13C pathway analysis of biofilm metabolism of Shewanella oneidensis MR-1
Abstract
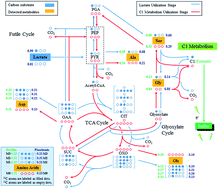
Biofilm metabolism of Shewanella was analyzed via 13C tracing experiments for the first time. The activity of C1 metabolism in the biofilm cells was found to be interestingly higher than that in planktonic cells, which could be related to utilizing C1 metabolites as electron donors when growing Shewanella in biofilms.

 Please wait while we load your content...
Please wait while we load your content...