | R4 | a | 19.5 ± 4.8 | 20.9 ± 3.8 | 5.87 |
We8–14 and others15–18 have shown that placing an N-methyl (N-Me) in combination with a D-amino acid around the pentapeptide ring provides: (a) structure rigidification, which can lead to enhancing compound potency and (b) improved pharmacokinetic properties. Thus, a single analog, compound 15, with a thiazole at R2 was chosen as a template because it was the most potent analog with the lowest C![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P to date. We performed an N-Me scan on compound 15 to evaluate the impact of this moiety on the biological activity. The N-Me substituent was rotated around this macrocycle, or removed completely, which produced a series of five additional compounds (37–41), (Table 3 and Fig. 3).
P to date. We performed an N-Me scan on compound 15 to evaluate the impact of this moiety on the biological activity. The N-Me substituent was rotated around this macrocycle, or removed completely, which produced a series of five additional compounds (37–41), (Table 3 and Fig. 3).
| Cmpd # | N-Me | GI50 (μM) HCT-116 | GI50 (μM) MiaPaCa-2 | C![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P P |
# of con-formers | Ratio |
|---|---|---|---|---|---|---|
| 15 | R3 | 3.0 ± 1.2 | 3.0 ± 0.8 | 7.40 | 1 | N/A |
| 37 | R1 | 24.1 ± 3.3 | 17.5 ± 4.2 | 7.40 | 2 | 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 30 |
| 38 | R2 | 14.4 ± 0.4 | 14.6 ± 1.2 | 7.40 | 1 | N/A |
| 39 | R4 | 14.9 ± 1.1 | 12.6 ± 2.9 | 7.40 | 1 | N/A |
| 40 | R5 | 18.1 ± 1.8 | 16.7 ± 1.5 | 7.40 | 2 | 69![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 31 31 |
| 41 | N/A | 8.9 ± 2.7 | 10.0 ± 1.7 | 6.75 | 2 | 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 20 |
These methylated analogues were evaluated for their: (a) cytotoxicity, (b) the number of conformations observed on the NMR timescale (calculated from the 1H NMR, Table 3). Interestingly, the lowest GI50 value was when an N-methyl was placed at R3, i.e. 15. Although compound 41 did not have an N-methyl moiety, it only had two conformers (previous work has shown that up to 9 conformers can exist when no N-Me is present in a cyclic pentapeptide). Furthermore, despite having multiple conformers, 41 was the second most potent molecule, with an GI50 = ∼9 and ∼10 μM respectively. The four N-methylated analogues, 37–40 displayed some potency but their GI50 values were higher than 15. However, consistent with our data and others, compound 38 and 39, which only had a single conformation had lower GI50 values than 37 and 40, which both had 2 conformations.
Cell growth inhibition and apoptosis induction analysis
It has been established that hsp90 inhibition suppresses cancer cell growth, and eventually leads to apoptotic cell death. Thus, we evaluated and compared the ability of NVP-AUY922 and the two most active compounds, 9 and 15, for their ability to inhibit cancer cell growth and induce apoptosis. Analysis of cell growth inhibition in HCT116 cells indicated that although at GI50 concentrations NVP-AUY922 showed the best inhibitory effect on cell growth, at high concentrations (5 and 10 fold over the GI50), 15 was the most effective at inducing cell death (Fig. 4). NVP-AUY922 significantly reduced the percentage of cell growth to ∼25% at concentrations of 5 fold over the GI50; however, doubling the concentration to 10 fold GI50 did not improve its effectiveness. In contrast, compound 15 decreased cell growth to ∼20% at 5 fold the GI50 concentration, and to 0% at 10 fold the GI50 (Fig. 4a).Running an Annexin V/7AAD-based apoptosis assay provided important evidence on the efficiency of three inhibitors regarding their ability to induce apoptotic cancer cell death. In order to compare the their abilities in suppressing cell growth and triggering cell death, the cancer cell line, drug concentrations and the treatment time in apoptosis analysis are the same as those used in cell growth inhibition assays. Treating cells for 24 h-treatment with compound 9 at 5 and 10 fold their GI50 showed that the percentages of apoptotic cells in the total population were 34% and 54%, respectively (Fig. 4). By comparison, compound 15 induced extremely high levels of early and late apoptosis (Fig. 4b and c) in HCT116 cells. Indeed, 96% of cells were apoptotic after 24 h-treatment of 15 with concentration of 10 fold GI50 (30 μM). Interestingly, NVP-AUY922 at high concentrations (5–10 fold of its GI50) failed to induce significant apoptotic cell death (Fig. 4b and c), even though it effectively inhibited cell growth (∼75% inhibition, Fig. 4a) at the same concentrations. Thus, these data are highly significant as they clearly demonstrate that 15 is an extremely effective molecule for suppressing cancer cell growth by inducing apoptosis (i.e. it is cytotoxic). In contrast, the clinical inhibitor efficiently stops cancer cell growth, but it does not kill them, (i.e. it is cytostatic). Such a significant difference between 15 and NVP-AUY922 is highly supportive of our hypothesis that 15 is not inducing a HSR and the cells die immediately upon treatment, whereas NVP-AUY922 is producing a HSR, which rescues the cells. Moreover, the lack of apoptotic cell death when cells are treated with NVP-AUY922 in increasing concentrations is an indicator of perhaps why this inhibitor has done so poorly in clinical trials.
