DNA conformational changes induced by cationic gemini surfactants: the key to switching DNA compact structures into elongated forms†
Elia Grueso*a,
Edyta Kuliszewskab,
Emilio Roldana,
Pilar Perez-Tejedaa,
Rafael Prado-Gotora and
Lothar Breckerc
aDepartment of Physical Chemistry, Faculty of Chemistry, University of Seville, C/ Profesor García González, s/n, 41012, Sevilla, Spain. E-mail: elia@us.es; Fax: +34-954557174; Tel: +34-954557175 ext. 215
bInstitute of Heavy Organic Synthesis-Ul, Energetykow 9, Kedzierzyn-Kozle 47-225, Poland
cInstitute of Organic Chemistry, University of Vienna, Währingerstrasse 38, A-1090 Wien, Austria
First published on 20th March 2015
Abstract
The DNA conformational changes induced by different members of the N,N′-bis(dimethyldodecyl)-α-ω-alkanediammonium dibromide series (m-s-m, m = 12, s = 3 and 6) and the analogous series of hexadecyl gemini surfactants (m = 16, s = 3 and 6) were investigated in aqueous media by means of circular dichroism (CD), zeta potential, dynamic light scattering (DLS), viscometry, and atomic force microscopy (AFM) methods. The measurements were carried out by varying the gemini surfactant–DNA molar ratio, R = Cm-s-m/CDNA. For the conditions investigated two significantly different conformational changes were observed, the second of them being worth noting. At low molar ratios, all methods concurred by showing that gemini surfactants were able to form ordered aggregates which precedes DNA compaction. The second effect observed, at high molar ratios, corresponds to the transition from the compact state to a new more extended conformation. The degree of decompaction and the morphologies of the visualized structures are different not only depending on the surfactant tail's length, but also on the spacer's length. The results obtained for the 16-3-16/DNA and 16-6-16/DNA systems point out that the compaction/decompaction processes are somewhat different to those previously visualized for the analogous monoquaternary chain surfactant CTAB.
1. Introduction
The understanding of the interaction between double stranded (ds) DNA and cationic gemini surfactants will have a broad bearing on many important applications from drug delivery1 to the translocation of DNA across the cell for the purpose of gene therapy.2 These applications require that genetic material is placed inside the cell in order to produce an effective medical treatment. However, the entry of naked DNA into the cell nucleus would need to resolve two fundamental problems: (1) the polyelectrolyte character of DNA polynucleotide gives rise to electrostatic repulsion when the DNA is approximated to the negatively charged cell membrane,3 (2) the steric restrictions inside the cell which are able to hinder the DNA transportation to the nucleous.4 To achieve these requirements, DNA first has to form a positively charged complex and then, needs to modify its conformation from an extended coil state to a compact globular state in order to adapt its structure to tine specific sites in the cell. The collapse process by which extended polynucleotide chains modify their structures into compact particles is called condensation, which is a common process for native genomes.5 The controlled DNA condensation has been demonstrated to be possible with a variety of reagents such as enzymes,6 multivalent ions,7,8 alcohols,9,10 polyamines,11 metal complexes,12 nanoparticles,13 and cationic surfactants.14–18 In this regard, the gemini surfactants have received a lot of attention recently due to its unique physicochemical properties, low cytotoxicity and potential bactericidal properties.19–21 Due to their positive charge, low toxicity and the strong capacity as DNA complexing agents, the cationic gemini surfactant–DNA systems are suitable to be properly transferred to living cells. Accordingly, motivated by the promising application of these cationic surfactants in gene delivery, several groups have explored the efficiency of these vectors on DNA compaction.16,17,20,21 Among a variety of gemini surfactants, those that possess a small spacer (s ≤ 3) present better DNA compaction efficiency.22–24 In spite of some progress having been made in understanding the DNA compaction processes induced by gemini surfactants, these are not yet well understood. In particular, the interactions of DNA with the 16-s-16 series have been less explored. Hence, a thorough study of the effect of the tail and spacer length on DNA compaction seems to be pertinent.Even though the compaction of DNA is relevant in drug delivery and gene therapy, the study of the dissociation of DNA/gemini surfactants complexes in the appropriate place is also crucial to ensure the final transfection.25 Accordingly, once the compact DNA molecule arrives in the nucleus, it needs