A water-soluble cationic porphyrin showing pH-dependent G-quadruplex recognition specificity and DNA photocleavage activity†
Ting Zhao‡
ab,
Ya-Ling Wang‡bd,
Li-Na Zhuc,
Yan-Fang Huoab,
Yong-Jian Wangbd and
De-Ming Kong*ab
aState Key Laboratory of Medicinal Chemical Biology, College of Chemistry, Nankai University, Tianjin 300071, China. E-mail: kongdem@nankai.edu.cn; Fax: +86-22-23502458; Tel: +86-22-23500938
bCollaborative Innovation Center of Chemical Science and Engineering (Tianjin), Tianjin 300071, China
cDepartment of Chemistry, Tianjin University, Tianjin, 300072, China
dKey Laboratory of Bioactive Materials (Ministry of Education), College of Life Sciences, Nankai University, Tianjin 300071, China
First published on 21st May 2015
Abstract
G-quadruplex ligands lacking recognition specificity against duplex DNA are considered to be unsatisfactory due to non-specific cytotoxicity. However, a G-quadruplex ligand, which can interact with duplex DNA under acidic conditions but not under neutral conditions, may be highly desirable for cancer therapy because it can kill cancer cells with high efficiency, with very few side effects on healthy cells. Herein, a new water-soluble cationic porphyrin derivative 5,10,15,20-tetra-{4-[2-(1-methyl-1-piperidinyl)ethoxy]phenyl}porphyrin (i-TMPipEOPP) was synthesized and characterized, and its interactions with G-quadruplex and duplex DNA were compared using ultraviolet visible absorption, fluorescence, circular dichroism and gel electrophoresis assays. The results show that i-TMPipEOPP has pH-dependent G-quadruplex recognition specificity. That is, it can interact with both G-quadruplex and duplex DNA under acidic conditions, but can only interact with G-quadruplex under neutral conditions. Because of the synergy between π–π stacking and electrostatic interactions, i-TMPipEOPP interacts with G-quadruplex with higher binding affinity under acidic conditions than under neutral conditions in which only π–π stacking interactions occur. More interestingly, i-TMPipEOPP can mediate pH-dependent DNA photocleavage of duplex DNA and shows pH-dependent phototoxicity to cells. These findings suggest that i-TMPipEOPP may be developed as a promising photodynamic therapy drug showing higher cytotoxicity towards acidic tumor cells than neutral healthy cells.
Introduction
B-form duplex is the canonical conformation of human genomic DNA.1 However, more and more non-canonical secondary DNA structures are being observed and many of them appear to play important roles in biological processes.2 One of the best examples is the DNA G-quadruplex,3 a unique nucleic acid secondary structure formed by G-rich DNA or RNA sequences.4 Nucleic acid sequences with G-quadruplex-forming potential are common in biologically important genomic regions.5 It is well known that the single-stranded G-rich telomeric regions in the termini of the linear chromosomes might fold into G-quadruplex structures. Ligands that can promote telomeric G-quadruplex formation and stabilize telomeric G-quadruplex are considered to be good candidates for anticancer drugs, because the telomeric sequences with G-quadruplex structures cannot be extended by telomerase any more, resulting in cancer cells losing the ability to proliferate.6To date, a large number of G-quadruplex ligands have been reported,7 but most of them are considered to be unsatisfactory due to non-specific cytotoxicity. That is, healthy cells may also be killed by these ligands. It is believed that non-specific cytotoxicity arises from the interactions between ligands and duplex DNA.8 As a result, searching for G-quadruplex ligands that can selectively bind to their target while having little interaction with duplex DNA has become a major focus of G-quadruplex ligand studies.9 However, these non-specific G-quadruplex ligands may also be developed as highly desirable anticancer drugs since they can prevent the proliferation of cancer cells in two ways: (1) by interacting with duplex DNA, and (2) by inhibiting telomerase activity through promoting the formation of telomeric G-quadruplex structures. However, a side effect is that these ligands may be somewhat cytotoxic to healthy cells and should therefore not be allowed to interact with duplex DNA in healthy cells.
Such G-quadruplex ligands can be designed according to the differences that exist between healthy and cancerous cells. It has been widely reported that the microenvironments in healthy and cancerous cells are of different pH values.10,11 The intramolecular pH in healthy cells is neutral (∼7.4). However, acidity is a common characteristic of the microenvironments surrounding cancer cells. In such cancerous microenvironments, it is not uncommon for the local pH to range from 5.5 to 7.0.12 Based on this pH difference, some remarkable cancer diagnostic and therapeutic protocols have been proposed.10 If a G-quadruplex ligand can interact with duplex DNA under acidic conditions, but not under neutral conditions, it may be able to kill cancer cells in one of the two ways mentioned above and prevent the transformation of healthy cells into cancer cells by promoting the formation of telomeric G-quadruplexes. In the present study, we synthesized a new water-soluble cationic porphyrin derivative called 5,10,15,20-tetra-{4-[2-(1-methyl-1-piperidinyl)ethoxy]phenyl}porphyrin (i-TMPipEOPP) (Scheme 1) with four large side arms and characterized its interactions with different structural DNAs using ultraviolet visible (UV-vis) absorption, fluorescence, circular dichroism (CD) and gel electrophoresis assays. Our results indicated that i-TMPipEOPP can bind monomeric and multimeric G-quadruplexes under both acidic and neutral conditions. However, it can only interact with duplex DNA in acidic conditions. Furthermore, i-TMPipEOPP shows pH-dependent photocleavage activity towards duplex DNA and pH-dependent phototoxicity to cells.
