New insights into the microbial degradation of polyurethanes
Neha Mahajan
a and
Pankaj Gupta
*b
aDepartment of Biotechnology, Govt Degree College Kathua, Higher Education Department, J&K, India 184104. E-mail: pankajrrl@rediffmail.com; pankajrrl@gmail.com; Fax: +91 1922 234315; Tel: +91 1921 224051 Tel: +91 9419251800
bDepartment of Chemistry, Govt Degree College Kathua, Higher Education Department, J&K, India 184104
First published on 22nd April 2015
Abstract
Frequent and frequently deliberate release of plastics leads to accumulation of plastic waste in the environment which is an ever increasing ecological threat. Plastic waste represents 20–30% of the total volume of solid waste contained in landfills because in addition to the large amount of waste generated, plastic waste is recalcitrant and remains deposited in these landfills for long periods of time. Paradoxically, the most preferred property of plastics – durability – exerts also the major environmental threat. Polyurethanes (PU) represent a class of polymers that have found widespread use in medical, automotive and industrial fields. The wide use of PUs in our society makes their biodegradation of equal importance to their manufacturing. The balance between creating stable polymers that resist degradation and minimize their potential long-term environmental impact continues to be one of the major issues with the general use of these materials. Despite their microbial resistance, they are susceptible to the attack of fungi and bacteria. Currently, as environmental concerns have become so significant great effort needs to be made to degrade these plastics under environmentally benign conditions. In the present report we seek to highlight the efforts made in the last few years for the degradation of polyurethanes using microorganisms or enzymes.
1. Introduction
Polyurethanes (PU) are some of the most versatile polymers in existence today. These are commercially available in various forms, ranging from flexible or rigid lightweight foams to tough, stiff, and strong elastomers. They represent a class of polymers that have found widespread use in medical, automotive and industrial fields. Among the synthesized plastics, polyurethanes, mostly in the form of large blocks of foams, ranked as the 6th most common type of plastic used worldwide, and they accounted for 6 to 7% (i.e. 11.5 million tonnes per year) of the total plastics produced.1,2 Polyurethanes were first synthesized by Dr Otto Bayer in 1937. The synthesis of polyurethane foam at the industrial scale began in the 1950s, and their use grew slowly until the 1990s. Polyurethane (PU) is a versatile plastic with several industrial applications in the modern life. Key raw materials used in manufacture of polyurethane are diisocyanates, polyols and chain extenders. The rebound in furniture, interiors and construction industry in North America and Europe, as well as rapid economic growth in Asia Pacific is expected to remain the major driving force for the polyurethane market. The global PU market, by product type is dominated by the rigid and flexible foams, which together accounts for over 65% of total PU demand in 2011. The other major product types include coatings, adhesives, sealants and elastomers (Fig. 1).Polyurethane is mainly employed in furniture and interiors, construction, electronics and appliances, automotive, footwear and packaging. Furniture and interior industry dominated the global market followed by construction industry. The electronics and appliances industry is expected to be the key growth market for polyurethanes over the next five years. Asia Pacific accounted for 40.4% of the total polyurethane market in 2011, followed by Europe and North America. As one of major rising economic powers in the world, India has witnessed rapid growth of its polyurethane industry with the output of 320![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 tons in 2011.
000 tons in 2011.
Polyurethanes are replacing older polymers for various reasons. The United States government is phasing out chlorinated rubber in marine and aircraft and coatings because they contain environmentally hazardous volatile organic compounds.3 Auto manufacturers are replacing latex rubber in car seats and interior padding with PU foam because of lower density and greater flexibility.4 Other advantages of PUs are that they have increased tensile strength and melting points making them more durable.5 Their resistance to degradation by water, oils, and solvents make them excellent for the replacement of plastics.6 In the medical arena, polyurethane is considered as one of the most bio and blood compatible materials known. They have played a major role in the development of many medical devices due to their structural properties, blood and tissue compatibility and resistance to macromolecular oxidation, hydrolysis and calcification.7
2. Synthesis, structure and nature of linkage in simple polyurethane
Polyurethanes (PU) properties depend both on the method of preparation and the monomers used. PUs are synthesized by the exothermic reactions between alcohols with two or more reactive hydroxyl (–OH) groups per molecule (diols, triols, polyols) and isocyanates that have one or more than one reactive isocyanate group (–NCO) per molecule (diisocyanates, polyisocyantes), generally, in presence of a chain extender, catalyst, and/or other additives. The synthesis of simple polyurethane is depicted in Scheme 1.The frequently used diisocyanates in the synthesis of polyurethanes are aliphatic diisocyanates (viz hexamethylene diisocyanate, isophorone diisocyanate, dicyclohexylmethane-4,4′-diisocyanate, m-tetramethylxylylene diisocyanate, 1,4-butane diisocyanate and trans-cyclohexane diisocyanate) and aromatic diisocyanates (viz diphenylmethane diisocyanate, 2,4-toluene diisocyanate, p-phenylene diisocyanate and naphthalene diisocyanate). Polyols comprise the largest volume of polyurethanes and the four main types of long-chain diols used in the production of polyurethanes are: polyesters, polycaprolactones, polyethers and polycarbonates. In general, there are two types of compounds that are generally used as chain extenders, diols or diamines, which can either be aliphatic or aromatic, depending on the required properties in the synthesized polyurethanes. New chain extenders, including amino acids have been also used during polyurethane synthesis as isocyanates can react vigorously with amine, alcohol, and carboxylic acids.8 These novel chain extenders have been used to synthesize biodegradable polyurethanes.9,10
PUs are classified in polyester or polyether polyurethanes depending on what substrates are used and the biological attack towards them is determined by the type of substrates used in the polymer synthesis despite its xenobiotic origin.11 The molecular structure of repeating units of the polyester and polyether polyurethane elastomer is described in Fig. 2. Variations in the spacing between urethane linkages, as well as the nature of the substitutions, can change the properties of the resulting polymer from linear and rigid to branched and flexible. The structure of the linear polymeric chain of thermoplastic polyurethane is in blocks, alternating two different types of segments linked together by covalent links, forming a block copolymer. These segments are: (a) hard segments: These are segments formed by the reaction of the diisocyanate and the short-chain diol. They have a high density of urethane groups of high polarity, and for this reason, they are rigid at room temperature (high hardness) and (b) soft segments: These are segments formed by the reaction of the diisocyanate and the long-chain diol. They have a low polarity as they have a very low density of urethane groups, and therefore, they are flexible at room temperature (very low hardness).
3. Degradation of plastics/polyurethanes
Different polymers like polyethylene, polypropylene, polyvinyl chloride (PVC), polystyrene, polyester, polyamide, polyethylene terephthalate and polyurethane have been designed for resistance to the environment.12,13 As a result they undergo degradation or bio-degradation, at very slow rate. The ecological problems related to the environmental pollution by synthetic polymers like plastics are one of the major concerns of the present days; especially because they are difficult to degrade easily and the entire process is time consuming. Plastic waste represents 20–30% of the total volume of solid waste contained in landfills because in addition to the large amount of waste generated, plastic waste is recalcitrant and remains deposited in these landfills for long periods of time.14 The increasing quantities of plastic waste and their effective and safe disposal have become a matter of public concern. When plastics are dumped in a field or in dumping areas, there is evidential proof that they are causing a great change in the pH of the soil followed by disturbance in the leaching of the rain water and moisture, making the land bare and unfertile. The biological degradation time is very high, and it takes thousands of years to degrade these long chain polymers into simple hydrocarbons. Latest reports confirmed that some plastic products are mimicking human hormones (e.g., thyroxin and sex hormones), causing human health hazards.15 It is also creating a major problem in marine ecosystem. For the last 30 years, scientists are trying to develop some alternative ways other than the natural destruction to degrade these high molecular synthetic polymers, but yet now, very few evidences are available where scientists are able to develop some alternative ways to enhance the mode of degradation and make it faster. Recent research suggests that there have been a notable number of microorganisms (especially some bacteria and fungi) which have the capacity to degrade these synthetic polymers in much faster way in comparison to the natural method by using some exoenzymes under stress conditions.Thermal degradation is another approach16 that has been extensively investigated due to their wide range of applications, however it is even more hazardous as it generates toxic gases like carbon monoxide and dioxin, and burning of plastic has shown to release heavy metals like cadmium and lead. Moreover, virgin plastic polymers are rarely used by themselves and typically the polymer resins are mixed with various additives to improve performance. These additives include inorganic fillers such as carbon and silica that reinforce the material, plasticizers to render the material pliable, thermal and ultraviolet stabilizers, flame retardants and colourings. Many such additives are used in substantial quantities and in a wide range of products.17 To reinforce polyurethanes against potential biotic break-down, some additives are included during the polyurethane polycondensation process. These additives have two distinct roles: (i) to increase polyol condensation and (ii) to enhance antimicrobial function. Tin derivative compounds such as dibutyl tin dilaurate (DBTDL) were proposed to inhibit polymer biodegradation by fungal activities.18 Additionally, a wide variety of compounds can be added during the polyol formation to increase polyol chain extension and to inhibit microbial degradation for the production of both polyether- and polyester-based polyols. Because these additives have a large set of origins, their toxicity and ecotoxicity range from glycerol (a safe product) to a highly toxic hazard such as DBTDL. Research has been initiated to elucidate whether additives to the chemical structure of PU could decrease biodegradation. Kanavel et al.19 observed that sulfur-cured polyester and polyether PU had some fungal inertness. However they noted that even with fungicides added to the sulfur- and peroxide-cured PU, fungal growth still occurred on the polyester PU and most fungicides had adverse effects on the formulations.
Despite recognition of the persistent pollution problems posed by plastic, global production is still increasing, with the largest increases expected in developing nations. The sheer volume of plastics produced each year presents a problem for waste disposal systems. The scale of this problem and the recalcitrance of some polymers to degradation necessitate investigation into effective methods for biodegradation of plastics.