Cell cycle arrest analysis
Since hsp90 inhibition has been indicated to cause cell cycle arrest, we studied and compared the impacts of NVP-AUY922, 9 and 15 on HCT116 cell cycle distribution. Treatment of cells with NVP-AUY922 (0.05 and 0.1 μM) for 24 h produced a dose-dependent increase in the population of G0/G1 phase-cells (Fig. 5). This was accompanied by a significant reduction of cells in S and G2/M phases. Given that >75% of cells were still alive (Fig. 4b), this cell cycle data is significant and indicates that the living cells that are arrested in G0/G1 phase are likely experiencing a rescue mechanism. That is, 75% of the cells are not being triggered to die upon NVP-AUY922, rather they are being rescued by the HSR. This data also indicates that because of the activated rescue mechanism, those arrested cells were trying to fix the damage before initiating the apoptotic program. The likely rescue and damage repair scenario that is occurring when cells are treated with NVP-AUY922 is consistent with the observation that the compound only stopped cell growth without killing them (Fig. 4).In comparison, after 24 h 15 μM of compound 15 also arrested cells in G0/G1 phase (Fig. 5). However, given that under the same conditions ∼70% of treated cells were already apoptotic, they could not be analyzed for the cell cycle study, which only provides data on living cells. Therefore the data of compound 15 in this assay is not informative, given that it represents only 2–4% of cells. However, the cell cycle data supports the hypothesis that there is no rescue mechanism activated by 15, which leads the treated cells to initiate apoptotic programing immediately when exposed to this compound. Interestingly, compound 9 has a distinct impact on cell cycle distribution. Even though only 50% of treated cells were still alive after the treatment with 30 μM of 9, the cells significantly increased the population of G2/M phase-cells, and produced a corresponding sharp drop in the number of cells in G0/G1 phase (Fig. 5). This result indicates that compound 9 and 15 have distinct mechanisms that impact cancer cells. Taken together, it is clear that 15 strongly inhibits cancer cell growth as well as arresting the cell cycle arrest via a rapid and intensive apoptosis induction, whereas the clinical drug NVP-AUY922 does not.
Inhibitory effect on hsp90–HOP protein binding event
As a major cellular chaperone, hsp90 folds unfolded or aggregated proteins in a cell. In its protein folding mechanism, the proteins that need to be folded are transferred from hsp70 to hsp90 through the Heat shock Organizing Protein (HOP), which is a tetratricopeptide (TPR)-containing co-chaperone that interact with hsp90's C-terminus. Without the hsp90–HOP interaction, the protein-folding event is stopped, inducing a large amount of unfolded proteins accumulated inside a cell, which eventually triggers apoptotic cell death. Therefore preventing the protein-folding event by blocking the hsp90–HOP binding is a good approach to killing cancer cells.19,20In order to evaluate the direct impact of compounds 9 and 15 on hsp90–HOP interaction, an in vitro protein-binding assay was performed by incubating native human hsp90 with different concentrations (0 up to 30 μM) of compound 9 and 15, or NVP-AUY922, followed by the addition of His-tagged HOP and immobilizing beads. Compound 28 and NVP-AUY922 were the negative controls, as we did not expect to observe a significant effect with these compounds. The results showed that only 63% and 48% of hsp90 bound to HOP in the presence of 15 μM of compound 9 and 15, respectively (Fig. 6). Gratifyingly, treatments with 30 μM of compound 15 efficiently blocked the binding between hsp90 and HOP by ∼77%, which is ∼3 fold more effective than 30 μM of compound 9 (Fig. 6).