Experimental section
Materials and reagents
The oligonucleotides listed in Table 1 were purchased from Sangon Biotech. Co. Ltd. (Shanghai, China). The concentrations of the oligonucleotides were represented as single-stranded concentrations. Single-stranded concentrations were determined by measuring the absorbance at 260 nm. Molar extinction coefficient was determined using a nearest neighbour approximation (http://www.idtdna.com/analyzer/Applications/OligoAnalyzer), and the calculated molar extinction coefficients of these oligonucleotides were listed in Table 1. Calf thymus DNA (Ct DNA) was purchased from Sigma. The concentration of Ct DNA was represented as the base concentration, which was determined by the absorbance at 260 nm using the molar absorption coefficient of 6600 M−1 cm−1. The Ct DNA solution gave a ratio of UV absorbance at 260 and 280 nm of 1.83![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, which indicates that the DNA was sufficiently free of protein. pBR322 plasmid DNA was purchased from Takara. Na2EDTA (disodium ethylenediamine tetraacetic acid), Tris (tris(hydroxymethyl)aminomethane) was obtained from Sigma. CH3OH, N,N-dimethylformamide (DMF) and CH2Cl2 were obtained from Jiang Tian Chemical Factory (Tianjin China). N-(2-Chloroethyl)-methylpiperidine hydrochloride was bought from Kang Man Lin Chemical Factory (Nanjing China). DMF and CH2Cl2 were distilled over before use. Other chemical reagents were of analytical grade and used without further purification. Deionized and sterilized water (resistance = 18.25 MΩ cm−1) was used throughout the experiments. The details of synthesis and characterization of 5,10,15,20-tetra-{4-[2-(1-methyl-1-piperidinyl)ethoxy]phenyl}porphyrin (i-TMPipEOPP) are available in ESI (Fig. S1–S3†).
1, which indicates that the DNA was sufficiently free of protein. pBR322 plasmid DNA was purchased from Takara. Na2EDTA (disodium ethylenediamine tetraacetic acid), Tris (tris(hydroxymethyl)aminomethane) was obtained from Sigma. CH3OH, N,N-dimethylformamide (DMF) and CH2Cl2 were obtained from Jiang Tian Chemical Factory (Tianjin China). N-(2-Chloroethyl)-methylpiperidine hydrochloride was bought from Kang Man Lin Chemical Factory (Nanjing China). DMF and CH2Cl2 were distilled over before use. Other chemical reagents were of analytical grade and used without further purification. Deionized and sterilized water (resistance = 18.25 MΩ cm−1) was used throughout the experiments. The details of synthesis and characterization of 5,10,15,20-tetra-{4-[2-(1-methyl-1-piperidinyl)ethoxy]phenyl}porphyrin (i-TMPipEOPP) are available in ESI (Fig. S1–S3†).
| DNA | Sequence (from 5′ to 3′) | Extinction coefficient [L mol−1 cm−1] | Structure |
|---|---|---|---|
| Hum24 | (TTAGGG)4 | 215![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
G-quadruplex (monomeric) |
| KRAS | AG3CGGTG3AAGAG3AAGAG5AGG | 322![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 200 |
G-quadruplex (monomeric) |
| M3Q | GAG3AG3AG3AGAG3A | 222![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 500 |
G-quadruplex (monomeric) |
| Hum48 | (TTAGGG)8 | 489![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
G-quadruplex (multimeric) |
| Hum54 | (TTAGGG)9 | 550![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
G-quadruplex (multimeric) |
| AT | (AT)6 | 133![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 300 |
Short duplex |
| LD | GCGCAATTGCGC | 108![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 700 |
Short duplex |
| Ct DNA | Long duplex |
UV-vis absorption spectroscopy
UV-vis absorption spectra were measured on a Cary 60 UV-vis spectrophotometer (Agilent Technologies) with 1 cm-path-length micro quartz cell (40 μL, Starna Brand, England). Each sample contained individual DNAs with designated concentration, 10 mM Tris–HCl buffer (pH = 7.4 or 5.5), 50 mM KCl and 1 mM Na2EDTA. Each sample was heated to 95 °C for 5 min to remove any aggregates, then cooled rapidly to 25 °C and incubated at this temperature for 30 min. After incubated at 4 °C overnight, 5 μM of i-TMPipEOPP was added. The absorption spectra in the range of 350–800 nm were recorded.Continuously changing the DNA concentration but keeping i-TMPipEOPP concentration at 5 μM in constant, absorption titration experiments were carried out. The absorption spectra in the range of 350–800 nm were recorded. The data from the absorption titration experiments were analysed according to the independent-site mode by non-linear fitting to eqn (1):9k,13
 | (1) |
Job plot analysis was carried out by systematic variation of the molar fraction of i-TMPipEOPP and DNA while keeping a constant total concentration of 10 μM. The sample was prepared as above, and the absorption signals at 700 and 687 nm were recorded for G-quadruplex and duplex DNA, respectively.
Fluorescence spectroscopy
Fluorescence spectra were measured on a SHIMADZU RF-5301PC spectrofluorimeter with 1 cm-path-length micro quartz cell (40 μL, Starna Brand, England). Each sample contained individual DNAs with designated concentration, 10 mM Tris–HCl buffer (pH = 7.4 or 5.5), 50 mM KCl and 1 mM Na2EDTA. Each solution was heated to 95 °C for 5 min to remove any aggregates, then cooled rapidly to 25 °C and incubated at 25 °C for 30 min. After incubation at 4 °C overnight, 5 μM of i-TMPipEOPP was added. Fixing the excitation wavelength at 455 nm, the emission spectra in the range of 600–850 nm were collected at room temperature. The excitation and emission slit width were both set at 5 nm.Continuously changing the DNA concentration but keeping i-TMPipEOPP concentration at 5 μM in constant, fluorescence titration experiments were carried out. The sample was prepared as above. Setting the excitation wavelength at 455 nm, the fluorescence spectra were recorded in the range of 600–850 nm.