4. Microbial degradation of polyurethane
The wide use of polyurethanes (PU) in our society makes their biodegradation of equal importance as their manufacturing.20 The balance between creating stable polymers that resist degradation and minimize their potential long-term environmental impact continues to be one of the major issues with the general use of these materials. The degradation of plastics have been reviewed and documented several times in the past few years covering various aspects.21,22 In these days when environmental concerns have become so significant great efforts need to be developed to degrade these plastics under environmental benign conditions. Microbes are known to survive in environments where recalcitrant materials, like polyurethane, are present, so it is possible that these microorganisms can use this material and be useful tools in biodegradation.23 In the present report comprising more than 125 references, we seek to highlight the efforts made in the last one decade for the biodegradation of polyurethanes using microorganisms since microorganisms are involved in the deterioration and degradation of both synthetic and natural polymers.Depending on the type of long chain diol used in polyurethane production, the three main types of polyurethanes are polyester, polycaprolactone and polyether urethanes. It is well known that ether groups have much better hydrolysis resistance than ester groups. The hydrolysis reaction of an ester group follows the three-centre mechanism and is catalyzed by both acids and bases; since a free acid is liberated as a result of the hydrolysis of ester bonds, this reaction becomes autocatalytic. Therefore polyethers URs have much better hydrolysis resistance than polyester and polycaprolactone URs. In other words: microbial stability order of different PUs is:
| Polyether URs ⋙ polycaprolactone URs = polyester URs |
With regard to diisocyanate, it has been suggested that aliphatic diisocyanates are more susceptible to biodegradation than those having aromatic groups24 as aliphatic diisocyanates are more flexible and accessible.
4.1 Biodegradation mechanism
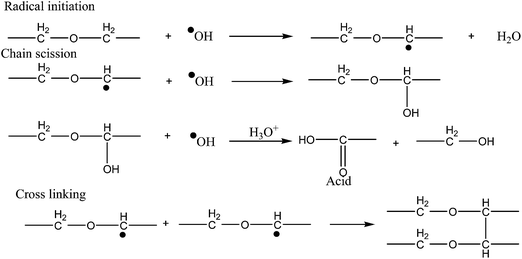
The degradation mechanism of polyurethanes depends on not only the PU chemistry structure but also the degradation environment, i.e. in the presence of water, acidic, alkaline or oxidative conditions, or in the presence of enzymes. Generally, the characterization of the by-products during the degradation of the polyurethane is the key to understand the mechanisms of degradation. Biodegradation of PUs occurs in two different mechanisms: (a) biological oxidation and (b) biological hydrolysis. In general, polyesterurethanes are susceptible to hydrolytic degradation because of ester groups in the soft segments while polyether urethanes are susceptible to oxidative degradation.25 Furthermore, the presence of metallic ions such as cobalt accelerates the process of oxidative degradation.26–28 Tanzi et al.29 studied the oxidative degradation of polyether (Pellethane 2363 80A) and polycarbonate (Corethane 80A, Bionate 80A and Chronoflex AL 80A) urethanes in 0.5 N nitric acid (acidic) and sodium hypochlorite (4% Cl2, alkaline) up to 14 days at 50 °C and under constant strain (100%). It was found that PEU were more degraded under alkaline oxidation (HClO) mainly in the absence of applied strain while poly(carbonate urethane) (PCU) was more affected by HNO3.Oxidative degradation has been generally associated with poly(ether-urethane)s, since many studies have determined that these polymers degrade by mean alpha-hydrogen abstraction adjacent to oxygen in polyethers and polycarbonates.30,31 In contrast, few works related to oxidative degradations on polyester polyurethanes has been done, and even less has studied the mechanism of degradation of polyurethane ureas. It is pertinent to mention here that, in the biological systems, the reactive oxygen species released by adherent leucocytes initiated degradation of polyether urethanes through oxidative attack of the soft segment. These reactive oxygen species abstracts an alpha-methylene hydrogen atom from polyether soft segment. Addition of a hydroxyl radical to the carbon radical forms a hemiacetal, which oxidises to an ester. Acid hydrolysis of the ester results in chain scission of the soft segment and formation of acid end groups. Significant chain scission results in the solubilisation and extraction of low molecular weight degradation products. This mechanism is illustrated in Fig. 3.32
A similar oxidative mechanism was proposed for hard segment degradation.33 Oxygen radicals abstract an alpha-methylene hydrogen atom from the chain extender at the urethane. Additional hydroxyl radicals combine with the chain radical to form a highly reactive carbonyl hemiacetal. Oxidative hydrolysis of the carbonyl hemiacetal results in chain scission and formation of an unstable carbamic acid and carboxylic acid end groups. The carbamic acid decarboxylates readily to form a free amine (Fig. 4).
The biological hydrolysis mechanism of PU includes three steps in the presence of hydrolase type enzymes. Firstly, chemical dissolution of ester and amide bonds in the polymer chain; secondly, decreasing molecular weight and viscosity; and finally, ending by cleaving all polymer chains. Therefore hydrolase type enzymes such as lipase, esterase and protease could degrade polymer films.22,34,35 In 2012, Trevino et al. proposed three possible sites of breakdown,36 depending on the enzyme (urease, protease or esterase) that acts directly on the urethane bond (Fig. 5).
Enzymes implicated in polyurethane biodegradation are localized in two main compartments and are either secreted or membrane-bound during the first step of polyurethane biodegradation.37 This model has been partially confirmed by recombinant expression approaches with extracellular enzymes and biochemical assays with the membrane-bound enzymes, and a mechanism of biodegradation has been described.38,39 The first step of polyurethane biodegradation is initiated by the adhesion of the membrane-bound enzyme to the polyurethane surface. Thereafter, the urethane bond is hydrolyzed by the membrane enzyme bound to the substrate, and monomers and the building-blocks of the polyurethane are released.40 This process allows the microorganism to generate a large amount of compounds released from the polyurethane cleavage in its vicinity (Fig. 6).41 Nakajima-Kambe et al. (1999) proposed a mechanism to explain the function of the membrane-bound polyurethanase of D. acidovorans. The role of this membrane-bound enzyme appears to be critical for the ability of the microorganism to grow using polyurethane as a substrate. In this view, the importance of free polyurethanases released into the medium may be underestimated, but their role is also essential. Indeed, as the membrane-bound enzyme degrades the polyurethane, a high amount of polyurethane monomers remains bound to the enzymes. Moreover, these enzymes have only a low surface area of recovery. In this sense, extracellular polyurethanases enable efficient polymer degradation and serve primarily to finish the job initiated by the membrane-bound enzymes (Fig. 6).
 | ||
| Fig. 6 Mechanistic scheme of microbial polyurethane degradation, with the path 1 corresponding to a biodegradation issued from cell adhesion and the path 2 corresponding to a biodegradation issued from free enzyme activity. (Reprinted from Cregut et al.41 with permission from Elsevier.) | ||
Hafeman et al.42 investigated the effects of esterolytic and oxidative conditions on scaffold degradation by incubating in 1 U mL−1 cholesterol esterase (CE), 1 U mL−1 carboxyl esterase (CXE), and 10 U mL−1 lipase (L) hydrogen peroxide (20 wt% hydrogen peroxide (H2O2) in 0.1 M cobalt chloride (CoCl2), and buffer alone (0.5 M monobasic sodium phosphate buffer with 0.2% w/w sodium azide) and measured the mass loss for 10 weeks at 37 °C. Polyurethane scaffolds were prepared by one-shot reactive liquid molding of hexamethylene diisocyanate trimer (HDIt) or lysine triisocyanate (LTI) and a polyol as hardener. Trifunctional polyester polyols of 900 Da molecular weight were prepared from a glycerol starter and 60% ε-caprolactone, 30% glycolide, and 10% D,L-lactide monomers (6C), (t1/2 = 20 days) and 70% caprolactone, 20% glycolide, and 10% lactide (7C) (t1/2 = 225 days) and stannous octoate catalyst. Incubation with esterases slightly accelerated degradation relative to Phosphate Buffer Saline (PBS). Differences in degradation between the three candidate enzymes at any given time point were not significant. In contrast, incubation with medium that created an oxidative microenvironment had a more significant effect on the polyurethane degradation rate, especially for the LTI-based materials, except the 6C/HDIt (hexamethylene diisocyanate trimer) + PEG, which interestingly degraded faster in the presence of cholesterol and carboxyl esterase than in oxidative medium.
Biodegradable poly(ether-ester-urethane)s (PEEUs) found applications in tissue repair43 or drug delivery.44 Chiellini et al. reported the preparation as well as hydrolytic and microbial degradation of multi-block poly(ether-ester-urethane)s (PEEUs) based on poly(3-caprolactone)/poly(ethylene glycol) segments. By varying the ratio of PCL and PEG and the molecular weight of the resultant copolymer, a modulation of bulk and surface hydrophilicity, as well as the degradation rate, were observed. Total mineralization of the copolymers was achieved in liquid media in presence of microorganisms from Arno River with different kinetics.45
Three types of PU degradation have been identified in literature: fungal biodegradation, bacterial biodegradation and degradation by polyurethanase enzymes. Both bacteria and fungi have demonstrated activity against polyurethanes under laboratory conditions. In comparison, more fungi than bacteria have been isolated.46 Two bacterial colonies capable of degrading polyurethane in contrast with mold countings of 3 × 105 colony forming units (CFU) were isolated.47 However, it is pertinent to mention here that most work in the area of PU biodegradation has focused on bacteria with several strains studied to determine the mechanistic pathway of degradation.
4.2 Biodegradation of polyurethane by bacteria
Although there are few reports on PU degrading bacteria, both Gram positive and Gram negative bacteria have the ability to degrade PU.48 Kay et al., in 1991 have isolated 15 bacterial strains capable of degrading ester-based polyurethanes and have also reported the results of degradation profiles examined for Corynebacterium strains having a strong degradation ability.49 The other bacterial isolates that can degrade PU includes Comamonas acidovorans,50 Pseudomonas flourescens,51 P. chlororaphis52 and Bacillus subtilis.53 Bacillus pumilus strain NMSN-Id isolated from polyurethane contaminated water can grow in high salt concentration (NaCl 10%, w/v) and degrade Impranil-DLN, water-dispersible polyurethane.54 Comamonas acidovorans TB-35 has been recovered by Nakajima and co-workers which can utilize solid PU as a sole carbon and nitrogen source.55 The authors disclosed that the strain does not utilize polyether PU, but utilizes polyester PU containing polydiethyleneglycol adipate as the sole source of carbon. Later on, a membrane bound esterase was isolated from Comomonas acidovorans by Akutsu et al., (1998), that absorbed hydrophobically on to the surface of PU followed by hydrolysis. The enzyme consists of two domains, a surface binding domain (SBD) and a catalytic domain. Both the domains are in close proximity with the hydrophobic SBD enabling binding of the enzyme to PU surface.37 Another bacterial strain Pseudomonas chlororaphis56 can show their activity against automotive waste polyester polyurethane (PU) foams and produce ammonia, nitrogen and diethylene glycol.In another significant achievement, two bacterial strains (BQ1 and BQ8) of Alicycliphilus cultured in polyester polyurethane showed exclusively esterase activity in their culture supernatant with polyester polyurethane as sole carbon source.57 The authors emphasized that the first strain, BQ8 showed 25% more esterase activity than BQ1 strain at 18 h of culture when p-nitrophenyl acetate was used as substrate for the enzymatic reaction. The capacity of Alicycliphilus sp. to degrade PU was demonstrated by changes in the PU IR spectrum and by the numerous holes produced in solid PU observed by scanning electron microscopy after bacterial culture. Signals in the regions for ether, methyl, methylene and terminal alcohols (93–1450 cm−1) suggested the hydrolysis of the ester region of the polyurethane. Also the increase of the amide signal (1530–1550 cm−1) discarded the possibility of the action of protease or urease enzymes.