 | ||
| Fig. 6 The inhibitory activity of hsp90 inhibitors on the binding between hsp90 and its co-chaperone, HOP. | ||
Interestingly, NVP-AUY922, this classic inhibitor slightly disrupted the binding between HOP to hsp90 (∼26%), at 0.1 μM 10-fold over NVP-AUY922's GI50 value. However, 10 fold over the IC50 compound 15 disrupted the binding by 77%, demonstrating that despite its relatively high GI50 against cancer cells, it effectively blocks binding to the C-terminus. These results illustrate the limitations associated with N-terminal inhibitors' hsp90 mechanism of action. Specifically they inhibit hsp90's ATPase activity, which likely plays a role in inducing the HSR, but they do not affect the binding between hsp90 and the TPR-containing protein, HOP. Thus, they cannot inhibit the protein folding or other roles regulated by HOP and hsp90. Furthermore, the data suggest that NVP-AUY922 is likely acting via multiple mechanisms, given that its GI50 = 10 nM but its IC50 binding affinity for hsp90 is ∼5 μM. In addition, the other negative control, a structurally similar molecule 28 did not inhibit the binding event at concentration of 30 μM, which is consistent with its poor cytotoxicity in cancer cells (Fig. 5). This data clearly indicates that the binding impact is structure dependent and not a factor related to the hydrophobic impact.
Inhibitory effect on hsp90's protein folding function
In order to assess the direct inhibition of the hsp90 protein folding function by compounds 9 and 15, we utilized the hsp90-dependent luciferase-refolding assay in rabbit reticulocyte lysate (RRL) protein-folding system. A luminescence signal is detected only if the denatured luciferase protein is refolded by hsp90 that is functionally active. On the other hand, when hsp90 is inhibited, a decreased signal will be observed. NVP-AUY922, 9, and 15 were tested in this assay and the data is shown below (Fig. 7). Compound NVP-AUY922 only inhibits protein folding function by 5–10% at 10 fold over its IC50 value (Fig. 7). These data strongly indicate that infact NVP-AUY922 is not impacting any protein folding function associated with hsp90. Compound 15 is ∼2 fold more effective than 9 at inhibiting hsp90's ability to refold the heat-denatured luciferase at 30 μM concentration, where 15 and 9 decreased the luciferase signal by ∼70% versus ∼35%, respectively (Fig. 7). Compound 28 was used as a negative control, and as expected, it failed to inhibit hsp90's protein folding function even at high concentrations. | ||
| Fig. 7 The inhibitory impact of compounds on hsp90's protein folding function, which was evaluated by using the hsp90-dependent luciferase-refolding assay. | ||
As reported by others, classical inhibitors had poor inhibitory activity against the hsp90-mediated refolding of denatured luciferase. Even with concentrations of 100 fold over the GI50 (17-AAG at 5 μM,21 and NVP-AUY922 at 1 μM), the classic inhibitors did not effectively inhibit the hsp90-dependent protein folding event (Fig. 7). These results clearly showed that compound 9 and 15 suppressed hsp90 protein-folding function in a concentration-dependent manner, which was more effective than the clinical candidates.
Impact on heat shock protein expression
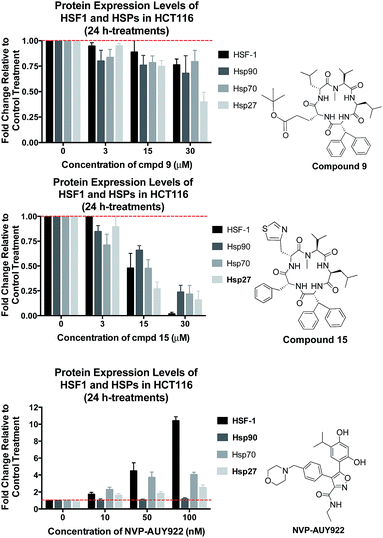
In addition to cell growth inhibition, apoptosis induction, and the suppressive impact on hsp90–HOP protein binding and hsp90-mediated protein folding machinery, it is important to prove that the active compounds 9 and 15 do not induce the HSR as a rescue mechanism.The HSR activation was detected by evaluating the protein expression levels of specific heat shock proteins (hsp70, hsp27, hsp90 and HSF-1) in treated HCT116 cells. Assessment of 9 and 15 for their ability to reduce hsp27, hsp70, hsp90, and HSF-1 protein levels was accomplished by treating HCT116 cells with compounds at three different concentrations. Evaluation of these 4 protein levels after 24 h indicates the activation of heat shock response: increased levels of these proteins indicate that a HSR has occurred, and the decreased levels signify the absence of a HSR.
Treating HCT116 cells with compound 9 for 24 h only reduced HSF-1 levels by a small amount (Fig. 8). However, cells treated with 30 μM of 15 eradicated the HSF-1 levels. This is unprecedented, and no drug has produced this kind of response on HSF-1. Given that HSF-1 is a key regulator of oncogenic pathways and it protects the cell, deleting this protein using a small molecule is an extremely positive outcome. In contrast, treatment with NVP-AUY922 significantly increased HSF-1 expression in a concentration-dependent manner. Specifically, at 10 fold over its GI50 value (100 nM) NVP-AUY922 dramatically increased HSF-1 levels by more than 10 fold.