Melting temperature (T1/2) detection of G-quadruplexes
Melting temperature (T1/2) detection of G-quadruplexes was carried out on a Cary-60 UV-vis spectrophotometer equipped with a temperature control accessory. The G-quadruplex (3 μM) sample was prepared in 10 mM Tris–HCl buffer (pH = 7.4 or 5.5) containing 50 mM KCl and 1 mM Na2EDTA. The sample was heated to 95 °C for 5 min, then cooled rapidly to 25 °C and incubated at this temperature for 30 min. After incubation at 4 °C overnight, 0 or 3 μM i-TMPipEOPP was added. Then, the absorption signal at 295 nm (400 nm as control wavelength) was recorded at about 20 °C. When the absorption signal became constant, the temperature was increased in steps of 1 °C and the absorption signal was recorded at each temperature until the signal did not decrease any more. Then, the absorption intensity–temperature plot was constructed, and the temperature at which the absorption signal is midway between the minimal and maximal levels is the T1/2 of G-quadruplex structure.Circular dichroism (CD) study
2 mL sample was prepared in 10 mM Tris–HCl buffer (pH = 7.4 or 5.5) containing 2 μM Hum24 (or 1 μM Hum48 or Hum54), 2 μM i-TMPipEOPP, 50 mM KCl and 1 mM Na2EDTA. The sample was heated at 95 °C for 5 min, cooled slowly to 25 °C and incubated at this temperature for 30 min. After incubation at 4 °C overnight, CD spectrum of the sample was recorded between 220 and 350 nm in 1 cm-path-length cuvettes on a Jasco J-715 spectropolarimeter. Spectra were averaged from 3 scans, which were recorded at 200 nm min−1 with a response time of 1 s and a bandwidth of 1.0 nm.DNA photocleavage study
The reaction mixtures (25 μL total volume) contained 10 ng μL−1 of supercoiled plasmid DNA (pBR322 DNA), 10 mM Tris–HCl buffer (pH = 7.4 or 5.5), different concentrations of i-TMPipEOPP. Samples were incubated at 30 °C for 1.5 h in dark or under sunlight. After addition of 5 μL loading buffer (0.25% bromophenol blue, 0.25% xylene cyanol, 30% glycerol, 10 mM EDTA). 20 μL of the mixtures were loaded onto a 1% agarose gel containing ethidium bromide (1 μg mL−1) in TBE buffer (90 mM Tris–borate, pH 8.0, 20 mM EDTA). The gel was run at 80 V for 30 min and photographed under UV light.In vitro cytotoxicity study
In vitro cytotoxicity of i-TMPipEOPP was estimated by a standard 3-(4,5-dimethylthiazol-z-yl)-2,5-diphenyltetrazolium (MTT) assay. Briefly, cells were seeded in 96-well culture plates at a density of 1000 cells per well and allowed to grow over 12 h (the cells reached 70–80% confluence). The pH of DMEM culture medium was adjusted between 7.4 and 5.5 with Tris–HCl buffer. Different concentrations of i-TMPipEOPP in the DMEM medium without phosphate buffer solution (FBS) was added to each well separately. The cells were then incubated in dark for 24 h or incubated under sunlight for 1 h. Then, the cells were washed with PBS buffer, and fresh culture media was added. Afterwards, MTT solution (20 μL, 0.1 mg) was added to each well and the cells were incubated for another 4 h. N,N-Dimethyl sulfoxide (DMSO) (150 μL) was used to completely liberate the formazan crystals. The absorbance at 490 nm was measured for the calculation of the cell survival rate. The cells treated with PBS were taken as the control group and six parallel samples were tested in each group. Three independent experiments were performed under identical conditions.Results and discussion
Design, synthesis and characterization of i-TMPipEOPP
Porphyrin is a large family of G-quadruplex ligands. A well-known member in this family is TMPyP4 (Scheme 1). However, a serious disadvantage of this porphyrin derivative is that it can interact with G-quadruplexes as well as duplex DNA in both acidic and neutral pH conditions.8a,14 It has been reported that even TMPyP4 bound preferentially to duplex DNA.8a One reason might be attributed to its relatively small side arm substituents, which cannot provide enough steric hindrance to prevent the intercalation of TMPyP4 into the base planes or grooves of duplex DNA. Herein, i-TMPipEOPP was developed and characterized as a new porphyrin derivative. i-TMPipEOPP has four large positively charged side arms, which might efficiently prevent interactions between i-TMPipEOPP and duplex DNA, thus greatly increasing its G-quadruplex recognition specificity against duplex DNA. Cationic side arms can also improve the water-solubility of this porphyrin derivative. This is very important for its application in biomedical fields. The details of synthesis and characterization of i-TMPipEOPP are available in the Experimental and ESI sections (Fig. S1–S3†).G-quadruplex recognition specificity of i-TMPipEOPP against duplex DNA under different pH conditions
Firstly, UV-vis absorption spectroscopy was used to investigate the discriminating abilities of i-TMPipEOPP towards G-quadruplex and duplex DNA under acidic or neutral conditions. As shown in Fig. 1, free i-TMPipEOPP shows similar absorption spectra under acidic (pH 5.5) and neutral (pH 7.4) conditions. That is, it gives a strong absorption peak centered at around 422 nm and three weak peaks centered at around 524, 564 and 670 nm, respectively. With the addition of G-quadruplexes (Table 1), significant changes were observed for i-TMPipEOPP absorption spectra under both acidic and neutral conditions. Two new absorption peaks emerged at around 455 and 700 nm, respectively, thus suggesting that strong interactions occur between i-TMPipEOPP and G-quadruplexes. This also suggests that i-TMPipEOPP may be considered a potential G-quadruplex ligand. Such a ligand can not only bind to monomeric G-quadruplexes (simple G-quadruplex formed by short G-rich sequence and contains only one G-quadruplex unit), but it can also interact with multimeric ones, which consist of two or more G-quadruplex units combined. The studies on multimeric G-quadruplex ligands have attracted increasing attention in the past years since it has been reported that the G-rich single-stranded telomeric region that exists at the end of linear chromosomes might fold into multimeric G-quadruplexes.15 | ||
| Fig. 1 UV-vis absorption spectra of i-TMPipEOPP in the absence or presence of monomeric G-quadruplex (Hum24), multimeric G-quadruplex (Hum48) or duplex (LD) under acidic and neutral conditions. [Porphyrin] = 5 μM; [Hum24] = [LD] = 20 μM; [Hum48] = 10 μM. The effects of other DNAs on the absorption spectrum of 1 were shown in ESI Fig. S4.† | ||
Duplex DNAs (Table 1) exhibit different effects on the absorption spectrum of i-TMPipEOPP under different pH conditions. Under acidic conditions (pH 5.5), the emergence of the two new peaks can also be observed in the presence of duplex DNA, though their peaks are blue-shifted to 452 and 687 nm, respectively, thus indicating that binding interactions also occur between i-TMPipEOPP and duplex DNA. However, under neutral conditions (pH 7.4), the addition of duplex DNA has almost no effect on the absorption spectrum of i-TMPipEOPP, suggesting that i-TMPipEOPP loses the ability to interact with duplex DNA. From these findings, we elucidated that i-TMPipEOPP can bind with both G-quadruplex and duplex DNA under acidic conditions, but it can only interact with G-quadruplex DNA under neutral conditions. Such a pH-dependent G-quadruplex recognition specificity against duplex DNA might be attributed to the charge change in i-TMPipEOPP side arms. Under acidic conditions, the four cationic side arms give positive net charge to i-TMPipEOPP. As a result, the electrostatic attraction between positively charged i-TMPipEOPP and negatively charged DNA phosphate backbones promote the interactions between i-TMPipEOPP and duplex DNA. As pH increases, the pyridine nitrogens in the side arms can receive hydroxide ions, thus weakening the electrostatic interactions between i-TMPipEOPP and DNA. In addition, the presence of four large side arms is not conducive to the intercalation of i-TMPipEOPP into base planes or grooves of duplex DNAs.16 Therefore, i-TMPipEOPP shows a greatly improved G-quadruplex-binding specificity against duplex DNA. This is the case even with long duplex DNA, such as that derived from calf thymus (Ct DNA) (Fig. S4†).