Jiang et al. in 2007, synthesized a new family of water borne polyurethanes (WBPU) using isophorone diisocyanate (IPDI), polycaprolactone (PCL), polyethylene glycol (PEG) and BD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lysine (1
lysine (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) as the chain extender. The polyurethane was then enzymatically degraded in PBS (pH = 7.4) with a solution mixture including PBS 60.0 mL, 0.1% MgCl2 15.0 mL and Lipase AK (10 mg mL−1) 15.0 mL and then incubated with shaking for certain time at 55 °C, which was the optimum temperature for enzyme activities of Lipase AK.58 An increased degradation was observed on decreasing the amount of PEG in soft segments of WBPU, as judged from the change of tensile properties with time, owing to Lipase AK only interacting with PCL soft segments in these polymers structures. This result reveals that the degradation rate is proportional to the PCL content, and inverse proportion to the PEG content in the WBPUs.
1) as the chain extender. The polyurethane was then enzymatically degraded in PBS (pH = 7.4) with a solution mixture including PBS 60.0 mL, 0.1% MgCl2 15.0 mL and Lipase AK (10 mg mL−1) 15.0 mL and then incubated with shaking for certain time at 55 °C, which was the optimum temperature for enzyme activities of Lipase AK.58 An increased degradation was observed on decreasing the amount of PEG in soft segments of WBPU, as judged from the change of tensile properties with time, owing to Lipase AK only interacting with PCL soft segments in these polymers structures. This result reveals that the degradation rate is proportional to the PCL content, and inverse proportion to the PEG content in the WBPUs.
Adhikari and Sarkar demonstrated the preparation and biodegradation studies of lactic acid and polyethylene glycol based polyester urethanes under soil burial conditions and by cultural bacteria at different temperatures. They found that after 30 days of exposure of the polyester urethane films to cultured Pseudomonas aeruginosa, around 33–36% degradation in terms of weight loss was observed. Also the rate of degradation in cultured bacteria is faster than that of soil burial conditions.59
Later on, Shah et al. in 2008 reported the isolation of bacteria from soil with the ability to degrade plastic polyurethane. The authors described that the bacterial strains attached on the polyurethane film, after soil burial for six months, were identified as Bacillus sp. AF8, Pseudomonas sp. AF9, Micrococcus sp. AF10, Arthrobacter sp. AF11 and Corynebacterium sp. AF12. SEM and FT IR showed certain changes on the surface of PU film and formation of new intermediate products after polymer breakdown.60
A polyurethane was synthesized with lysine diisocyanate (LDI), polycaprolactone (PCL), and 1,4-butanediol (BD) in the presence of dilaurate as catalyst by Han et al. (Han et al., 2009) and then degraded in phosphate buffer solution (PBS) with a solution mixture including 4.0 mL PBS, 1.0 mL 0.1 wt.% MgCl2 and 1.0 mL Lipase AK (10 mg mL−1) in water at 50 °C. It was found that mass loss decreased with increasing the PCL soft segment content in hydrolytic degradation in PBS. Because PCL is hydrophobic in comparison with the polar hard segment, increasing its content would decrease water uptake of PU films, and then decrease mass loss. In contrast, in the presence of Lipase AK the mass loss was observed to be increased with increasing the PCL soft segment content.61
Polyurethane diol (PU-diol), a synthetic polymer, is widely used as a modifier for water-soluble resins and emulsions in wood appliances and auto coatings. Mukherjee et al., isolated a soil bacterium that can survive using PU-diol as sole carbon source. The ribotyping and metabolic fingerprinting analysis showed that this organism is a strain of Pseudomonous aeruginosa (P. aeruginosa). It has also been observed that this strain is able to degrade Impranil DLN™, a variety of commercially available PU.62
Khan in 2011, isolated the bacterial stains having the ability to utilize PU as a sole carbon source after soil burial through enrichment in liquid medium. Maximum activity of the enzymes (lipases and esterases) was observed at 37 °C, pH 9 and in the presence of 5% glucose as an additional carbon source.63 Later on, a polyester polyurethane (PU) degrading bacteria, Pseudomonas aeruginosa designated as strain MZA-85 (ref. 64) and Bacillus subtilis designated as strain MZA-75,65 having the potential to reduce PU-related waste burden, were isolated from soil through enrichment. The degradation of PU film pieces by P. aeruginosa strain MZA-85 was investigated by scanning electron microscopy (SEM), Fourier transformed infra-red spectroscopy (FT-IR) and gel permeation chromatography (GPC). SEM micrographs of PU film pieces, treated with strain MZA-85, revealed changes in the surface morphology. FTIR spectrum showed increase in organic acid functionality and corresponding decrease in ester functional group. GPC results revealed increase in polydispersity, which shows that long chains of polyurethane polymer are cleaved into shorter chains by microbial action.
Estenoz and coworkers,66 studied the biodegradation of PU foams based on castor oil modified with maleic anhydride (MACO) by Pseudomonas sp. strain (DBFIQ-P36). During investigation it was found that the materials with high MACO content exhibited a considerable increase of the degradation rate associated to the hydrophilicity of the polymeric structures due to the presence of ester groups and to the effect of amide groups on the hard segment.
Several reports revealed that for most bacteria, the use of polyurethane as a carbon and nitrogen source abolishes or decreases the level of degradation.67 Therefore, nitrogen sources, generally supplied by yeast extracts, were added to the medium to sustain polyurethane degradation by bacteria.68 A soil microorganism, designated as P7 and identified as Acinetobacter gerneri, was characterized and investigated for its ability to degrade polyurethane (PU) by Howard et al.40 The ability of this organism to degrade polyurethane was characterized by the measurement of growth, SEM observation, measurement of electrophoretic mobility and the purification and characterization of a polyurethane degrading enzyme. The purified protein has a molecular weight of approximately 66 kDa as determined by SDS-PAGE.
In another significant development, three novel PU degrading bacteria were isolated from farm soils and activated sludge. Their identities were determined by 16S ribosomal RNA gene sequence blast and the robust activity was observed in Pseudomonas putida.69 It spent 4 days to degrade 92% of Impranil DLN™ for supporting its growth. The optimum temperature and pH for DLN removal by P. putida were 25 °C and 8.4, respectively. It is pertinent to mention here that the polyurethanolytic activities were presented both in the extracellular fraction and in the cytosol. Later on, Biffinger and co workers developed a quantitative assay for the direct measurement of polymer film degradation from bacterial colonies on agar plates. Small (1 mm diameter) colonies of Pseudomonas protegens Pf-5 (formerly Pseudomonas fluorescens Pf-5) were used for this work. Interactions between the Pf-5 colonies and thin polyurethane (PU) coatings on ZnSe coupons were evaluated for degradation using infrared spectroscopy.70
Very recently, Nakkabi et al. studied the biodegradation of polyurethane sold under the name Impranil DLN by bacteria isolated from decayed cedar wood.71 In this study the degradation of polyurethane by Bacillus subtilis has been chemically demonstrated by infrared spectroscopy. The bacterium bacillus subtilis was added to the media 0.3% and 0.6% of polyurethane. A progressive reduction in the relative intensity of the peak at 1730 cm−1 was observed. By the time the culture has become visually transparent, there was a complete loss of the absorbance peak at 1735 cm−1. The loss of this peak is consistent with hydrolysis of the ester bond in the urethane linkage. Some bacterial species reported as polyurethane degraders are summarized in Table 1.
| S. no. | Bacterial species | Source | Ref. |
|---|---|---|---|
| 1 | Acinetobacter gerneri P7 | Soil, USA | Howard et al., 2012 (ref. 40) |
| 2 | Acinetobacter calcoacetics | Oil contaminated soil, USA | El-Sayed et al., 1996 (ref. 72) |
| 3 | Arthrobacter globiformis | ||
| 4 | Aeromonas salmonicida | Soil buried PU samples | Kay et al., 1991 (ref. 49) |
| 5 | Alcaligenes denitrificans | ||
| 6 | Alicycliphilus spp. | Decomposed soft foam | Oceguera-Cervantes et al., 2007 (ref. 73) |
| 7 | Alicycliphilus denitrificans | ||
| 8 | Bacillus pumilus | PU contaminated water from industrial waste sites | Nair & Kumar 2007 (ref. 54) |
| 9 | Bacillus subtilis | Decayed cedar wood | Nakkabi et al., 2015 (ref. 71) |
| 10 | Comamonas acidovorans | Soil Japan | Nakajima-Kambe et al., 1995 (ref. 55) |
| 11 | Corynebacterium sp. | Soil buried PU samples Activated sludge | Shah et al., 2008 (ref. 22) |
| 12 | Enterobacter agglomerans | Soil buried PU samples | Kay et al., 1991 (ref. 49) |
| 13 | Methanotrix sp. | Degraded PU foam | Varesche et al., 1997 (ref. 74) |
| 14 | Micrococcus sp. | Activated sludge, Pakistan | Shah et al., 2008 (ref. 22) |
| 15 | Pseudomonas aeruginosa | Soil buried PU samples | Kay et al., 1991 (ref. 49) |
| 16 | P. Aeruginosa strain MZA-85 | Soil samples | Shah et al., 2013 (ref. 64) |
| 17 | Pseudomonas chlororaphis | Soil, USA | Howard and Blake 1998 (ref. 51) |
| 18 | Pseudomonas fluorescens | Soil, USA | Howard and Hilliard 1999 (ref. 52) |
| 19 | Pseudomonas putida | Activated sludge, Taiwan | Peng et al., 2014 (ref. 69) |
| 20 | Rhodococcus equi | Soil, Japan | Akutsu-Shigeno et al., 2006 (ref. 34) |
| 21 | Serratia rubidaea | Soil buried PU samples | Kay et al., 1991 (ref. 49) |
Genetic and biochemical research have also been performed on polyurethane degraders. Recombinant esterases, proteases, and ureases could digest PU through cleaving the ester or peptide bonds.75 Purified PUases presented either protease or esterase activity and would be blocked by serine hydrolase inhibitor, soybean trypsin inhibitor, or bivalent cation chelator. Therefore, the putative PUases did not restrict to a single type of enzyme. The functions of PUases were illustrated only when the responsible genes were identified. The genetics of the polyurethane biodegradation pathways have been characterised for a few relevant bacteria, including P. fluorescens, P. chlororaphis ATCC 55729, Bacillus subtilis, and D. acidovorans TB-35. So far, only four genes encoding PUases have been cloned and characterized from environmental microorganisms: Pul from P. fluorescens,76 PueA and PueB from P. chlororaphis,39,77 and PudA from C. acidovorans TB-35.55
Two genes encoding polyurethanase activity from P. chlororaphis have been cloned in E. coli.38,39 Both genes were expressed in E. coli. The PueA enzyme was secreted in the recombinant E. coli and displayed a beta-zone of clearing on polyurethane agar plates while PueB was not secreted in the recombinant E. coli and displayed an alpha-zone of clearing of polyurethane agar plates. Both enzymes are temperature stable up to 100 °C. In addition, PueB has been noted to display esterase activity towards p-nitrophenylacetate, p-nitrophenylpropionate, p-nitrophenylbutyrate, p-nitrophenylcaproate and p-nitrophenylcaprylate while PueA has been reported to display esterase activity only towards p-nitrophenylacetate and p-nitrophenylpropionate.