to be accessible to the cell enzymatic machinery. However, this final step is only possible if decompaction occurs or if the degree of compaction of the DNA is not too high.26,27 Therefore, from an application viewpoint, controlling the balance between compacted/decompacted DNA structures with different gemini surfactant concentrations could be a matter of critical interest. In this regard, recent investigations have demonstrated that reversible compaction is possible in DNA–CTAB systems, without using additional chemical agents, by using a high surfactant content.15 The mechanism of DNA decompaction induced by CTAB was explained by postulating the existence of sphere-rod transitions in CTAB micelles, the latter being favored by an increase of bromide anions in solution.28 However, the factors that control the DNA–CTAB decompaction remain unclear. Different from conventional monomeric surfactants, the aggregation behavior of gemini surfactants in solution is controlled by both inter-molecular and intra-molecular interactions of these surfactants, along with their interactions with solvents.29 In comparison with their conventional counterparts, gemini surfactants, composed of two hydrophobic chains and two hydrophilic head groups covalently attached through a spacer, present strong self-aggregation ability.29 In addition, variety in spacer length generates a great tendency to form a variety of aggregate structures, such as tubule aggregates,29,30 elongated vesicles31 and rod-like or worm-like micelles32 in aqueous solutions. Atkin et al. postulated that in the gemini surfactants 12-s-12 with a short spacer (s ≤ 3) the head groups are close in proximity and they can assemble, forming flattened ellipsoidal aggregates structures in aqueous solution, when the surfactant concentration is increased.33 These aggregate structures, however, are strikingly different from those observed in gemini surfactants with longer spacers (12-s-12 with s ≥ 6). Due to the increase in the head group area, the surfactant chains interact with each other promoting the formation of more flattened aggregates of higher curvature. We believe that these aggregation properties can be applied to effectively influence the conformational behavior of m-s-m/DNA systems. Therefore, the purpose of this study was to investigate whether gemini surfactants are able to induce reversible DNA compaction/decompaction phenomena in aqueous solutions at different molar ratios. The choice of 12-3-12, 12-6-12, 16-3-16 and 16-6-16 gemini surfactants (see Fig. S1†) was made to explore the effect of varying the size of the tail together with the spacer's length on the conformational changes of the polynucleotide. The results obtained here show that switching DNA conformation is possible using the appropriate gemini structure (as a complexing agent), as well as a suitable surfactant concentration to achieve the critical Cm-s-m/CDNA molar ratio.
2. Experimental section
2.1. Materials
Calf thymus DNA was purchased from Pharmacia and used without further purification since preliminary experiments showed that purification did not make a difference in the experimental results. An agarose gel electrophoresis test using ethidium bromide indicated that the average number of base pairs per DNA molecule was greater than 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 bp.34 Polynucleotide concentrations, given in phosphate groups, were determined spectrophotometrically from the molar absorptivity (6600 M−1 cm−1 at 260 nm).35 Sodium cacodylate and 3-aminopropyltriethoxilane (APTES) were purchased from Sigma-Aldrich. All reagents were used as purchased. The water used in the preparation of solutions had a conductivity of less than 10−6 S m−1. All measurements were performed at pH 7.0 (0.01 M NaCac buffer where NaCac = sodium cacodylate = (CH3)2AsO2Na) and 298.2 ± 0.1 K. The total concentrations of the polynucleotide and the gemini surfactants in a working solution will now be referred to as CDNA and Cm-s-m, respectively; the molar ratio Cm-s-m/CDNA will be designated as R.
000 bp.34 Polynucleotide concentrations, given in phosphate groups, were determined spectrophotometrically from the molar absorptivity (6600 M−1 cm−1 at 260 nm).35 Sodium cacodylate and 3-aminopropyltriethoxilane (APTES) were purchased from Sigma-Aldrich. All reagents were used as purchased. The water used in the preparation of solutions had a conductivity of less than 10−6 S m−1. All measurements were performed at pH 7.0 (0.01 M NaCac buffer where NaCac = sodium cacodylate = (CH3)2AsO2Na) and 298.2 ± 0.1 K. The total concentrations of the polynucleotide and the gemini surfactants in a working solution will now be referred to as CDNA and Cm-s-m, respectively; the molar ratio Cm-s-m/CDNA will be designated as R.