Although i-TMPipEOPP shows binding interactions with both G-quadruplex and duplex DNA under acidic conditions, the effect of duplex DNA on the i-TMPipEOPP absorption spectrum is obviously weaker than that of G-quadruplex DNA. This suggests that other modes of binding, besides electrostatic interactions, may occur between G-quadruplex DNA and i-TMPipEOPP. It is possible that the porphine core of i-TMPipEOPP stacks onto the terminal G-quartet(s) of G-quadruplex DNA by π–π stacking interactions as the side arms interact with the loops or extend into the grooves of G-quadruplex.17 Under acidic conditions, the electrostatic interaction between positively charged side arms and negatively charged phosphate backbones of loops or grooves of G-quadruplex undoubtedly increases the binding affinity between i-TMPipEOPP and G-quadruplex. This is the reason why the effect of G-quadruplex DNA on the i-TMPipEOPP absorption spectrum is much stronger under acidic conditions than neutral conditions. As shown in Fig. 1, in the presence of 20 μM monomeric or 10 μM multimeric G-quadruplexes, the absorption peak at 422 nm disappears entirely, and the intensities of the new peaks at 455 and 700 nm significantly increase under acidic conditions. Conversely, under neutral conditions, the absorption peak at 422 nm can still be observed, and the intensities of the two new peaks are obviously lower than those under acidic conditions.
Binding stoichiometries of i-TMPipEOPP to different DNAs
To further investigate the interactions between i-TMPipEOPP and DNA under different pH conditions, Job plot analysis was used to determine the binding stoichiometry of i-TMPipEOPP to different DNAs. Absorption spectra of the mixtures of i-TMPipEOPP and DNA (at different i-TMPipEOPP/DNA ratios, while keeping the total concentration constant) were recorded, and the signals at 700 nm and 687 nm were used to construct Job plots for G-quadruplex and duplex DNA, respectively (Fig. 2). At the pH of 7.4, i-TMPipEOPP bound with all of the G-quadruplexes, including monomeric and multimeric G-quadruplexes, with the same binding stoichiometry of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (Table 2). That is, one i-TMPipEOPP molecule could bind with two G-quadruplex molecules. Similar binding stoichiometry was observed in the studies of G-quadruplex and other porphyrin derivatives,9a,16a thus suggesting that i-TMPipEOPP might interact with G-quadruplex DNA through an end-stacking modality forming a sandwich-like G-quadruplex/i-TMPipEOPP/G-quadruplex complex (Scheme 2). The binding stoichiometries obtained at a pH of 5.5 were much higher than those obtained at a pH of 7.4. In addition, DNA length-dependent binding stoichiometries were observed at a pH of 5.5. Specifically, the binding stoichiometries of i-TMPipEOPP to multimeric G-quadruplexes (Hum48 and Hum54) were obviously higher than those of i-TMPipEOPP to monomeric ones (Hum24, KRAS and M3Q). These results suggested that besides end-stacking, electrostatic interactions may also occur between i-TMPipEOPP and DNA (Scheme 2). No interactions were observed between i-TMPipEOPP and duplex DNA at neutral conditions. Under acidic conditions, however, i-TMPipEOPP could bind with LD and AT with stoichiometries of 0.6
2 (Table 2). That is, one i-TMPipEOPP molecule could bind with two G-quadruplex molecules. Similar binding stoichiometry was observed in the studies of G-quadruplex and other porphyrin derivatives,9a,16a thus suggesting that i-TMPipEOPP might interact with G-quadruplex DNA through an end-stacking modality forming a sandwich-like G-quadruplex/i-TMPipEOPP/G-quadruplex complex (Scheme 2). The binding stoichiometries obtained at a pH of 5.5 were much higher than those obtained at a pH of 7.4. In addition, DNA length-dependent binding stoichiometries were observed at a pH of 5.5. Specifically, the binding stoichiometries of i-TMPipEOPP to multimeric G-quadruplexes (Hum48 and Hum54) were obviously higher than those of i-TMPipEOPP to monomeric ones (Hum24, KRAS and M3Q). These results suggested that besides end-stacking, electrostatic interactions may also occur between i-TMPipEOPP and DNA (Scheme 2). No interactions were observed between i-TMPipEOPP and duplex DNA at neutral conditions. Under acidic conditions, however, i-TMPipEOPP could bind with LD and AT with stoichiometries of 0.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1.0
1 and 1.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, respectively, thus further suggesting that electrostatic interactions may exist between i-TMPipEOPP and DNAs.
1, respectively, thus further suggesting that electrostatic interactions may exist between i-TMPipEOPP and DNAs.