Later on, in a significant development, Howard and co workers identified a gene cluster containing seven open reading frames resembling a binding-protein-dependent ATP binding cassette (ABC) transport system in Pseudomonas chlororaphis in connection with PueA and PueB, which are involved in polyurethane degradation. The authors created kanamycin insertion mutations of PueA and PueB to study growth of the P. chlororaphis mutants on polyurethane. Immunodetection revealed that the PueA and PueB proteins are present in the wild-type P. chlororaphis and that the PueA protein was inactivated in the PueA mutant P. chlororaphis and the PueB protein was inactivated in the PueB mutant P. chlororaphis. Thus, growth studies were performed to compare the effects of the PueA-deficient strain and the PueB deficient strain with the wild type strain in polyurethane utilization. Analysis from the creation of knock-out mutants in PueA and PueB suggests that degradation of polyurethane by P. chlororaphis may be more dependent on PueA than on PueB.78
Comamonas acidovorans TB-35, which had been isolated as a solid PUR-degrading bacterium,55 was found to produce two kinds of esterases: one is secreted to the culture broth (CBS esterase)79 and the other is bound to the cell surface (PUR esterase).37 However only the cell bound esterase (PUR esterase) was shown to be able to degrade PUR. This enzyme, which is a kind of esterase, degraded solid polyester PUR, with diethylene glycol and adipic acid released as the degradation products. The optimum pH for this enzyme was 6.5, and the optimum temperature was 45 °C. The structural gene (Pud A) which encodes PUR esterase has also been cloned in Escherichia coli and its primary structure has also been analyzed by Nomura et al.80 The amino acid sequence of PudA revealed no significant homology to sequences of PHA [poly(3-hydroxyalkanoate)] depolymerases except within the PHA surface-binding domain. The open reading frame (ORF) consists of 1644 base pairs with a putative ATG initiation codon, and encodes a 548 amino acid enzymes. The recombinant protein expressed in E. coli can degrade solid polyurethane. The amino acid sequence of this enzyme shows only about 30% homology to the acetylcholine esterase from Torpedo californica (T AChE)81 and the lipase from Geotrichum candidum (GcL1)82 respectively. From computerized searches of databases, it was shown that PudA possessed a high degree of homology with the T AChE/GcL1 serine hydrolase family only with the catalytic regions of the serine hydrolase family proteins which contain the Ser-His-Glu catalytic triad with a glutamate residue replacing the usual aspartate residue. Comparison of the positions of each residue in the Ser-His-Glu catalytic triad reveals that the amino acid residues for PudA are similar with T AChE, GcL1 and human choline esterase (H ChE). This infers that prokaryotic esterases possess the Ser-His-Glu catalytic triad as the active site. To confirm Glu instead of Asp as necessary for activity in prokaryotic esterate, site directed mutagenesis was performed. Results from this have demonstrated that each residue in the Ser-His-Glu catalytic triad is in fact essential for enzymatic activity.
4.3 Biodegradation of polyurethane by fungi
Several reports have appeared in the literature on the susceptibility of PUs to fungal attack.83 These reports revealed that polyester type PUs in particular are known to be vulnerable to microbial attack than other forms, as they contain ester and urethane linkages that are naturally vulnerable to enzymatic degradation.84 Polyurethane degradation activity was found mainly in the Ascomycota phylum for fungi in the genera Aspergillus, Pestalotiopsis and Gliocladium.85 Many fungi isolated from the soil have shown potential for the biodegradation of plastics, including polyurethane foams.86 Gliocladium roseum was isolated from polyester PU buried for 21 days in soil,87 whilst a number of isolates from the genera Aspergillus, Emericella, Fusarium, Penicillium, Trichoderma and Gliocladium were recovered from the surface of polyester PU foam buried for 28 days.88 Crabbe et al. (1994) isolated four fungi, Curvularia senegalensis, Fusarium solani, Aureobasidium pullulans and Cladosporium sp.89 Filamentous fungi are known for their ability to degrade many organic substances. These abilities depend on efficient colonisation of the substrate and the secretion of numerous enzymes.90Barratt and co-workers investigated the relationship between soil water holding capacity (WHC) and biodegradation of polyester polyurethane.91 The authors demonstrated that the PU degradation is dependent on the soil water holding capacity as the tensile strength of polyester PU was reduced by up to 60% over 20–80% soil WHC but no reduction occurred at 15, 90 or 100% soil WHC. Moreover, three morphologically distinct fungal colony types capable of degrading polyester polyurethane agar were identified as viz Geomyces pannorum (peach colonies), Nectria gliocladioides (white colonies) and Penicillium ochrochloron (green colonies).
In 2007, Cosgrove et al., reported on involvement of soil fungal communities in the biodegradation of polyester polyurethane. Fungal communities on the surface of the PU were compared to the native soil communities using culture based and molecular techniques. Putative PU degrading fungi were common in both the soils, as <45% of the fungal colonies cleared the colloidal PU dispersion Imranil on solid medium. Denaturing gradient cell electrophoresis revealed that fungal communities associated with PU coupons were less diverse than in the soil, and only a few species in the PU communities were detectable in the soil indicating that only a small sub-set of the soil fungal communities colonized the PU. Geomyces pannorum and a Phoma sp. were the dominant species recovered by culturing from the PU buried in the acidic and neutral soils respectively.46 The effect of biostimulation and bioaugmentation of soil microorganisms on degradation of PU was also investigated and the results showed that biostimulation with yeast extract alone or in conjunction with Impranil causes a 62% increase in PU degradation compared to the degradation in untreated control soil and was associated with 45% increase in putative PU degraders colonizing PU. Further bioaugmentation with Nectria haematococca, Penicillium viridicatum, Penicillium ochrochloron, or an unidentified Mucormycotina sp. increased PU degradation a further 30 to 70%, suggesting that biostimulation and bioaugmentation were operating in concert to enhance PU degradation.92
Fungus Chaetomium globosum are known for the biodegradation of PEG/castor oil-based polyurethane which is mostly used as biomaterial and is synthesized by using polyethylene glycol 1500 (PEG1500) as chain extender in the hard segment and castor oil in the soft segment of the polymer.93 It was found that the sorption of water and degradability of polyurethane is directly proportional to the amount of PEG1500 in the hard segment of the polyurethane. Morphological changes of the polyurethane surface films were determined by SEM, after washing with distilled water and displayed in Fig. 7.
 | ||
| Fig. 7 Scanning electron micrographs of polyurethane films before and after being exposed to rot fungus Chaetomium globosum (after washing the exposed surface). | ||
Various other fungal strains such as Aspergillus flavus,24 Aspergillus fumigates,87 Aspergillus terreus,94 and Fusarium solani95 were reported from time to time by different workers as biodegraders of polyurethane. Oprea and Doroftei studied the biodegradation features of a novel blend of polyurethane acrylate–acrylated epoxidized soybean oil-based cross-linked polyurethane elastomers in the presence of the soft-rot fungus Chaetomium globosum. The biodegradation results show that samples with a high content of acrylated epoxidized soybean oil are more biodegradable than mere polyurethane acrylate.96
In studies conducted by Ibrahim and coworkers,97 two novel fungal strains such as Alternaria solani and Spicaria sp. were reported for the degradation of polyester-polyurethane (PS PU). Strobel and co workers screened several dozen endophytic fungi for the degradation of synthetic polymer polyester polyurethane (PU).98 They hypothesized that one of the serine hydrolases secreted by Pestalotiopsis microspora was the enzyme behind the degradation of polyester PU. Capable to grow in an anaerobic environment, this Amazon fungus relied on polyester PU as a sole carbon source by cleaving ester bonds in the PU substrate. Mathur and Prasad in 2012 reported the polyurethanolytic activity of Aspergillus flavus (ITCC 6051) isolated from the soil and they suggested that the esterase detected in its extracellular fluid may be the reason behind its activity; a 60.6% ± 0.3% reduction in the weight of the PU was noted.99 In a study conducted on 22 fungal strains capable of growing using PU as a carbon source, Aguilar et al. suggested that in all the strains the most common enzymatic activity related with PU biodegradation was the protease activity.100
Wong and Ma revealed that different polyurethane products may require varying esterase for biodegradation since the esterase from Aspergillus flavus is able to breakdown water based PU but not TPU (a granular form of thermoplastic PU).101 This suggests that either this particular strain was simply unable to produce esterase on certain types of substrates, or that perhaps more than one type esterase is needed for the degradation of various types of polyester polyurethane.
Zhang et al. reported the hydrolysis of the ester and urethane bonds in PUs made from liquefied wood-based polyols.102 In another significant development, Robson et al. reported that a number of fungal isolates, including thermotolerant and thermophilic fungi, are able to degrade Impranil (liquid dispersion of PU).103 The most dominant fungi identified from the surfaces of PU coupons by pyrosequencing was Fusarium solani at 25 °C, while at both 45 °C and 50 °C, Candida ethanolica was the dominant species. In this study, they demonstrated the potential of the composting process to deteriorate PU by comparing the rate of biodegradation when buried in compost at different temperatures representing the mesophilic and thermophilic stages. The effect of compost burial on the surface of PU coupons visualized by environmental scanning electron microscopy is depicted in Fig. 8. Later on, the same group investigated the biodegradation of polyester polyurethane during the maturation stage of a commercial composting.104 Fungal communities colonising polyester PU coupons were compared with the native compost communities using culture based and molecular techniques. Studies suggested that putative polyester PU degrading fungi were ubiquitous in compost and rapidly colonized the surface of polyester PU coupons with significant deterioration. Also the rate of degradation is enhanced under thermophilic and early maturation stage of commercial composting and that thermophilic and thermotolerant fungi have the capacity to cause significant polyester PU degradation.