In previous work we had shown that inhibition of hsp90 using SM122, SM145, or the clinical candidate 17-AAG (which also acts via the classical mechanism) does not affect hsp90 protein levels.3–5 As seen with HCT116 cells treated with 9 (Fig. 8) does not show significant impact on hsp90 levels either. However, treating the cells with 5 fold and 10 fold over its GI50 value, compound 15 showed a significant decrease in hsp90 level by approximately 25% (=∼1.5 fold decrease in hsp90) and 75% (=∼3.0 fold decrease in hsp90), respectively. Treatment of cells with NVP-AUY922 at concentrations of 5 and 10 fold over its GI50 value demonstrated a slight increase in hsp90 protein levels. Thus, compound 15 is highly effective at reducing hsp90 protein levels, in contrast to the clinical candidate, which increases their levels.
Like hsp90, hsp70 expression levels were not impacted when cells were treated by compound 9 after 24 h. However, similar to SM122 and SM145, treatment of 15 with 15 or 30 μM concentration reduced the amount of the cytoprotective protein hsp70 by ∼2 fold and 3 fold respectively relative to the non-treated control. NVP-AUY922 treatment with concentrations of 5 and 10 fold over its GI50 value produced a ∼4-fold increase in hsp70 protein levels (Fig. 8). These results are consistent with those published by others using NVP-AUY922, and indicate the clinical molecule is indeed producing a heat shock response.
The expression of hsp27 in HCT116 cells was effectively suppressed by treating cells with compound 9 and 15 at 30 μM (10 fold over GI50), versus the 2.5-fold increase seen in cells treated with 100 nM of NVP-AUY922 (10 fold over GI50). Since hsp27 plays an important role in the development of chemo-resistance by preventing apoptosis and protein aggregation, the depletion of this protein makes our compounds highly relevant as chemotherapeutics.
In summary, 9 and 15 inhibit hsp90 activity without inducing the HSR, which makes them novel hsp90 inhibitors that overcome the resistance mechanisms associated with current and past clinical hsp90 inhibitors. Furthermore, 15 shows a unique profile that is highly advantageous as a drug treatment against cancer, in that it completely eliminates the HSF-1 levels, and strongly depletes the other two cytoprotective proteins hsp27 and hsp70.
In conclusion, we report the synthesis of 35 new compounds, including N-methylated derivatives. In contrast to other reports, the N-methyl was optimal when placed at only one position, R3, or removed from the structure. We report here that the thiazole structure increased the molecule's potency by ∼2 fold over lead SM249 when incorporated at positions R1–R3, but not at R4. Interestingly an imidazole at R1 or R2 completely obliterated activity, which may be due to the basicity of the imidazole moiety or the recognition of the histidine by histidases in cell leading to degradation. We also report that dual modification to molecules was less effective than a single modification. This data strongly supports our hypothesis that a specific conformation is critical for binding to hsp90 effectively.
However, without dual modifications to the macrocyclic structure, we were unable to produce highly soluble molecules and could only drop the C![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values from 9 to 7.5, which is still significantly more hydrophobic than desired. On the other hand, these molecules show a highly specific SAR, which indicates that despite their hydrophobicity they are acting via a defined mechanism of action. That is, if they were only cytotoxic due to their hydrophobic nature, then all compounds with similar C
P values from 9 to 7.5, which is still significantly more hydrophobic than desired. On the other hand, these molecules show a highly specific SAR, which indicates that despite their hydrophobicity they are acting via a defined mechanism of action. That is, if they were only cytotoxic due to their hydrophobic nature, then all compounds with similar C![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values would generate the same data. The inability of these compounds to be cytotoxic at GI50 levels lower than 3 μM may be related to their hydrophobicity, which limits their solubility.
P values would generate the same data. The inability of these compounds to be cytotoxic at GI50 levels lower than 3 μM may be related to their hydrophobicity, which limits their solubility.
The two most effective compounds, 9 and 15, were tested in cell growth inhibition assays, apoptosis induction studies, and cell cycle distribution analysis, in order to evaluate their impact on cancer growth or death. Our data clearly indicate that as the most active compound in all those assays, 15 effectively triggers cancer cell death through a rapidly initiated apoptotic pathway, which is consistent with its strongly suppressive impact on the HSR rescue mechanism. As potential hsp90 inhibitors, the ability of 9 and 15 to inhibit hsp90 function was also well studied by: (1) detecting their effect on disrupting the interaction between hsp90 and its C-terminal co-chaperone HOP, which is a critical event in hsp90-dependent protein folding machinery, and (2) by detecting their activity in directly inhibiting hsp90's protein folding function.