2.2. Synthesis and characterization of gemini surfactants
For the synthesis of the gemini surfactant compounds a mixture of α,ω-dibromoalkane (0.05 mol) and N,N-dimethylalkylamine (0.12 mol) in acetonitrile (100 mL) was stirred under reflux for 20 h. Upon cooling a white solid was recovered by filtration. All products were recrystallized from ethyl acetate up to five times and dried under vacuum.36 Characterization of the gemini surfactants was based on mass spectrometry results with detection of fragments in the third quadrupole,37 nuclear magnetic resonance spectroscopy experiments, elemental analysis and surface tension experiments. The critical micelle concentrations (CMC) of the gemini surfactants were measured by the surface tension technique. The measured CMC values were collected in Table S1,† which are in good agreement with the previous values registered by Zana et al.38 The characterization techniques used for gemini surfactants are described in the ESI† section of the paper.2.3. Methods
 | (1) |
3. Results
3.1. Viscosity measurements
In Fig. 1A, the viscosity ratio, (η/η0)1/3, was plotted as a function of the R molar ratio for the 12-3-12/DNA and 12-6-12/DNA systems. The complexity of the analysed systems is confirmed by the viscosity trend that changes its features depending on the reagents ratio. In such a way that the viscosity trend at first is ascending (Cm-s-m/CDNA < 0.025), subsequently it descends until reaching a minimum (Cm-s-m/CDNA = 0.35–0.50), and finally increases again for the higher R ratios. Importantly, as displayed in Fig. 1B, the position of the (η/η0)1/3 maxima for the 16-3-16/DNA and the 16-6-16/DNA systems correlate well with those obtained from the dodecyl series of surfactants.3.2. CD measurements
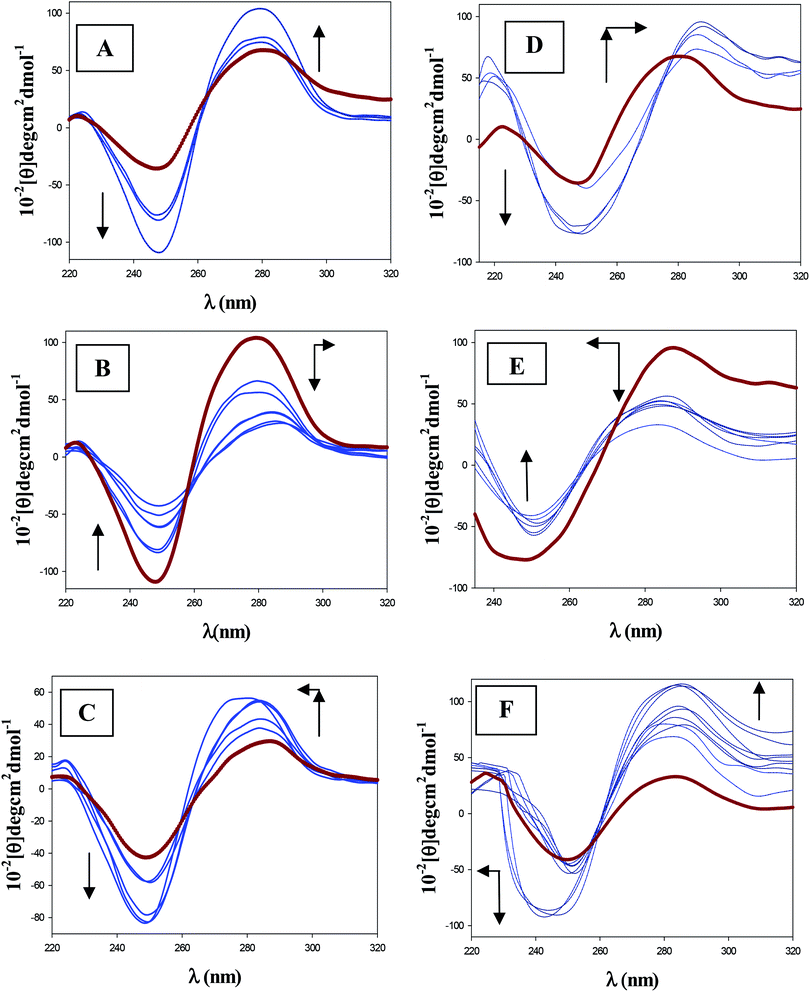
The CD technique was used to investigate the influence of gemini surfactants on the DNA conformation. As is known, the backbone conformation of DNA shows a characteristic CD spectrum of the right-handed B form in the far UV region (220–320 nm). Structural alterations of the DNA caused by its interaction with ligands are reflected in changes in this intrinsic CD spectrum.39,40 Fig. 2 shows that the secondary structure of DNA is perturbed by the four surfactants studied here, this perturbation being different for the 12-s-12 series (Fig. 2A) and the 16-s-16 series (Fig. 2B). Besides, the addition of different surfactant concentrations to the DNA solution registers complex behavior according to the value of the molar ratio R. For the 12-s-12 series, the measured dichroic bands are found to be dramatically different (with respect to sign, position, shape and intensity) exhibiting two inflection points: one of them at C12-s-12/CDNA ∼ 0.03 (red curves of Fig. 3B and S2B†) and the other at C12-s-12/CDNA ≈ 0.5–0.6 (red curves of Fig. 3C and S2C†). On the other hand, the 16-s-16 series also display characteristic changes in CD bands upon DNA binding, showing two inflection points. The first maximum is coincident in position with the one obtained for the 12-s-12 series, C16-s-16/CDNA ∼ 0.03 (red curves of Fig. 3E and S2E†); while, the position of the minimum is slightly displaced to low R ratios, C16-s-16/CDNA ∼ 0.4 (red curves of Fig. 3F and S2F†).3.3. Dynamic light scattering (DLS) measurements