While significant achievements in the production of PUs from bio-based polyols have been made,105 there are still numerous uncertainties about the interaction of these polymers with the environment. Li, Michel Jr. and coworkers compared the relative biodegradation of PUs produced from petroleum- and crude glycerol-based polyols under composting, anaerobic digestion and soil conditions.106 They observed that PU foams made from 100% crude glycerol-based polyols were mineralized during 320 days of soil incubation at rates faster than those observed for the petroleum-based analogs. However, no significant differences in soil mineralization rates were observed between PU foams made from blend polyols, which contained 50% crude glycerol based polyols and 100% petroleum-based polyols.
Adhikari and Basak reported the biodegradation of the polyester urethane (PEU) in simulated body fluid (SBF) at 37 °C for an extended period of time. Moreover the effect of soft segment to hard segment ratio on the degradation rate of PEU matrix has also been assessed.107
Barreiro and co workers investigated the biodegradation of rigid polyurethane (RPU) foams synthesized from lignin based polyols. The lignin-based polyols used here were obtained by oxypropylation of four distinct lignins (Alcell, Indulin AT, Curan 27-11P, and Sarkanda). A 100% commercial polyol-based (Lupranol® 3323) RPU foam was also prepared and used as the reference. Studies revealed that biodegradation seems to be, particularly, favored if using Indulin AT-based polyols mixed with Lupranol® 3323.108
Recently Kamini et al., explored the utilization of fish meal and fish oil for the production of lipase from Cryptococcus sp. MTCC 5455. The authors disclosed that the crude concentrated enzyme hydrolyzed polyurethane efficiently and the hydrolysis was 94% at 30 °C in 96 h. SEM studies of the degraded polymer showed significant increase in size of the holes from 24 to 72 h of incubation.109
Although traditional biochemical enzymatic degradation of polyurethane appeared relatively inefficient, the development of new, specific arsenal in a selected organism will offer alternative methods for the production of side compounds with commercial value while degrading the pollutant. Shibasaki et al. in 2009, investigated the biodegradability of polyurethanes containing dulcitol units that involves the use of CALB-displaying “arming yeast”. The authors developed a strain of S. cerevisiae molecularly engineered to dispose of lipases localised on its whole cell membrane to degrade polyester polyurethane and this was the first report to illustrate this possibility.110 Moreover, the arming yeasts were applicable to evaluate the degradation of the film state of polyurethane. This S. cerevisiae issued from engineering methods employing yeast-arming technologies initiates that new degradation strategies can be developed based on the use of strains designed with a defined genetic arsenal against polyurethanes. Some fungal species reported as PU degraders are summarized in Table 2.
| S. no. | Fungal species | Source | References |
|---|---|---|---|
| 1 | Acremonium sp. | Degraded PU cable in marine environment | Stranger-Johannessen, 1985 (ref. 111) |
| 2 | Aspergillus sp. | ||
| 3 | Alternaria alternata | Soil, Germany | Pommer and Lorenz, 1985 (ref. 112) |
| 4 | Alternaria dauci | Ecuadorian rainforest | Russell et al., 2011 (ref. 98) |
| 5 | Alternaria spp. | ||
| 6 | Alternaria sp. strain 18-2 | Garden soil, Manchester, UK | Cosgrove et al., 2007 (ref. 46) |
| 7 | Alternaria solani | Soil, Jordan | Ibrahim et al., 2009 (ref. 113) |
| 8 | Aspergillus flavus | Soil from waste disposal site, India | Mathur and Prasad, 2012 (ref. 99) |
| 9 | Aspergillus fischeri | Soil John Innes no. 2 compost | Bentham et al., 1987 (ref. 88) |
| 10 | Aspergillus fumigatus | Soil, Birmingham, UK | Pathirana and Seal, 1984 (ref. 84) |
| 11 | Aspergillus niger | Not specified | Amaral et al., 2012 (ref. 114) |
| 12 | Aspergillus terreus | Not specified | Wales and Sagar, 1985 (ref. 94) |
| 13 | Aspergillus versicolor | Not specified | Darby & Kaplan, 1968 (ref. 24) |
| 14 | Aureobasidium pullulans | Soil, Washington DC | Crabbe et al., 1994 (ref. 89) |
| 15 | Bionectria spp. | Ecuadorian rainforest | Russell et al., 2011 (ref. 98) |
| 16 | Cladosporium sp. | Garden soil, Washington DC | Crabbe et al., 1994 (ref. 89) |
| 17 | Curvularia senegalensis | ||
| 17 | Chaetomium globosum | Not specified | Darby & Kaplan, 1968 (ref. 24) |
| 18 | Cryptococcus sp. MTCC | Air, India | Kamini et al., 2015 (ref. 109) |
| 19 | Cylindrocladiella parva | Garden soil, Manchester, UK | Cosgrove et al., 2007 (ref. 46) |
| 20 | Exophiala jeanselmei REN-11A | Soil | Owen et al., 1995 |
| 21 | Edenia gomezpompae | Ecuadorian Amazonian rainforest | Russell et al., 2011 (ref. 98) |
| 22 | Fusarium solani | Garden soil, Washington DC USA | Crabbe et al., 1994 (ref. 89) |
| 23 | Geomyces pannorum | Soil John Innes no. 2 compost | Barratt et al., 2003 (ref. 91) |
| 24 | Gliocladium roseum | Soil, Birmingham, UK | Pathirana and Seal, 1984 (ref. 84) |
| 25 | Lasiodiplodia sp. E2611A | Ecuadorian rainforest | Russell et al., 2011 (ref. 98) |
| 26 | Nectria spp. | Garden soil, Manchester, UK | Cosgrove et al., 2007 (ref. 46) |
| Ecuadorian rainforest | Russell et al., 2011 (ref. 98) | ||
| 27 | Nectria gliocladiodes | Soil John Innes no. 2 compost | Barratt et al., 2003 (ref. 91) |
| 28 | Neonectria ramulariae | Garden soil, Manchester, UK | Cosgrove et al., 2007 (ref. 46) |
| 29 | Penicillium inflatum | ||
| 30 | Penicillium notatum | ||
| 31 | Penicillium viridicatum | ||
| 32 | Phoma fimenti | Not specified | Pommer and Lorenz, 1985 (ref. 112) |
| 33 | Pestalotiopsis microspora | Ecuadorian Amazonian rainforest | Russell et al., 2011 (ref. 98) |
| 34 | Pestalotiopsis sp. | ||
| 35 | Phaeosphaeria spp. | ||
| 36 | Plectosphaerella spp. | ||
| 37 | Pleosporales spp. | ||
| 38 | Rhizopus stolonifer | Not specified | Wales and Sagar, 1985 (ref. 94) |
| 39 | Scopulariopsis brevicaulis | Soil, Birmingham, UK | Pathirana and Seal, 1984 (ref. 84) |
| 40 | Talaromyces spp | Not specified | Pommer and Lorenz, 1985 (ref. 112) |
| 41 | Trichoderma viride | Soil, Birmingham, UK | Pathirana and Seal, 1984 (ref. 84) |
4.4 Degradation of polyurethanes by enzymes derived from animal/botanical origins
The effect of proteolytic enzymes on the degradation has been studied for amino acid based polyurethanes. Phenyl alanine based polyurethanes have shown α-chymotrypsin mediated degradation in vitro.115,116 These studies show that the presence of specific sites, e.g. hydrophobic aromatic side chains enhances the tendency toward enzymatic degradation of these PUs. Sarkar et al., in 2007 studied the enzymatic degradation of L-tyrosine based PU using proteolytic enzyme α-chymotrypsin in phosphate buffer solution (pH 7.4) at 37 °C. The authors disclosed that the presence of an amino acid moiety in the polymer structure enhances the degradability of the PU in the presence of enzyme. The mechanistic pathway for enzymatic degradation shows that L-tyrosine based PU is degraded initially by surface erosion which is followed by bulk degradation.10Yamamoto et al. degraded with different thiol proteases (papain, bromelain, and ficin), protease K and chymotrypsin, lysine diisocyanate (LDI) based poly(urethanes) and segmented poly(urethane ureas).117 For this, 1 mg of enzyme was added into the test tube coated with the polymer at 37 °C and the total organic carbon (TOC) measured. From 1H NMR results, it was evident that the pendant methyl ester group in LDI was rapidly hydrolyzed, followed by slow hydrolysis of urethane bonds in the backbone chain while the susceptibility of urea bonds to papain was very low. Before 50 h almost 30% of the PU has been degraded, with ethylene glycol exhibiting the highest rate of degradation; thiol proteases were most effective for all segmented poly(urethane urea)s.
Biodegradable polyurethanes were prepared by Wang et al., using poly(lactic acid) (PLA)–PEG–PLA as soft segment, and L-lysine ethyl ester diisocyanate (LDI) and 1,4-butanediol (BD) as rigid segment.118 These polymers were degraded in phosphate buffer solution(PBS) (0.1 M PBS with 0.9% NaCl and 0.02% NaN3, pH 7.4, 6 and 5) and enzymatic (0.1 mg mL−1 lipase from porcine pancreas in 0.1 M PBS with 0.9% NaCl and 0.02% NaN3, pH 7.4) solutions at 37 °C to simulate in vivo dynamic tissue environment. PU samples demonstrated rapid degradation in 96 h (more than 90%) which might be attributed to hydrophilicity of PEG segments, low number-average molecular weight and microphase separation degree of these polyurethanes and enzyme functions. The enzymatic degradation rate was higher than hydrolytic degradation rate, verifying that lipase from porcine pancreas can accelerate hydrolysis on these polyurethanes.