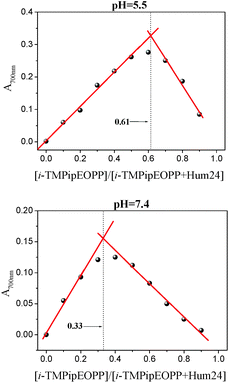 | ||
| Fig. 2 Job plot analysis of the interactions between i-TMPipEOPP and Hum24 under acidic and neutral conditions. [i-TMPipEOPP] + [Hum24] = 10 μM. Job plot analysis results of the interactions between i-TMPipEOPP and other DNAs were shown in ESI Fig. S5.† | ||
| pH = 5.5 | pH = 7.4 | |||
|---|---|---|---|---|
| Stoichiometrya | Ka (×106 M−1) | Stoichiometrya | Ka (×106 M−1) | |
| a Bind stoichiometry of i-TMPipEOPP to DNA. | ||||
| Hum24 | 1.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
6.70 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
0.78 |
| KRAS | 1.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
2.93 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
1.25 |
| M3Q | 0.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
6.16 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
1.01 |
| Hum48 | 3.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
1.72 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
0.83 |
| Hum54 | 4.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
2.82 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
1.05 |
| LD | 0.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
0.73 | Undetected | Undetected |
| AT | 1.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
0.60 | Undetected | Undetected |
Binding constants of i-TMPipEOPP to different DNAs
To further investigate the binding behaviors of i-TMPipEOPP to DNA, the UV-vis absorption titration spectra of i-TMPipEOPP were measured by fixing i-TMPipEOPP concentration at 5 μM while varying the DNA concentration (Fig. 3). DNA concentration-dependent absorption spectrum changes were observed for both G-quadruplex and duplex DNA at the pH of 5.5. That is, with the increase of DNA concentration, the absorption signal intensity at 422 nm decreased continuously, accompanied by the appearance of two new bands that exhibited continuous increases in their signal intensities. However, at the pH of 7.4, only G-quadruplex DNA could cause obvious absorption spectrum change of i-TMPipEOPP. | ||
| Fig. 3 DNA concentration-dependent absorption spectrum changes of i-TMPipEOPP in the presence of Hum24 or LD under acidic or neutral conditions. The concentrations of Hum24 are (arrow direction): 0, 0.3, 0.6, 1.0, 1.3, 1.6, 2.0, 2.3, 2.6, 3.0, 3.3, 3.6, 4.0, 4.5, 5.0, 5.5, 6.0, 6.5, 7.0, 7.5, 8.0, 8.5, 9.0, 10, 15, 20, 25, 30, 40 and 50 μM. The concentrations of LD are labeled in the figures. DNA concentration-dependent spectrum changes in the presence of other DNAs can be found in ESI Fig. S6.† | ||
Data from each absorbance titration were analysed to obtained the apparent binding constants (Ka) between i-TMPipEOPP and individual DNAs (Table 2).9k,13 As for the interactions between i-TMPipEOPP and G-quadruplexes, the Ka values obtained at acidic conditions were significantly higher than those obtained at neutral conditions. This is not surprising because, besides π–π stacking between the porphine core of i-TMPipEOPP and the G-quartet planes of G-quadruplex DNAs, the electrostatic interactions between the positively charged side arms of i-TMPipEOPP and negatively charged phosphate backbones or grooves of G-quadruplex DNA further enhances the binding stability between i-TMPipEOPP and G-quadruplexes under acidic conditions. i-TMPipEOPP showed better G-quadruplex recognition specificity against duplex DNA at neutral conditions than at acidic conditions. However, even at the pH of 5.5, i-TMPipEOPP also had a higher binding affinity towards G-quadruplex than towards duplex DNA, which was reflected by the significantly different Ka values of i-TMPipEOPP towards these two kinds of DNA structures (Table 2).
Effects of i-TMPipEOPP on G-quadruplex stability
The experiments discussed above strongly suggest that i-TMPipEOPP has a pH-dependent G-quadruplex binding affinity. To demonstrate this further, the effects of i-TMPipEOPP on the stability of G-quadruplex DNA were studied by determining the change in melting temperature (T1/2) of G-quadruplexes before and after the addition of i-TMPipEOPP. Obvious T1/2 increases could be observed under both acidic and neutral conditions, indicating that i-TMPipEOPP could stabilize G-quadruplex structures under both conditions. However, the extent of the T1/2 increase under acidic conditions was much higher than that under neutral conditions (Table S1†). Taking Hum24 as an example (Fig. 4), around 13.5 °C and 5.2 °C T1/2 changes were observed at pH values of 5.5 and 7.4, respectively, suggesting that i-TMPipEOPP displayed higher G-quadruplex-stabilizing ability under acidic conditions. This result is also consistent with the previously mentioned binding affinity study. | ||
Fig. 4 Melting temperature (T1/2) detection of Hum24 in the absence ( ) and presence ( ) and presence ( ) of 3 μM i-TMPipEOPP at pH of 7.4 (red) or 5.5 (green). T1/2 detection of other G-quadruplexes are shown in ESI Fig. S7.† ) of 3 μM i-TMPipEOPP at pH of 7.4 (red) or 5.5 (green). T1/2 detection of other G-quadruplexes are shown in ESI Fig. S7.† | ||
Effects of DNA concentration on the fluorescence spectrum of i-TMPipEOPP