Fig. S3 and S4† show the intensity-weighted size distributions of a DNA solution in the presence and absence of the 12-s-12 and 16-s-16 series of gemini surfactants, working at different molar ratios. Besides, the hydrodynamic diameters of the DNA molecules, taken from the position of the peaks of the intensity distribution function, are collected in Table 1. Results reflect two different trends in the systems studied varying the R ratios. At the low R ratios, in all the four surfactants systems, the hydrodynamic diameter decreases reaching a minimum, between a R ratio of 0.4 and 0.6, depending on the surfactant studied. The positions of the minimum correlate well with the minimum observed by viscosity and CD measurements (see Fig. 1A and 2). Then, as the R ratio increases after passing the minimum, a drastic change in the hydrodynamic diameter occurs. However, a closer examination of Fig. S3 and S4† highlighted that this behavior is different for the 12-s-12 and the 16-s-16 series of gemini surfactants.| 12-3-12 | 12-6-12 | ||||
|---|---|---|---|---|---|
| R | d1 (nm) | d2 (nm) | R | d1 (nm) | d2 (nm) |
| 0.00 | 951 (100%) | 0.00 | 951 (100%) | ||
| 0.03 | 566 (81%) | 84 (19%) | 0.03 | 505 (64%) | 90 (36%) |
| 0.10 | 476 (78%) | 78 (22%) | 0.05 | 434 (88%) | 48 (18%) |
| 0.25 | 203 (100%) | 0.40 | 252 (53%) | 61 (47%) | |
| 0.60 | 82 (100%) | 0.60 | 156 (100%) | ||
| 1.00 | 239 (100%) | 1.50 | 920 (100%) | ||
| 2.50 | 789 (100%) | 4.00 | 1221 (100%) | ||
| 4.00 | 1179 (100%) | 5.00 | 1531 (100%) | ||
| 5.00 | 1526 (100%) | ||||
| 16-3-16 | 16-6-16 | ||||
|---|---|---|---|---|---|
| R | d1 (nm) | d2 (nm) | R | d1 (nm) | d2 (nm) |
| 0.03 | 615 (93%) | 35 (7%) | 0.03 | 534 (85%) | 52 (15%) |
| 0.05 | 330 (92%) | 35 (8%) | 0.14 | 294 (27%) | 74 (73%) |
| 0.25 | 255 (88%) | 45 (12%) | 0.25 | 282 (48%) | 52 (52%) |
| 0.50 | 120 (100%) | 0.40 | 182 (100%) | ||
| 3.50 | 157 (100%) | 0.60 | 222 (100%) | ||
| 4.00 | 293 (31%) | 60 (69%) | 2.50 | 637 (2%) | 88 (98%) |
| 5.00 | 460 (21%) | 90 (79%) | 5.00 | 615 (39%) | 130 (61%) |
3.4. Zeta potential measurements
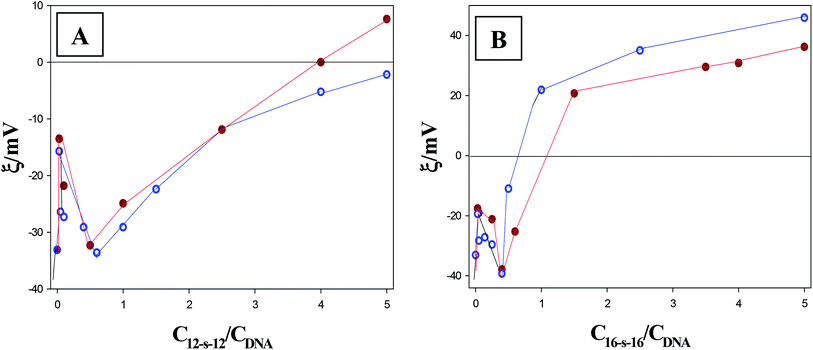
In an applied electric field, charged species are attracted to the electrode of the opposite polarity resulting in an electrostatic potential called zeta potential. The zeta potential value of the 12-s-12/DNA and 16-s-16/DNA complexes is determined by the charge ratio between the gemini surfactants and the DNA. Fig. 4A and B show the zeta potential values obtained for the interaction of DNA with the 12-s-12 and 16-s-16 series of gemini surfactants, respectively. Two inflection points are observed from the figures, whose position is in agreement with that observed from the rest of the techniques.3.5. AFM measurements
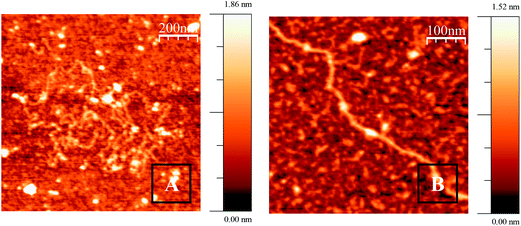