Most importantly, neither compound 9 nor 15 induced the HSR rescue mechanism. Treating HCT116 cells with 9 showed a slight reduction of HSF-1, hsp90, and hsp27 protein expression levels. However, treatment of cells with 15 produced an unprecedented result: HSF-1 protein levels were completely obliterated. This has never been seen before when using an hsp90 inhibitor, where all clinical inhibitors have shown a significant increase in HSF-1 protein levels. Treatment with 15 also led to strongly decreased expression levels of hsp70 and hsp27 in cancer cells, which play critical roles in preventing apoptotic cell death. Taken all together, compound 15, reported here the first time, exhibits extraordinary properties as an hsp90 inhibitor, behavior that will be extremely advantageous in a cancer therapy.
Experimental
General information
All moisture sensitive reactions were performed under nitrogen gas and were monitored by thin-layer chromatography (TLC) and liquid-chromatography mass spectrometry (LC/MS). TLC was performed on aluminium silica gel sheets 250 μm Whatman® (4861-820) using UV light (λ = 254 nm) as visualizing method. The developing agents for TLC include potassium permanganate (general purpose) and ninhydrin (for amine group detection). Silica gel was used for flash chromatography. LC/MS analyses were performed on a LCMS system connected to a trap running in positive electrospray ionization (ESI+) mode. The mobile phase consist of DDI water with 0.1% (v/v) formic acid (solvent A), and HPLC grade acetonitrile with 0.1% (v/v) formic acid (solvent B) at a flow rate of 0.5 mL min−1, starting at 70% solvent A, 30% solvent B. The gradient elution were as follow: flow rate 2 mL min−1; initial 70% solvent A, 30% solvent B hold for 35 min; at 35 min 100% solvent B hold for 18 min; at 53 min 70% solvent A, 30% solvent B hold for 7 min. 1H and 13C NMR spectra were obtained and recorded at 25 °C on Bruker Avance III 500 MHz and 600 MHz.General solid phase peptide synthesis
Stepwise solid phase peptide synthesis (SPPS) was performed in a polypropylene solid-phase extraction cartridge fitted with a 20 μM polyethylene frit and pre-loaded 2-chlorotrityl resins with an approximately 0.5 mmol g−1 loading scale were used. The resin was weight, transferred to the cartridge and was swelled in DMF for 30 minutes prior to peptide coupling in the corresponding sequence.Fmoc-protected amino acids coupling reaction were performed in DMF solution at 0.2 M, consisting 3.0 equiv. of amino acid, 3.0 equiv. of 1-hydroxybenzotriazole and 6.0 equiv. of diisopropylcarbodiimide. Coupling was allowed to shake for a minimum of four hours on a shaker and checked via ninhydrin test to confirm completion. Once completed, the coupling reaction solution was drained, and the resin was subjected to Fmoc removal. (Note: for the peptide coupling between Fmoc and N-methyl amino terminus, 1-hydroxybenzotriazole was replaced by 1-hydroxy-7-azabenzotriazole and the coupling process was allowed to run overnight.)
General N-terminal solid phase amine deprotection
After the peptide coupling process was completed, removal of Fmoc protecting group was performed according to the following steps: DMF wash (3 × 1 minute), 20% piperidine/DMF (1 × 5 minutes), 20% piperidine/DMF(1 × 10 minutes), DMF wash (2 × 1 minute), IPA wash (1 × 1 minute), DMF was (1 × 1 minute), IPA (1 × 1 minute), DMF (3 × 1 minute).Cleavage of linear peptide
The eventual cleavage of linear pentapeptide from resin was carried out by swelling the resin in a mixed solution of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) 2,2,2-trifluoroethanol (TFE)/CH2Cl2 (10 millilitres per gram of dried resin) and was allow to stir for 24 hours. The suspension was then filtered through a Büchner filter, and the resin was washed repeatedly with additional CH2Cl2 to fully extract the cleaved peptide. The filtrate was then evaporated and dried in vacuo overnight. The dried solid was eventually re-dissolved in CH2Cl2, co-evaporated with CH2Cl2 several times to remove the entrapped TFE residue completely and was dried in vacuo overnight.