pH-dependent binding selectivity of i-TMPipEOPP towards G-quadruplex and duplex DNA could also be reflected by the fluorescence titration spectra of i-TMPipEOPP. Considering that a new 455 nm band emerged in the absorption spectrum of i-TMPipEOPP with the addition of DNAs, 455 nm was used as the excitation wavelength while the changes in the fluorescence spectrum of i-TMPipEOPP with the addition of various concentrations of DNA were investigated at different pH conditions (Fig. 5). At the pH of 5.5, titration of i-TMPipEOPP using G-quadruplex DNA caused a complex fluorescence spectrum change. With the addition of G-quadruplex DNA, two distinct fluorescence-changing steps were observed. For example, the fluorescence signal first decreased and then increased as the concentration of Hum24 increased. This resulted in an obvious emission wavelength shift (about 12 nm) between the two steps. These results suggest that i-TMPipEOPP interacts with G-quadruplex DNA by complex binding modalities, including electrostatic and π–π end-stacking interactions. Titration of i-TMPipEOPP with duplex DNA also gave two fluorescence-changing steps, but no wavelength shift was observed, which is consistent with the suggestion that only electrostatic interactions occur between i-TMPipEOPP and duplex DNA. The initial fluorescence of i-TMPipEOPP is caused by its self-dimerization in water.18 With the addition of DNA, the electrostatic interactions between i-TMPipEOPP and DNA cause the disassembly of dimer to monomer, accompanied by the decrease of the fluorescence signal. Because i-TMPipEOPP has four positively charged side arms, one i-TMPipEOPP molecule can interact with several DNA strands, and one DNA strand can also interact with several i-TMPipEOPP molecules. Through such a cross-linking interaction, i-TMPipEOPP is surrounded by DNA strands, thus preventing the fluorescence quenching of water to i-TMPipEOPP. As a result, the fluorescence signal increased again with increasing DNA concentrations. Because sandwich-like end-stacking interaction can provide a more hydrophobic environment for i-TMPipEOPP, the fluorescence peak shifts to a longer wavelength region in the presence of G-quadruplex DNA. | ||
| Fig. 5 Hum24 or LD concentration-dependent fluorescence spectrum changes of i-TMPipEOPP under acidic or neutral conditions. The concentrations of Hum24 are (arrow direction): 0, 0.2, 0.4, 0.6, 0.8, 1, 1.2, 1.4, 1.6, 1.8, 2, 3, 4, 6, 10, 14, 20, 30, 40 and 50 μM. The concentrations of LD at pH 5.5 are (arrow direction): 0, 0.5, 1, 1.5, 2, 2.5, 3, 3.5, 4, 4.5, 5, 6, 7, 8, 10, 20, 30 and 50 μM. The concentrations of LD at pH 7.4 are labeled in the figure. Fluorescence titration spectra of i-TMPipEOPP by other DNAs are shown in ESI Fig. S8.† | ||
Relatively simple fluorescence titration spectra were obtained at the pH of 7.4. Since no dimerization occurs under this condition, the fluorescence of i-TMPipEOPP remained at a low level. With the addition of duplex DNA, no obvious change in fluorescence was observed, thus further demonstrating that no electrostatic interactions occur between i-TMPipEOPP and DNA. Conversely, the addition of G-quadruplex DNA continuously increased the fluorescence intensity. These results further demonstrate that relatively simpler interaction modalities occur between i-TMPipEOPP and G-quadruplex DNA under neutral conditions than under acidic conditions. That is, under neutral conditions, end-stacking interaction is the dominant binding mode between i-TMPipEOPP and G-quadruplex DNA while forces of electrostatic interaction are nearly neglected. The fluorescence titration results further demonstrated the improved G-quadruplex recognition specificity of i-TMPipEOPP against duplex DNA under neutral conditions.
Effects of i-TMPipEOPP on the CD spectrum of G-quadruplex DNA
CD spectroscopy, which has been widely used in G-quadruplex studies,19 was used to further investigate the interactions between i-TMPipEOPP and G-quadruplex DNA. As shown in Fig. 6, the CD spectrum of Hum24 gave a positive peak at around 290 nm and a negative peak at around 240 nm, indicating that a mixed parallel/antiparallel hybrid G-quadruplex structure was formed.20 Addition of i-TMPipEOPP caused a significant CD spectrum change by shifting the positive peak to around 265 nm, which is characteristic of parallel G-quadruplexes, thus suggesting that the presence of i-TMPipEOPP could promote the formation of parallel G-quadruplexes. Such a G-quadruplex structure conversion might be attributed to above-mentioned end-stacking interactions between i-TMPipEOPP and G-quadruplexes.9k Antiparallel or hybrid G-quadruplex DNA contains at least one lateral or diagonal loop on the top and bottom G-quartet surfaces, which would certainly result in significant steric hindrance to prevent i-TMPipEOPP end-stacking on the G-quartet surfaces.21 However, the three loops of parallel G-quadruplexes all distribute on the side faces of G-quadruplexes, thus making it easier for i-TMPipEOPP to access the end quartet surfaces. The similar effects of i-TMPipEOPP on G-quadruplex CD spectra under acidic and neutral conditions indicated that i-TMPipEOPP may be used as an efficient G-quadruplex ligand under both conditions. | ||
| Fig. 6 Effects of i-TMPipEOPP on the CD spectra of Hum24 under acidic and neutral conditions. (1) Hum24; (2) Hum24 + 50 mM K+; (3) Hum24 + 50 mM K+ + 5 μM i-TMPipEOPP. CD spectra of other G-quadruplexes are shown in ESI Fig. S9.† | ||
i-TMPipEOPP-triggered pH-dependent photocleavage of duplex DNA
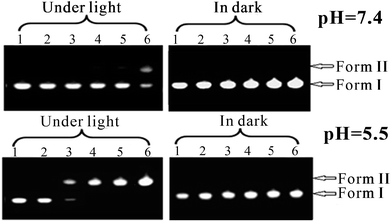
It is believed that non-specific cytotoxicity of most G-quadruplex ligands arises from the interactions between ligands and duplex DNA. Our experiments showed that i-TMPipEOPP can bind to duplex DNA under acidic conditions but not under neutral conditions. To further investigate the interaction between i-TMPipEOPP and duplex DNA, DNA cleavage abilities of i-TMPipEOPP were compared under different conditions (pH 5.5 or 7.4, with or without sunlight radiation) using pBR322 plasmid DNA. Without radiation from sunlight, DNA cleavage was neither observed under acidic nor neutral conditions. In contrast, under sunlight radiation, i-TMPipEOPP concentration-dependent DNA cleavage was observed (Fig. 7). That is, with increasing i-TMPipEOPP concentration, more and more supercoiled pBR322 DNA (Form I) was cleaved to nicked DNA (Form II). This result suggested that i-TMPipEOPP has great potential as a new sensitizer for photodynamic therapy (PDT), which may be related to i-TMPipEOPP's ability to absorb light and its high electron excitation energy for strong exergonic electron transfer.22 Moreover, i-TMPipEOPP-triggered DNA photocleavage occurred in a pH-dependent manner. The DNA cleavage efficiency at a pH of 5.5 was much higher than that at 7.4. This is consistent with our results that i-TMPipEOPP can bind with duplex DNAs at the pH of 5.5, but cannot at the pH of 7.4, thus suggesting that such a potential PDT drug might show higher cytotoxicity towards acidic tumor cells than neutral healthy cells.pH-dependent phototoxicity to cells