The use of enzyme cocktail solutions in the biodegradation of polyurethanes was first reported by Ozsagiroglu et al. in 2012. Esterase, protease DSM and Pellucit FS enzymes were mixed for making different enzyme cocktails. Moreover, the effects of enzyme types and its mixtures on PU degradation were also studied. It was found that protease type enzymes could erode polymer films and were more effective than esterase enzymes. In addition enzyme cocktail solutions showed that enzymes could compete with each other and one enzyme could suppress the activity of another enzyme. Thus ternary enzyme cocktail solutions did not accelerate degradation rates of PU chains because of competition whereas binary enzyme solutions comprising of protease DSM and pellucit FS were more effective.119
Kang et al., isolated a novel thermostable esterase, estCS2, which belongs to family VII, from a compost metagenome library. EstCS2 has high stability in organic solvents; it can degrade polyurethane which is a hydrophobic synthetic polymer. EstCS2 formed a clear zone on an indicator plate containing poly(diethylene glycol adipate), which is one of components of polyester polyurethane.120
5. Microbial degradation of polyether urethanes
As mentioned above lot of reports are there in the literature for the microbial degradation of polyester polyurethanes since ester bonds in the structure are susceptible to microbial attack. Ether-type PU (ether-PU) was developed in late 1950s as a durable plastic. Ether-PU (–R1–NHCOO–(CH2)m–O–(CH2)m–O–R2–) is widely used in automobile interiors, building insulation and home electronics.121 Poly(tetramethylene oxide) (PTMO) is the most common polyether in conventional medical formulations (Silverstri et al., 2011).122 Thus, for example, the Pellethane® 2363 80A and Elasthane™ 80A are poly(ether-urethane)s obtained by the reaction of PTMO, MDI and BD monomers; Tecoflex® by Thermedics is also a poly(ether-urethane) synthesized by the reaction of PTMO, HMDI and BD monomers.The production and utilization of ether-PU are increasing, making landfill disposal and incineration treatment of ether-PU into a serious problem all over the world.123 Polyether-based polyurethanes (PBP) are extremely problematic polymers in the environment due to their unique corrosion resistance, hydrolysis resistance, resistance to bending and good adhesion. Now, new recycling and disposal systems of ether-PU are desired. The degradation of polyurethanes by candidate enzymes has been the subject of numerous studies over the past decade. However very limited reports are there in the literature about microbial degradation of ether type polyurethanes.
In some reports, efforts were made to isolate ether type polyurethane-degrading micro-organisms from various environmental sources and to identify and characterize the isolated ether-PU-degrading micro-organism.41 In vitro degradation of poly(ether urethanes) (PEU) and poly(carbonate urethanes) (PCU) by hydrolytic enzymes was reported in several studies.124 These studies identified the hydrolytic enzyme, cholesterol esterase (CE), as the most active enzyme in polyurethane degradation.125 Labow et al. reported that cholesterol esterase cleaved polyetherurethanes at the most probable site susceptible to hydrolytic cleavage, which is the urethane bonds, resulting in the release of free amine.126 However, Hiltner and co workers examined the effect of cholesterol esterase on the degradation of commercial poly(ether urethane) and poly(carbonate urethane), and compared the results with in vivo degradation.127 Although the study used a concentration of cholesterol esterase that was considerably higher than the estimated physiological level in order to accelerate the effect, only a small weight loss and a small loss in surface soft segment content by ATR-FTIR were observed after 36 days. It appeared that any action of CE was confined to the immediate surface, and the magnitude of the effect was too small to account for the changes observed on implanted films. Degradation processes initiated by CE did not penetrate into the bulk and cause deterioration of bulk properties as is observed with oxidation. It was concluded that in comparison to oxidation, hydrolytic enzymatic degradation of PEU and PCU in vivo is negligible.
Later on, a fungus, identified as Alternaria sp. and designated as strain PUDK2, capable of changing the configuration of ether-PU, was isolated. The enzyme(s) from PUDK2 degraded urethane and urea bonds to convert the high molecular weight structure of ether-PU to small molecules; and then the fungus seems to use the small molecules as an energy source.128 Obruca and coworkers investigated the degradation process of polyether–polyol-based polyurethane (PU) elastomeric films in the presence of a mixed thermophilic culture as a model of a natural bacterial consortium. The authors suggested that the modification of PU by proper biopolymers is a promising strategy for reducing potential negative effects of waste PU materials on the environment and enhancing their biodegradability.129
6. Conclusion
Polyurethane is the most widely used polymer in the world. However, the ecological problems related to the environmental pollution by these types of synthetic polymers are one of the major concerns of the present days; especially because they are difficult to degrade easily and the entire process is time consuming. Hence, under such circumstances degradation of plastic by microbes is one of the eco-friendly and innovative methods. This review article has covered the microbial degradation of ester and ether type polyurethanes reported in the last one and half decade. Another area that was examined is the importance of polyurethanes in day today life. This article may give brief information regarding the nature and biodegradation of polyurethanes. It is expected that this work will encourage researchers to find out one or more microbial strain(s) from nature for the potential biodegradation of polyurethanes since a diverse group of microorganisms including bacteria and fungi capable of PU degradation can be isolated from the nature. Also learning more about the pathways for degradation and the genes involved in PU degradation is essential in developing either recombinant derivatives or enriching for indigenous PU-degrading microorganisms for bioremediation.Acknowledgements
Authors are highly thankful to Principal, Govt Degree College Kathua and Higher Education Department, Jammu and Kashmir, India for their support.References
- J. M. Cangemi, A. Dos Santos, S. C. Neto and G. O. Chierice, Polym. Sci. Technol., 2008, 18, 201–206 CAS
.
- PlasticsEurope, Plastics – the Facts 2011 – an analysis of European plastics production, demand and recovery for 2010, Ave E. van Nieuwenhuyse 4/3, Bruxelles, Belgium: Association of Plastics Manufacturers, 2011.
-
(a) C. R. Hegedus, D. F. Pulley, S. J. Spadafora, A. T. Eng and D. J. Hirst, J. Coat. Technol., 1989, 61, 31–42 CAS
; (b) M. S. Reisch, Chem. Eng. News, 1990, 17, 39–68 CrossRef PubMed
.
- H. Ulrich, Polyurethane, in Modern Plastics Encyclopedia, McGraw-Hill, New York, 1983, vol. 60, pp. 76–84 Search PubMed
.
- O. Bayer, Polyurethanes, Mod. Plast., 1947, 24, 149–152 CAS
.
- J. H. Saunders and K. C. Frisch, Polyurethanes: Chemistry and Technology, Part II: Technology, Interscience Publishers, New York, 1964 Search PubMed
.
- J. P. Santerra, K. Woodhouse, G. Laroche and R. S. Labow, Biomaterials, 2005, 26, 7457–7470 CrossRef PubMed
.
- T. Thomson, in Polyurethanes as Specialty Chemicals: Principles and Applications, CRC Press, Boca Raton, FL, 2004 Search PubMed
.
- A. Marcos-Fernández, G. A. Abraham, J. L. Valentín and J. S. Roman, Polymer, 2006, 47(3), 785–798 CrossRef PubMed
.
- D. Sarkar and S. T. Lopina, Polym. Degrad. Stab., 2007, 92(11), 1994–2004 CrossRef CAS PubMed
.
- R. E. Vega, T. Main and G. T. Howard, Int. Biodeterior. Biodegrad., 1999, 43, 49–55 CrossRef CAS
.
- S. Lal, S. Kumar, M. Kumar and S. Arora, Arch. Appl. Sci. Res., 2011, 3(4), 309–318 CAS
.
- M. Urgun-Demirtas, D. Singh and K. Pagilla, Polym. Degrad. Stab., 2007, 92, 1599–1610 CrossRef CAS PubMed
.
- T. Ishigaki, W. Sugano, A. Nakanishi, M. Tateda and M. Ike, Chemosphere, 2004, 54, 225–233 CrossRef CAS
.
- J. Hao, J. Wang, W. Zhao, L. Ding, E. Gao and W. Yuan, Department of Epidemiology, School of Public Health, Shanxi Medical University, Taiyuan, China, Wei Sheng Yan Jiu, 2011, 40(3), 312–319.
-
(a) T. Ohkita and S. H. Lee, J. Appl. Polym. Sci., 2006, 100, 3009–3017 CrossRef CAS PubMed
; (b) N. B. Vogt and E. A. Kleppe, Polym. Degrad. Stab., 2009, 94(4), 659–663 CrossRef CAS PubMed
; (c) J. M. Cervantes-Uc, J. I. M. Espinosa, J. V. Cauich-Rodriguez, A. Avila-Ortega, H. Vazquez-Torres, A. Marcos-Fernandez and J. San Roman, Polym. Degrad. Stab., 2009, 94(10), 1666–1677 CrossRef CAS PubMed
.
- J. D. Meeker, S. Sathyanarayana and S. H. Swan, Philos. Trans. R. Soc., B, 2009, 364, 2097–2113 CrossRef CAS PubMed
.
- A. M. Kaplan, R. T. Darby, M. Greenberger and M. R. Rodgers, Dev. Ind. Microbiol., 1968, 82, 362–371 Search PubMed
.
- G. A. Kanavel, P. A. Koons and R. E. Lauer, Rubber world, 1966, 154, 80–88 Search PubMed
.
- K. M. Zia, H. N. Bhatti and I. A. Bhatti, React. Funct. Polym., 2007, 67, 675–692 CrossRef CAS PubMed
.
-
(a) J.-M. Restrepo-Florez, A. Bassi, R. Michael and M. R. Thompson, Int. Biodeterior. Biodegrad., 2014, 88, 83–90 CrossRef CAS PubMed
; (b) A. Sivan, Curr. Opin. Biotechnol., 2011, 22, 422–426 CrossRef CAS PubMed
.
- A. A. Shah, F. Hasan, A. Hameed and S. Ahmed, Biotechnol. Adv., 2008, 26, 246–265 CrossRef CAS PubMed
.
- K. Mukherjee, P. Tribedi, A. Chowdhary, T. Ray, A. Joardar, S. Giri and A. K. Sik, Biodegradation, 2011, 22, 377–388 CrossRef CAS PubMed
.
- R. T. Darby and A. M. Kaplan, Appl. Microbiol., 1968, 16, 900–905 CAS
.
- S. J. Stachelek, I. Alferiev, H. Choi, C. W. Chan, B. Zubiate, M. Sacks, R. Composto, I. W. Chen and R. J. Levy, J. Biomed. Mater. Res., Part A, 2006, 78(4), 653–661 CrossRef PubMed
.
- P. A. Gunatillake, G. F. Meijs, E. Rizzardo, R. C. Chatelier, S. J. McCarthy, A. Brandwood and K. Schindhelm, J. Appl. Polym. Sci., 1992, 46(2), 319–328 CrossRef CAS PubMed
.
- Polymeric Biomaterials, ed. S. Dumitriu, Marcel Dekker, New York, 2nd edn, 2002 Search PubMed
.
- J. P. Santerre, K. Woodhouse, G. Laroche and R. S. Labow, Biomaterials, 2005, 26(35), 7457–7470 CrossRef CAS PubMed
.
- M. C. Tanzi, S. Fare and P. Petrini, J. Biomater. Appl., 2000, 14(4), 325–348 CrossRef CAS PubMed
.
- E. Christenson, M. Dadsetan, M. Wiggins, J. Anderson and A. Hiltner, J. Biomed. Mater. Res., Part A, 2004, 69(3), 407–416 CrossRef PubMed
.
- X. Xie, R. Wang, J. Li, L. Luo, D. Wen, Y. Zhong and C. Zhao, J. Biomed. Mater. Res., Part B, 2009, 89(1), 223–241 CrossRef PubMed
.
