Enzyme immobilization on silicate glass through simple adsorption of dendronized polymer–enzyme conjugates for localized enzymatic cascade reactions†
Abstract
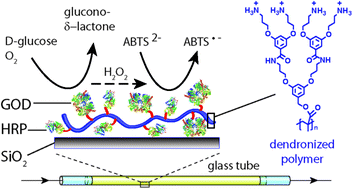
A methacrylate based, water soluble, polycationic second generation dendronized polymer (denpol de-PG2) was used for the preparation of de-PG2–enzyme conjugates. Aspergillus sp. glucose oxidase (GOD) and horseradish peroxidase isoenzyme C (HRP) were covalently bound to the denpol, using a UV/vis traceable bis-aryl hydrazone (BAH) linker, to form two different de-PG2–BAH–enzyme hybrid structures, each carrying several copies of either GOD or HRP on a single denpol chain (on average ≈ 50 GOD or 108 HRP bound per denpol chain). In addition, a conjugate with several copies of both types of enzymes on the same polymer chain was synthesized (≈25 GOD and 78 HRP). These denpol–BAH–enzyme conjugates were found to be useful for the immobilization of the enzymes on unmodified silicate glass surfaces via simple adsorption of the conjugates from solution in one single step. The adsorbed conjugates strongly adhered to the glass surface due to multiple interactions between the conjugates and the surface. The conjugate adsorption was characterized with the transmission interferometric adsorption sensor (TInAS) and by AFM imaging, which showed formation of a homogenous thin layer of the conjugates. Additionally, the catalytic activity and stability of the immobilized enzymes were determined and the conjugates were used for the simple fabrication of enzymatic flow reactors for catalyzing a cascade reaction which involved both enzymes, either through a sequential immobilization of the two enzymes, or through a co-immobilization. In both cases, the two enzymes remained highly active during continuous operation at room temperature for at least several hours without any desorption of the enzymes from the surface. Overall, the methodology presented can be considered as a promising platform for a desired (co-)localization of active enzymes on solid supports.

 Please wait while we load your content...
Please wait while we load your content...