pH-dependent cytotoxicity of i-TMPipEOPP was estimated by standard MTT assays. As shown in Fig. 8, without the radiation of sunlight, >85% cell viability was observed even when the cells was incubated with 5 μM i-TMPipEOPP for 24 h, indicating that i-TMPipEOPP was not cytotoxic in dark under both acidic and neutral conditions. However, radiation with sunlight for 1 h could greatly decrease the cell viability, suggesting that i-TMPipEOPP has phototoxicity to cells. In addition, such a phototoxicity was also pH-dependent. That is, i-TMPipEOPP showed higher phototoxicity under acidic conditions than under neutral conditions. Specially, when the concentration of i-TMPipEOPP was lower than 2 μM, no obvious phototoxicity was observed under neutral conditions, and similar cell viabilities were observed with or without sunlight radiation. However, under acidic conditions, greatly enhanced cell phototoxicity was observed, about 50% cells were killed under the sunlight radiation for 1 h. These results further demonstrated that i-TMPipEOPP might be used as PDT drug for acidic tumor cells. | ||
| Fig. 8 Cell viability of MDA-MB-231 cells incubated with different concentrations of i-TMPipEOPP in dark for 24 h or under sunlight for 1 h at the pH of 5.5 or 7.4. | ||
Conclusions
In summary, we proposed a new strategy for anticancer drug design. That is, searching for G-quadruplex ligands that can interact with duplex DNA under acidic conditions but not under neutral conditions. As an example, a new water-soluble cationic porphyrin derivative i-TMPipEOPP was synthesized, characterized and its interactions with G-quadruplex and duplex DNA were investigated. The four positively charged side arms confer this porphyrin derivative with not only water solubility, but also pH-dependent G-quadruplex recognition specificity against duplex DNA. Under acidic conditions (pH 5.5), i-TMPipEOPP binds with G-quadruplex DNA by end-stacking and electrostatic interactions. It also binds with duplex DNA with only electrostatic interaction. Under neutral conditions (pH 7.4), i-TMPipEOPP binds with G-quadruplex DNA by end-stacking interaction, however, it cannot bind with duplex DNA. Because of the synergy between π–π end-stacking and electrostatic interactions, the G-quadruplex binding affinity of i-TMPipEOPP under acidic conditions is higher than that under neutral conditions, in which only π–π stacking interactions occur. More interestingly, i-TMPipEOPP shows pH-dependent DNA photocleavage activity towards duplex DNA and pH-dependent phototoxicity to cells, thus it may be developed as a promising photodynamic therapy drug showing higher cytotoxicity towards acidic tumor cells than neutral healthy cells.Acknowledgements
This work was supported by the National Basic Research Program of China (no. 2011CB707703), and the Natural Science Foundation of China (no. 21322507, 21371130, 21175072, 51173083).Notes and references
- (a) O. Doluca, J. M. Withers and V. V. Filichev, Chem. Rev., 2013, 113, 3044 CrossRef CAS PubMed; (b) J. Choi and T. Majima, Chem. Soc. Rev., 2011, 40, 5893 RSC.
- Y. Du and X. Zhou, Chem. Rec., 2013, 13, 371 CrossRef CAS PubMed.
- M. L. Bochman, K. Paeschke and V. A. Zakian, Nat. Rev. Genet., 2012, 13, 770 CrossRef CAS PubMed.
- Z. Yu and H. Mao, Chem. Rec., 2013, 13, 102 CrossRef CAS PubMed.
- V. T. Mukundan and A. T. Phan, J. Am. Chem. Soc., 2013, 135, 5017 CrossRef CAS PubMed.
- M. Ruden and N. Puri, Cancer Treat. Rev., 2013, 39, 444 CrossRef CAS PubMed.
- Q. Li, J. F. Xiang, H. Zhang and Y. L. Tang, Curr. Pharm. Des., 2012, 18, 1973 CrossRef CAS.
- (a) W. Tuntiwechapikul, T. Taka, M. Béthencourt, L. Makonkawkeyoon and T. R. Lee, Bioorg. Med. Chem. Lett., 2006, 16, 4120 CrossRef CAS PubMed; (b) J. Cuesta, M. A. Read and S. Neidle, Mini-Rev. Med. Chem., 2003, 3, 11 CrossRef CAS; (c) P. Alberti, L. Lacroix, L. Guittat, C. Helene and J. L. Mergny, Mini-Rev. Med. Chem., 2003, 3, 23 CrossRef CAS.
- (a) L.-N. Zhu, B. Wu and D.-M. Kong, Nucleic Acids Res., 2013, 41, 4324 CrossRef CAS PubMed; (b) X.-X. Huang, L.-N. Zhu, B. Wu, Y.-F. Huo, N.-N. Duan and D.-M. Kong, Nucleic Acids Res., 2014, 13, 8719 CrossRef PubMed; (c) J. Alzeer, B. R. Vummidi, P. J. C. Roth and N. W. Luedtke, Angew. Chem., Int. Ed., 2009, 48, 9362 CrossRef CAS PubMed; (d) C. Romera, O. Bombarde, R. Bonnet, D. Gomez, P. Dumy, J. F. Gwan, J. H. Lin, E. Defrancq and G. Pratviel, Biochimie, 2011, 93, 1310 CrossRef CAS PubMed; (e) A. C. Bhasikuttan, J. Mohanty and H. Pal, Angew. Chem., Int. Ed., 2007, 46, 9305 CrossRef CAS PubMed; (f) D.-M. Kong, Y.-E. Ma, J. Wu and H.-X. Shen, Chem.–Eur. J., 2009, 15, 901 CrossRef CAS PubMed; (g) M.-P. Teulade-Fichou, C. Carrasco, L. Guittat, C. Bailly, P. Alberti, L. N. Mergny, A. David, J. M. Lehn and W. D. Wilson, J. Am. Chem. Soc., 2003, 125, 4732 CrossRef CAS PubMed; (h) H. Arthanari, S. Basu, T. L. Kawano and P. H. Bolton, Nucleic Acids Res., 1998, 26, 3724 CrossRef CAS PubMed; (i) J.-H. Guo, L.-N. Zhu, D.-M. Kong and H.-X. Shen, Talanta, 2009, 80, 607 CrossRef CAS PubMed; (j) J. Zhou, V. Le, D. Kalia, S. Nakayama, C. Mikek, E. A. Lewis and H. O. Sintim, Mol. BioSyst., 2014, 10, 2724 RSC; (k) B. Jin, X. Zhang, W. Zheng, X. Liu, J. Zhou, N. Zhang, F. Wang and D. Shangguan, Anal. Chem., 2014, 86, 7063 CrossRef CAS PubMed; (l) C.-C. Chang, J.-Y. Wu, C.-W. Chien, W.-S. Wu, H. Liu, C.-C. Kang, L.-J. Yu and T.-C. Chang, Anal. Chem., 2003, 75, 6177 CrossRef CAS PubMed; (m) D.-M. Kong, Y.-E. Ma, J.-H. Guo, W. Yang and H.-X. Shen, Anal. Chem., 2009, 81, 2678 CrossRef CAS PubMed.