- E. Christenson, J. Anderson and A. Hiltner, J. Biomed. Mater. Res., Part A, 2004, 70, 245–255 CrossRef PubMed
.
- A. Volynskii, S. Bazhenov, O. Lebedeva and N. Bakeev, J. Mater. Sci., 2000, 35, 547–554 CrossRef CAS
.
- Y. Akutsu-Shigeno, Y. Adachi, C. Yamada, K. Toyoshima, N. Nomura, H. Uchiyama and T. Nakajima-Kambe, Appl. Microbiol. Biotechnol., 2006, 70, 422–429 CrossRef CAS PubMed
.
- F. Kedzierewicz, P. Thouvenot, I. Monot, M. Hoffman and P. Maincent, J. Biomed. Mater. Res., 1998, 39, 588 CrossRef CAS
.
- A. Loredo-Trevino, G. Gutierrez-Sanchez, R. Rodriguez-Herrera and C. N. Aguilar, J. Polym. Environ., 2012, 20, 258–265 CrossRef CAS
.
- Y. Akutsu-Shigeno, T. Nakajima-Kambe, N. Nomura and T. Nakahara, Appl. Environ. Microbiol., 1998, 64, 62–67 Search PubMed
.
- R. S. Stern and G. T. Howard, FEMS Microbiol. Lett., 2000, 185, 163–168 CrossRef CAS PubMed
.
- G. T. Howard, B. Crother and J. Vicknair, Int. Biodeterior. Biodegrad., 2001, 47, 141–149 CrossRef CAS
.
- G. T. Howard, W. N. Norton and T. Burks, Biodegradation, 2012, 23(4), 561–573 CrossRef CAS PubMed
.
- M. Cregut, M. Bedas, M.-J. Durand and G. Thouand, Biotechnol. Adv., 2013, 31, 1634–1647 CrossRef CAS PubMed
.
- A. E. Hafeman, K. J. Zienkiewicz, A. L. Zachman, H. J. Sung, L. B. Nanney, J. M. Davidson and S. A. Guelcher, Biomaterials, 2011, 32(2), 419–429 CrossRef CAS PubMed
.
-
(a) J. Guan, K. L. Fujimoto, M. S. Sacks and W. R. Wagner, Biomaterials, 2005, 26(18), 3961–3971 CrossRef CAS PubMed
; (b) A. B. Saim, Y. Cao, Y. Weng, C. N. Chang, M. A. Vacanti and C. A. Vacanti, Laryngoscope, 2000, 110, 1694–1697 CrossRef CAS PubMed
; (c) D. Chon, D. Lando, A. Sosnik, S. Garty and A. Levi, Biomaterials, 2006, 27(9), 1718–1727 CrossRef PubMed
.
-
(a) X. J. Loh, S. H. Goh and J. Li, Polym. Prep., 2006, 47(2), 170–171 CAS
; (b) L. E. Bromberg and E. S. Ron, Adv. Drug Delivery Rev., 1998, 31, 197–222 CrossRef CAS
.
- S. Cometa, I. Bartolozzi, A. Corti, F. Chiellini, E. De Giglio and E. Chiellini, Polym. Degrad. Stab., 2010, 95, 2013–2021 CrossRef CAS PubMed
.
- L. Cosgrove, P. L. McGeechan, G. D. Robson and P. S. Handley, Appl. Environ. Microbiol., 2007, 73, 5817–5824 CrossRef CAS PubMed
.
- S. R. Barrat, A. R. Ennos, M. Greenhalgh, G. D. Robson and P. S. Handle, J. Appl. Microbiol., 2003, 95, 78–85 CrossRef
.
- K. T. Nakajima, Y. Shigeno-Akutsu, N. Nomura, F. Onuma and T. Nakahara, Appl. Microbiol. Biotechnol., 1999, 51, 134–140 CrossRef
.
- M. J. Kay, L. H. G. Morton and E. L. Prince, Int. Biodeterior., 1991, 27, 205–222 CrossRef CAS
.
- T. Nakajima-Kambe, F. Onuma, Y. Akutsu and T. Nakahara, J. Ferment. Bioeng., 1997, 83, 456–460 CrossRef CAS
.
- G. T. Howard and R. C. Blake, Int. Biodeterior. Biodegrad., 1998, 42, 213–220 CrossRef CAS
.
- G. T. Howard and N. P. Hilliard, Int. Biodeterior. Biodegrad., 1999, 43, 7–12 CrossRef CAS
.
- L. Rowe and G. T. Howard, Int. Biodeterior. Biodegrad., 2002, 50, 33–40 CrossRef CAS
.
- S. Nair and P. Kumar, World J. Microbiol. Biotechnol., 2007, 23, 1441–1449 CrossRef CAS
.
- K. T. Nakajima, F. Onuma, N. Kimpara and T. Nakahara, FEMS Microbiol. Lett., 1995, 129, 39–42 CrossRef PubMed
.
- R. Gautam, A. S. Bassi, E. K. Yanful and E. Cullen, Int. Biodeterior. Biodegrad., 2007, 60, 245–249 CrossRef CAS PubMed
.
- A. Oceguera-Cervantes, A. Carrillo-Garcia, N. Lopez, S. Bolanos-Nunez, M. J. Cruz-Gomez, C. Wacher and H. Loza-Tavera, Appl. Environ. Microbiol., 2007, 6214–6223 CrossRef CAS PubMed
.
- X. Jiang, J. Li, M. Ding, H. Tan, Q. Ling, Y. Zhong and Q. Fu, Eur. Polym. J., 2007, 43(5), 1838–1846 CrossRef CAS PubMed
.
- S. Sarkar and B. Adhikari, Indian J. Chem. Technol., 2007, 14, 221–228 CAS
.
- A. A. Shah, F. Hasan, J. I. Akhter, A. Hameed and S. Ahmed, Ann. Microbiol., 2008, 58(3), 381–386 CrossRef CAS
.
- J. Han, B. Chen, L. Ye, A. Zhang, J. Zhang and Z. Feng, Front. Mater. Sci. China, 2009, 3(1), 25–32 CrossRef PubMed
.
- K. Mukherjee, P. Tribedi, A. Chowdhary, T. Ray, A. Joardar, S. Giri and A. K. Sik, Biodegradation, 2011, 22, 377–388 CrossRef CAS PubMed
.
- S. Khan, Elixir Bio-Tech., 2011, 37, 3767–3772 Search PubMed
.
- Z. Shah, F. Hasan, L. Krumholz, D. F. Aktas and A. A. Shah, Int. Biodeterior. Biodegrad., 2013, 77, 114–122 CrossRef CAS PubMed
.
- Z. Shah, L. Krumholz, D. F. Aktas, F. Hasan, M. Khattak and A. A. Shah, Biodegradation, 2013, 6, 865–877 CrossRef PubMed
.
- M. Sponton, N. Casis, P. Mazo, B. Raud, A. Simonetta, L. Ríos and D. Estenoz, Int. Biodeterior. Biodegrad., 2013, 85, 85–94 CrossRef CAS PubMed
.
- C. Ruiz, N. Hilliard and G. T. Howard, Int. Biodeterior. Biodegrad., 1999, 43, 7–12 CrossRef
.
-
(a) C. Ruiz, T. Main, N. Hilliard and G. T. Howard, Int. Biodeterior. Biodegrad., 1999, 43, 43–47 CrossRef CAS
; (b) G. T. Howard and R. C. Blake, Int. Biodeterior. Biodegrad., 1998, 42, 213–220 CrossRef CAS
.
- Y. H. Peng, Y. H. Shih, Y. C. Lai, Y. Z. Liu, Y. T. Liu and N. C. Lin, Environ. Sci. Pollut. Res., 2014, 21(16), 9529–9537 CrossRef CAS PubMed
.
- J. C. Biffinger, D. E. Barlow, R. K. Pirlo, D. M. Babson, L. A. Fitzgerald, S. Zingarelli, L. J. Nadeau, W. J. Crookes-Goodson Jr and J. N. Russell, Int. Biodeterior. Biodegrad., 2014, 95, 311–319 CrossRef CAS PubMed
.
- A. Nakkabi, M. Sadiki, M. Fahim, N. Ittobane, S. IbnsoudaKoraichi, H. Barkai and S. El abed, Int. J. Environ. Res., 2015, 9(1), 157–162 CAS
.
- A. H. M. M. Halim El-Sayed, W. M. Mohmoud, E. M. Davis and R. W. Coughlin, Int. Biodeterior. Biodegrad., 1996, 37, 69–79 CrossRef
.
- A. Oceguera-Cervantes, A. Carrillo-García, N. Lopez, S. Bolaños-Nuñez, M. J. Cruz-Gómez, C. Wacher and H. Loza-Tavera, Appl. Environ. Microbiol., 2007, 73, 6214–6223 CrossRef CAS PubMed
.
- M. Varesche, M. Zaiat, L. G. T. Vieira, R. F. Vazoller and E. Foresti, Appl. Microbiol. Biotechnol., 1997, 48, 534–538 CrossRef CAS
.
- Y. V. Zachinyaev, I. I. Miroshnichenko and A. V. Zachinyaeya, Russ. J. Appl. Chem., 2009, 82, 1321–1323 CrossRef CAS
.
- R. Vega, T. Main and G. T. Howard, Int. Biodeterior. Biodegrad., 1999, 43, 49–55 CrossRef CAS
.
- R. S. Stern and G. T. Howard, FEMS Microbiol. Lett., 2000, 185, 163–168 CrossRef CAS PubMed
.
- G. T. Howard, R. I. Mackie, I. K. O. Cann, S. Ohene-Adjei, K. S. Aboudehen, B. G. Duos and G. W. Childers, J. Appl. Microbiol., 2007, 103, 2074–2083 CrossRef CAS PubMed
.
- Y. Akutsu-Shigeno, T. Nakajima-Kambe, N. Nomura and T. Nakahara, J. Ferment. Bioeng., 1998, 88, 484–487 Search PubMed
.
- N. Nomura, Y. Shigeno-Akutsu, T. Nakajima-Kambe and T. Nakahara, J. Ferment. Bioeng., 1998, 86(4), 339–345 CrossRef CAS
.
- M. Schumacher, S. Camp, Y. Maulet, M. Newton, K. Macphee-Guigley, S. S. Taylor, T. Friedmann and P. Taylor, Nature, 1986, 319, 407–409 CrossRef CAS PubMed
.
- Y. Shimada, A. Sugihara, T. Uzumi and Y. Tominaga, J. Biochem., 1990, 107, 426–430 Search PubMed
.