1 (v/v) 2,2,2-trifluoroethanol (TFE)/CH2Cl2 (10 millilitres per gram of dried resin) and was allow to stir for 24 hours. The suspension was then filtered through a Büchner filter, and the resin was washed repeatedly with additional CH2Cl2 to fully extract the cleaved peptide. The filtrate was then evaporated and dried in vacuo overnight. The dried solid was eventually re-dissolved in CH2Cl2, co-evaporated with CH2Cl2 several times to remove the entrapped TFE residue completely and was dried in vacuo overnight.
Macrocyclization procedure (syringe pump)
Macrocyclization of the double deprotected linear pentapeptide using a combination of three coupling agents (DMTMM, HATU, and TBTU) at 0.8 equivalent each, together with DIPEA (8.0 equivalents) in 75% of a calculated volume of anhydrous CH2Cl2 to generate a 0.001 M overall concentration. The crude and dry double deprotected linear peptide (DDLP) was dissolved in the remaining amount of CH2Cl2. The DDLP solution was then added to the bulk solution drop-wise using a syringe pump, over 2 hours. After the addition of all DDLP, the reaction was stirred overnight and was monitored via LCMS and generally complete in 1–2 hours. Upon completion, the crude product was subjected to acid–base wash to remove excess DIPEA and coupling agents. The resulting crude product was first purified by flash column chromatography, followed by reversed-phase HPLC, using a gradient of acetonitrile and deionized water with 0.1% TFA to afford the final pure compounds.Synthesis
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOAc = 0.25
EtOAc = 0.25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.75); LC/MS: m/z called for C42H61N5O7Na1 (M + Na+) = 770.45, found 770.10. HRMS (ESI-TOF): M + Na+, found 770.4465, C42H61N5O7Na1 requires 770.4469.
0.75); LC/MS: m/z called for C42H61N5O7Na1 (M + Na+) = 770.45, found 770.10. HRMS (ESI-TOF): M + Na+, found 770.4465, C42H61N5O7Na1 requires 770.4469.1H NMR (600 MHz, CDCl3): δ 7.90 (d, J = 9.1 Hz, N![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ), 7.44–7.20 (m, 10H, D-Biphe), 6.87 (br, N
), 7.44–7.20 (m, 10H, D-Biphe), 6.87 (br, N![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ), 5.28 (br, N
), 5.28 (br, N![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ), 5.21 (t, J = 10.2 Hz, 1H, CHα D-Biphe), 4.96 (q, J = 7.7 Hz, 1H, CHα Val), 4.84 (d, J = 10.8 Hz, 1H CHα D-Glu), 4.62 (br, 1H, CHβ D-Biphe), 4.58 (br, N
), 5.21 (t, J = 10.2 Hz, 1H, CHα D-Biphe), 4.96 (q, J = 7.7 Hz, 1H, CHα Val), 4.84 (d, J = 10.8 Hz, 1H CHα D-Glu), 4.62 (br, 1H, CHβ D-Biphe), 4.58 (br, N![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ), 3.74 (m, 2H, CHα D-Leu, CHα Leu), 3.09 (s, 3H, NCH3), 2.37 (m, 2H, CH2β D-Glu), 2.26 (m, 1H, CHβ Val), 2.06 (m, 1H, CH2β D-Leu), 1.93 (br, 1H, CH2β D-Leu), 1.71 (m, 1H, CHγ Leu), 1.62 (m, 1H, CH2β Leu), 1.56 (m, 2H, CH2β Leu, CH2γ D-Glu), 1.46 (s, 9H), 1.32 (m, 1H, CH2γ D-Glu), 1.12 (m, 1H, CHγ D-Leu), 1.00–0.76 (m, 18H, CH2CH(C
), 3.74 (m, 2H, CHα D-Leu, CHα Leu), 3.09 (s, 3H, NCH3), 2.37 (m, 2H, CH2β D-Glu), 2.26 (m, 1H, CHβ Val), 2.06 (m, 1H, CH2β D-Leu), 1.93 (br, 1H, CH2β D-Leu), 1.71 (m, 1H, CHγ Leu), 1.62 (m, 1H, CH2β Leu), 1.56 (m, 2H, CH2β Leu, CH2γ D-Glu), 1.46 (s, 9H), 1.32 (m, 1H, CH2γ D-Glu), 1.12 (m, 1H, CHγ D-Leu), 1.00–0.76 (m, 18H, CH2CH(C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3)2, CHCH(C
3)2, CHCH(C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3)2).
3)2).
13C NMR (150 MHz, CDCl3): δ 172.82, 171.86, 171.83, 171.56, 171.28, 170.01, 140.47, 140.33, 128.91, 128.80, 128.61, 128.52, 128.39, 128.32, 128.24, 127.94, 127.26, 126.99, 80.35, 63.66, 57.89, 57.35, 57.02, 54.92, 47.65, 41.96, 38.09, 31.34, 30.16, 28.12, 26.38, 25.74, 24.84, 24.82, 23.92, 22.89, 22.65, 22.19, 19.42, 18.16.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOAc = 0.25
EtOAc = 0.25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.75).