- D. Ling, W. Park, S.-j. Park, Y. Lu, K. S. Kim, M. J. Hackett, B. H. Kim, H. Yim, Y. S. Jeon, K. Na and T. Hyeon, J. Am. Chem. Soc., 2014, 136, 5647 CrossRef CAS PubMed.
- (a) Z. Popović, W. Liu, V. P. Chauhan, J. Lee, C. Wong, A. B. Greytak, N. Insin, D. G. Nocera, D. Fukumura, R. K. Jain and M. G. Bawendi, Angew. Chem., Int. Ed., 2010, 49, 8649 CrossRef PubMed; (b) F. A. Gallagher, M. I. Kettunen, S. E. Day, D.-E. Hu, J. H. Ardenkjær-Larsen, R. i. Zandt, P. R. Jensen, M. Karlsson, K. Golman, M. H. Lerche and K. M. Brindle, Nature, 2008, 453, 940 CrossRef CAS PubMed; (c) Y. Urano, D. Asanuma, Y. Hama, Y. Koyama, T. Barrett, M. Kamiya, T. Nagano, T. Watanabe, A. Hasegawa, P. L. Choyke and H. Kobayashi, Nat. Med., 2008, 15, 104 CrossRef PubMed; (d) E. S. Lee, D. Kim, Y. S. Youn, K. T. Oh and Y. H. Bae, Angew. Chem., Int. Ed., 2008, 47, 2418 CrossRef CAS PubMed.
- (a) C. R. Justus, L. Dong and L. V. Yang, Front. Physiol., 2013, 4, 354 Search PubMed; (b) R. A. Gatenby and R. J. Gillines, Nat. Rev. Cancer, 2004, 4, 891 CrossRef CAS PubMed; (c) P. Vaupel, F. Kallinowski and P. Okunieff, Cancer Res., 1989, 49, 6449 CAS.
- (a) X. Xie, B. Choi, E. Largy, R. Guillot, A. Granzhan and M.-P. Teulade-Fichou, Chem.–Eur. J., 2013, 19, 1214 CrossRef CAS PubMed; (b) F. H. Stootman, D. M. Fisher, A. Rodger and J. R. Aldrich-Wright, Analyst, 2006, 131, 1145 RSC.
- X. W. Hui, N. Gresh and B. Pullman, Nucleic Acids Res., 1990, 18, 1109 CrossRef CAS PubMed.
- (a) H. Q. Yu, D. Miyoshi and N. Sugimoto, J. Am. Chem. Soc., 2006, 128, 15461 CrossRef CAS PubMed; (b) Y. Xu, T. Ishizuka, K. Kurabayashi and M. Komiyama, Angew. Chem., Int. Ed., 2009, 48, 7833 CrossRef CAS PubMed; (c) G. W. Collie, G. N. Parkinson, S. Neidle, F. Rosu, E. De Pauw and V. Gabelica, J. Am. Chem. Soc., 2010, 132, 9328 CrossRef CAS PubMed; (d) S. Haider, G. N. Parkinson and S. Neidle, Biophys. J., 2008, 95, 296 CrossRef CAS PubMed; (e) H. Yu, X. Gu, S.-i. Nakano, D. Miyoshi and N. Sugimoto, J. Am. Chem. Soc., 2012, 134, 20060 CrossRef CAS PubMed; (f) L. Xu, S. Feng and X. Zhou, Chem. Commun., 2011, 47, 3517 RSC.
- (a) L.-N. Zhu, S.-J. Zhao, B. Wu, X.-Z. Li and D.-M. Kong, PLoS One, 2012, 7, e35586 CAS; (b) L.-N. Zhu, Y.-F. Huo and B. Wu, Anal. Methods, 2014, 6, 5067 RSC.
- V. Dhaamodharan, S. Harikrishna, C. Jagadeeswaran, K. Halder and P. I. Pradeepkumar, J. Org. Chem., 2012, 77, 229 CrossRef PubMed.
- (a) K. Kano, H. Minamizono, T. Kitae and S. Negi, J. Phys. Chem. A, 1997, 101, 6118 CrossRef CAS; (b) Y.-Y. Liu, M. Wu, L.-N. Zhu, X.-Z. Feng and D.-M. Kong, Chem.–Asian J., 2015 DOI:10.1002/asia.201500106.
- (a) J. Kypr, I. Kejnovská, D. Renciuk and M. Vorlícková, Nucleic Acids Res., 2009, 37, 1713 CrossRef CAS PubMed; (b) M. Vorlíčková, I. Kejnovská, J. Sagi, D. Renčiuk, K. Bednářová, J. Motlová and J. Kypr, Methods, 2012, 57, 64 CrossRef PubMed.
- V. Viglasky, L. Bauer and K. Tluckova, Biochemistry, 2010, 49, 2110 CrossRef CAS PubMed.
- A. Laguerre, L. Stefan, M. Larrouy, D. Genest, J. Novotna, M. Pirrotta and D. Monchaud, J. Am. Chem. Soc., 2014, 136, 12406 CrossRef CAS PubMed.
- C. Zhou, Q. Liu, W. Xu, C. Wang and X. Fang, Chem. Commun., 2011, 47, 2982 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra05970d |
| ‡ These two authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2015 |