-
(a) A. M. Kaplan, R. T. Darby, M. Greenberger and M. R. Rodgers, Dev. Ind. Microbiol., 1968, 82, 362–371 Search PubMed
; (b) R. T. Darby and A. M. Kaplan, Appl. Microbiol., 1968, 16, 900–905 CAS
.
- R. A. Pathirana and K. J. Seal, Int. Biodeterior., 1984, 20, 163–168 Search PubMed
.
-
(a) B. Jansen, F. Schumacher-Perdreau, G. Peters and G. Pulverer, Zentralbl. Bakteriol., 1991, 276, 36–45 CrossRef CAS
; (b) M. J. Kay, R. W. McCabe and L. H. G. Morton, Int. Biodeterior. Biodegrad., 1993, 31, 209–225 CrossRef CAS
.
- S. Iannace, R. Alfani and L. Nicolais, Cell. Polym., 1999, 18, 21–33 CAS
.
- R. A. Pathiranar and K. J. Seal, Biodeterioration, 1983, 5, 679–689 Search PubMed
.
- R. H. Bentham, L. G. H. Morton and N. G. Allen, Int. Biodeterior., 1987, 23, 377–386 CrossRef CAS
.
- J. R. Crabbe, J. R. Campbell, L. Thompson, S. L. Walz and W. W. Schultz, Int. Biodeterior. Biodegrad., 1994, 33, 103–113 CrossRef
.
- S. Oprea, Polym. Degrad. Stab., 2010, 95, 2396–2404 CrossRef CAS PubMed
.
- S. R. Barratt, A. R. Ennos, M. Greenhalgh, G. D. Robson and P. S. Handley, J. Appl. Microbiol., 2003, 95, 78–85 CrossRef CAS
.
- L. Cosgrove, P. L. McGeechan, P. S. Handley and G. D. Robson, Appl. Environ. Microbiol., 2010, 810–819 CrossRef CAS PubMed
.
- S. Oprea, Polym. Degrad. Stab., 2010, 95, 2396–2404 CrossRef CAS PubMed
.
-
(a) D. S. Wales and B. F. Sagar, Proceedings of the Autumn Meeting of the Biodeterioration Society, Occasional Publication, Kew, 1985, pp. 56–69 Search PubMed
; (b) D. S. Wales and B. R. Sagar, Mechanistic aspects of polyurethane biodeterioration, in Biodeterioration, ed. D. R. Houghton, R. N. Smith and H. O. W. Eggins, Elsevier Applied Science, London, UK, 1988, pp. 351–358 Search PubMed
.
- R. H. Bentham, L. H. G. Morton and N. G. Allen, Novel test methods for the microbial deterioration of polyester polyurethanes, in Biodeterioration, R. Houghton, N. Smith and W. Eggins, Elsevier Applied Science, London, New York, 1988, pp. 562–567 Search PubMed
.
- S. Oprea and F. Doroftei, Int. Biodeterior. Biodegrad., 2011, 65(3), 533–538 CrossRef CAS PubMed
.
- I. N. Ibrahim, A. Maraqa, M. Khalid, K. M. Hameed, I. M. Saadoun and H. M. Maswadeh, Turk. J. Biol., 2011, 35, 551–557 Search PubMed
.
- J. R. Russell, J. Huang, P. Anand, K. Kucera, A. G. Sandoval, K. W. Dantzler, D. Hickman, J. Jee, F. M. Kimovec, D. Koppstein, D. H. Marks, P. A. Mittermiller, S. J. Nunez, M. Santiago, M. A. Townes, M. Vishnevetsky, N. E. Williams, M. P. N. Vargas, L.-A. Boulanger, C. Bascom-Slack and S. A. Strobel, Appl. Environ. Microbiol., 2011, 6076–6084 CrossRef CAS PubMed
.
- G. Mathur and R. Prasad, Appl. Biochem. Biotechnol., 2012, 167(6), 1595–1602 CrossRef CAS PubMed
.
- A. Loredo-Trevino, G. García, A. Velasco-Téllez, R. Rodríguez-Herrera and C. N. Aguilar, Adv. Biosci. Biotechnol., 2011, 2, 52–58 CrossRef CAS
.
- A. Ma and Q. Wong, Canadian Young Scientist Journal, 2013, 24–31, DOI:10.13034/cysj-2013-004
.
- H. R. Zhang, H. Pang, L. Zhang, X. Chen and B. Liao, J. Polym. Environ., 2012, 21(2), 329–334 CrossRef PubMed
.
- U. Zafar, A. Houlden and G. D. Robson, Appl. Environ. Microbiol., 2013, 79(23), 7313–7324 CrossRef CAS PubMed
.
- U. Zafar, P. Nzeram, A. Langarica-Fuentes, A. Houlden, A. Heyworth, A. Saiani and G. D. Robson, Bioresour. Technol., 2014, 158, 374–377 CrossRef CAS PubMed
.
- Z. S. Petrovic, Polym. Rev., 2008, 48(1), 109–155 CrossRef CAS PubMed
.
- E. F. Gómez, X. Luo, C. Li, F. C. Michel Jr and Y. Li, Polym. Degrad. Stab., 2014, 102, 195–203 CrossRef PubMed
.
- P. Basak and B. Adhikari, Polym.–Plast. Technol. Eng., 2013, 52(4), 358–367 CrossRef CAS PubMed
.
- C. A. Cateto, M. F. Barreiro, C. Ottati, M. Lopretti and A. E. Rodrigues, J. Cell. Plast., 2013, 50(1), 81–95 CrossRef PubMed
.
- K. Thirunavukarasu, S. Purushothaman, M. K. Gowthaman, T. Nakajima-Kambe, C. Rose and N. R. Kamini, J. Food Sci. Technol., 2015 DOI:10.1007/s13197-014-1697-8
.
- S. Shibasaki, A. Kawabata, T. Tanino, A. Kondo, M. Ueda and M. Tanaka, Biocontrol Sci., 2009, 14, 171–175 CrossRef CAS
.
- M. Stranger-Johannessen, Microbial degradation of polyurethane products in service, in Biodeterioration and Biodegradation of Polymers, K. J. Seal, Biodeterioration Society, New York, 1985, pp. 264–267 Search PubMed
.
- E. H. Pommer and G. Lorenz, The behaviour of polyester and polyether polyurethanes towards microorganisms, in Biodeterioration and Biodegradation of Polymers, K. J. Seal, Biodeterioration society, New York, 1985, pp. 77–86 Search PubMed
.
- I. N. Ibrahim, A. Maraqa, K. M. Hameed, I. M. Saadoun, H. M. Maswadeh and T. Nakajima-kambe, Adv. Environ. Biol., 2009, 3, 162–170 CAS
.
- J. S. Amaral, M. Sepúlveda, C. Cateto, I. P. Fernandes, A. E. Rodrigues, M. N. Belgacem and M. F. Barreiro, Polym. Degrad. Stab., 2012, 97, 2069–2076 CrossRef CAS PubMed
.
- S. J. Huang, D. A. Bansleben and J. R. Knox, J. Appl. Polym. Sci., 1979, 23, 429–437 CrossRef CAS PubMed
.
- G. A. Sharja and K. A. Woodhouse, J. Biomater. Sci., Polym. Ed., 2001, 12(8), 851–873 CrossRef PubMed
.
- N. Yamamoto, A. Nakayama, M. Oshima, N. Kawasaki and S.-I. Aiba, React. Funct. Polym., 2007, 67(11), 1338–1345 CrossRef CAS PubMed
.
- Z. Wang, L. Yu, M. Ding, H. Tan, J. Li and Q. Fu, Polym. Chem., 2011, 2(3), 601–607 RSC
.
- E. Ozsagiroglu, B. Iyisan and Y. A. Guvenilir, Pol. J. Environ. Stud., 2012, 21(6), 1777–1782 CAS
.
- C.-H. Kang, Ki-H. Oh, M.-H. Lee, T.-K. Oh, B. H. Kim and J.-H. Yoon, Microb. Cell Fact., 2011, 10, 41 CrossRef CAS PubMed
.
-
(a) K. Troev, G. Grancharov, R. Tsevi and A. Tsekova, Polymer, 2000, 41, 7017–7022 CrossRef CAS
; (b) Y. Zheng, E. K. Yanful and A. S. Bassi, Crit. Rev. Biotechnol., 2005, 25, 243–250 CrossRef CAS PubMed
.
- A. Silvestri, P. Serafini, S. Sartori, P. Ferrando, F. Boccafoschi, S. Milione, L. Conzatti and G. Ciardelli, J. Appl. Polym. Sci., 2011, 122(6), 3661–3671 CrossRef CAS PubMed
.
- E. J. Bouwer, Bioremediation of organic contaminants in the subsurface, in Environmental Microbiology, ed. R. Mitchell, New York, Wiley-Liss, 1992, pp. 287–318 Search PubMed
.
-
(a) H. Salacinski, N. Tai, R. Carson, A. Edwards, G. Hamilton and A. Seifalian, J. Biomed. Mater. Res., 2002, 59, 207–218 CrossRef CAS PubMed
; (b) Y. Tang, R. Labow and J. Santerre, J. Biomed. Mater. Res., 2001, 57, 597–611 CrossRef CAS
; (c) R. Labow, E. Meek, L. Matheson and J. Santerre, Biomaterials, 2002, 23, 3969–3975 CrossRef CAS
; (d) R. Labow, E. Meek and J. Santerre, Biomaterials, 2001, 22, 3025–3033 CrossRef CAS
; (e) L. Matheson, R. Labow and J. Santerre, J. Biomed. Mater. Res., 2002, 61, 505–513 CrossRef CAS PubMed
; (f) Y. Tang, R. Labow, I. Revenko and J. Santerre, J. Biomater. Sci., Polym. Ed., 2002, 13, 463–483 CrossRef PubMed
.
- R. Labow, E. Meek and J. Santerre, J. Biomater. Sci., Polym. Ed., 1999, 699–713 CrossRef CAS PubMed
.
- R. S. Labow, Y. Tang, C. B. Mccloskey and J. P. Santerre, J. Biomater. Sci., Polym. Ed., 2002, 13(6), 651–665 CrossRef CAS PubMed
.
- E. M. Christenson, S. Patel, J. M. Anderson and A. Hiltner, Biomaterials, 2006, 27, 3920–3926 CrossRef CAS PubMed
.
- Y. Matsumiya, N. Murata, E. Tanabe, K. Kubota and M. Kubo, J. Appl. Microbiol., 2010, 108(6), 1946–1953 CAS
.
- S. Obruca, I. Marova and L. Vojtova, Environ. Technol., 2011, 32(9–10), 1043–1052 CrossRef CAS PubMed
.
| This journal is © The Royal Society of Chemistry 2015 |