0.75).LC/MS: m/z called for C42H50N6O5S (M + 1) = 751.36, found 751.05. HRMS (ESI-TOF): M + Na+, found 773.3449, C42H50N6O5S1Na1 requires 773.3461.
1H NMR (500 MHz, DMSO): δ 8.96 (d, J = 2.0 Hz, 1H, D-Thia), 8.16 (d, J = 8.6 Hz, N![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ), 8.08 (d, J = 8.5 Hz, N
), 8.08 (d, J = 8.5 Hz, N![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ), 7.82 (d, J = 7.5 Hz, N
), 7.82 (d, J = 7.5 Hz, N![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ), 7.55 (d, J = 9.6 Hz, N
), 7.55 (d, J = 9.6 Hz, N![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ), 7.29–7.16 (m, 15H, D-BiPhe, D-Phe), 6.87 (m, 1H, D-Thia), 5.17 (m, 1H, CHα D-BiPhe), 4.96 (q, J = 7.6 Hz, 1H, CHα D-Phe), 4.53 (d, J = 11.4 Hz, 1H, CHα Val), 4.33 (d, J = 11.8 Hz, 1H, CHβ D-BiPhe), 4.05 (m, 1H, CHα D-Thia), 3.52 (q, J = 7.5 Hz, 1H, CHα Leu), 3.29 (m, 1H, CH2β2 D-Phe), 2.94 (dd, J = 6.4, 14.6 Hz, 1H, CH2β1 D-Phe), 2.73 (dd, J = 6.9, 14.0 Hz, 1H, CH2β2 D-Thia), 2.64 (s, 3H, NCH3), 2.55 (dd, J = 7.1, 14.0 Hz, 1H, CH2β1 D-Thia), 2.14 (m, 1H, CHβ Val), 1.48 (m, 1H, CH2β2 Leu), 1.38 (m, 1H, CH2β1 Leu), 1.21 (m, 1H, CHγ Leu), 0.81–0.61 (m, 12H, CHCH(C
), 7.29–7.16 (m, 15H, D-BiPhe, D-Phe), 6.87 (m, 1H, D-Thia), 5.17 (m, 1H, CHα D-BiPhe), 4.96 (q, J = 7.6 Hz, 1H, CHα D-Phe), 4.53 (d, J = 11.4 Hz, 1H, CHα Val), 4.33 (d, J = 11.8 Hz, 1H, CHβ D-BiPhe), 4.05 (m, 1H, CHα D-Thia), 3.52 (q, J = 7.5 Hz, 1H, CHα Leu), 3.29 (m, 1H, CH2β2 D-Phe), 2.94 (dd, J = 6.4, 14.6 Hz, 1H, CH2β1 D-Phe), 2.73 (dd, J = 6.9, 14.0 Hz, 1H, CH2β2 D-Thia), 2.64 (s, 3H, NCH3), 2.55 (dd, J = 7.1, 14.0 Hz, 1H, CH2β1 D-Thia), 2.14 (m, 1H, CHβ Val), 1.48 (m, 1H, CH2β2 Leu), 1.38 (m, 1H, CH2β1 Leu), 1.21 (m, 1H, CHγ Leu), 0.81–0.61 (m, 12H, CHCH(C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3)2, CH2CH(C
3)2, CH2CH(C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3)2).
3)2).
13C NMR (125 MHz, DMSO): δ 171.36, 170.34, 170.13, 169.95, 168.92, 154.04, 153.49, 141.73, 141.10, 137.54, 129.26, 129.06, 128.84, 128.74, 128.58, 128.44, 127.13, 126.93, 126.81, 115.34, 63.38, 57.22, 56.69, 53.69, 51.90, 49.90, 38.12, 37.67, 33.33, 30.30, 25.26, 24.51, 22.81, 22.58, 19.70, 18.78.
Experimental procedures and compound characterizations for compound 7–41 (excluding compound 9 and 15) were performed as described in ESI.†
Biological methods
Acknowledgements
We thank UNSW for support of S.R.M. and YCK. We also thank the Australian government for providing the Endeavour scholarship fund. Finally, we thank the NHMRC for funding (grant APP1043561).References
- U. Banerji, A. O'Donnell, M. Scurr, S. Pacey, S. Stapleton, Y. Asad, L. Simmons, A. Maloney, F. Raynaud, M. Campbell, M. Walton, S. Lakhani, S. Kaye, P. Workman and I. Judson, J. Clin. Oncol., 2005, 23, 4152–4161 CrossRef CAS PubMed.
- L. Whitesell and S. L. Lindquist, Nat. Rev. Cancer, 2005, 5, 761–772 CrossRef CAS PubMed.
- Y. Wang and S. R. McAlpine, Chem. Commun., 2015, 51, 1410–1413 RSC.
- J. M. McConnell, L. D. Alexander and S. R. McAlpine, Bioorg. Med. Chem. Lett., 2014, 24, 661–666 CrossRef CAS PubMed.
- Y. C. Koay, J. R. McConnell, Y. Wang, S. J. Kim and S. R. McAlpine, ACS Med. Chem. Lett., 2014, 5, 771–776 CrossRef CAS PubMed.
- V. C. Ardi, L. D. Alexander, V. A. Johnson and S. R. McAlpine, ACS Chem. Biol., 2011, 6, 1357–1367 CrossRef CAS PubMed.
- J. R. McConnell and S. R. McAlpine, Bioorg. Med. Chem. Lett., 2013, 23, 1923–1928 CrossRef CAS PubMed.
- R. P. Sellers, L. D. Alexander, V. A. Johnson, C.-C. Lin, J. Savage, R. Corral, J. Moss, T. S. Slugocki, E. K. Singh, M. R. Davis, S. Ravula, J. E. Spicer, J. L. Oelrich, A. Thornquist, C.-M. Pan and S. R. McAlpine, Bioorg. Med. Chem., 2010, 18, 6822–6856 CrossRef CAS PubMed.
- P. S. Pan, R. C. Vasko, S. A. Lapera, V. A. Johnson, R. P. Sellers, C.-C. Lin, C.-M. Pan, M. R. Davis, V. C. Ardi and S. R. McAlpine, Bioorg. Med. Chem., 2009, 17, 5806–5825 CrossRef CAS PubMed.
- K. Otrubova, G. H. Lushington, D. Vander Velde, K. L. McGuire and S. R. McAlpine, J. Med. Chem., 2008, 51, 530–544 CrossRef CAS PubMed.
- P. S. Pan, K. McGuire and S. R. McAlpine, Bioorg. Med. Chem. Lett., 2007, 17, 5072–5077 CrossRef CAS PubMed.
- K. Otrubova, K. L. McGuire and S. R. McAlpine, J. Med. Chem., 2007, 50, 1999–2002 CrossRef CAS PubMed.
- T. J. Styers, A. Kekec, R. A. Rodriguez, J. D. Brown, J. Cajica, P.-S. Pan, E. Parry, C. L. Carroll, I. Medina, R. Corral, S. Lapera, K. Otrubova, C.-M. Pan, K. L. McGuire and S. R. McAlpine, Bioorg. Med. Chem., 2006, 14, 5625–5631 CrossRef CAS PubMed.
- K. Otrubova, T. J. Styers, P.-S. Pan, R. Rodriguez, K. L. McGuire and S. R. McAlpine, Chem. Commun., 2006, 1033–1034 RSC.
- J. Chatterjee, C. Gilon, A. Hoffman and H. Kessler, Acc. Chem. Res., 2008, 41, 1331–1342 CrossRef CAS PubMed.
- C. Mas-Moruno, F. Rechenmacher and H. Kessler, Adv. Anticancer Agents Med. Chem., 2010, 10, 753–768 CrossRef CAS.
- A. C. Rand, S. S. F. Leung, H. Eng, C. J. Rotter, R. Sharma, A. S. Kalgutkar, Y. Zhang, M. V. Varma, K. A. Farley, D. B. Khunte, C. Limberakis, D. A. Price, S. Liras, A. M. Mathiowetz, M. P. Jacobsonb and R. S. Lokey, MedChemComm, 2012, 3, 1282–1289 RSC.
- J. Chatterjee, D. F. Mierke and H. Kessler, J. Am. Chem. Soc., 2006, 128, 15164–15172 CrossRef CAS PubMed.
- R. C. Vasko, R. A. Rodriguez, C. N. Cunningham, V. C. Ardi, D. A. Agard and S. R. McAlpine, ACS Med. Chem. Lett., 2010, 1, 4–8 CrossRef CAS PubMed.
- H. Wegele, S. K. Wandinger, A. B. Schmid, J. Reinstein and J. Buchner, J. Mol. Biol., 2006, 356, 802–811 CrossRef CAS PubMed.
- Y. Wang and S. R. McAlpine, Org. Biomol. Chem., 2015, 13, 2108–2116 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra07056b |
| This journal is © The Royal Society of Chemistry 2015 |