The AFM images show the existence of a series of structures which depend on the R molar ratio for each surfactant studied. The morphology of these structures depends notably on the alkyl chain length and, in a less significant way, on the spacer length. These structures are stable in a time scale of hours and some of them appear to be early intermediates in a compaction/decompaction pathway. Fig. 5 shows AFM topographic images of CT–DNA, adsorbed onto an APTES modify mica surface, in cacodylate buffer at pH = 7.0. As can be observed in the figure, the double stranded DNA is in extended-coil conformation in the absence of any added surfactant. When gemini surfactants were added to the DNA, at low R ratios, a compaction process from an elongated coil to a compact globule was observed. In particular, at a molar ratio of R = 0.03, Fig. 6 shows the formation of different aggregates of DNA. The morphology of these aggregates are different depending on the spacer length of the surfactant, longer in size for surfactants with spacers s = 3. Multiple intramolecular and intermolecular loops are formed, specially, for the gemini surfactants with spacers s = 3, only some random parts of the DNA chains being condensed. Subsequently, when the R ratio increases to 0.1, Fig. S5,† several intermediates of the compaction process with well-defined characteristics are seen. A partially condensed phase was observed with the formation of different globules protruding from the DNA chain where loops are increased in number. Another feature of these intermediates is the formation of some crossover points (see Fig. S5B and C†). With further addition of gemini surfactants, molar ratio was close to 0.6 for the 12-s-12 series and near 0.4 for the 16-s-16 series; only compact globule structures appear, pointing out the completion of the compaction process (Fig. 7). A closer examination of the structures reveals that globules of a larger size are formed for surfactants with a longer spacer length.Once the compaction process has been reached, the further addition of gemini surfactant entails a increment in the size of the complexes (see Fig. S6 and S7†). This behavior points out that a strictly different conformational change is beginning to be observed, which is consistent with the above mentioned results of viscosity and DLS. It is a remarkable fact that different stages of DNA decompaction phenomena are observed when it goes from molar ratio R = 1 to R = 5.0, for both series of gemini surfactants. Considering first the 12-s-12/DNA systems, at R = 1.0, the DNA globules interact with each other to form larger DNA/surfactant aggregates (see Fig. S6A and S6D†). Then, at molar ratio R = 2.5, extended DNA chains emerge from DNA globules (Fig. S6B and S6E†); and finally, at the 4.5 R ratio, the structures are almost fully decompacted (Fig. 6SC and 5SF†). We next examined the intermediates of decompaction for the 16-s-16 series. At first, starting from the molar ratio R = 1, DNA fibers emerging from the extreme of DNA globules can be seen (Fig. S7A and B and S7D†). Then, at the molar ratio R = 2.5, characteristic features called “pearling” structures, in which spherical small globules are generated on a single polymer chain, are seen (see Fig. S7C and S7E and F†).41 It is important to mention here that the “pearling” structures were not detected as intermediates in the compaction process. Consequently, for the 16-s-16/DNA systems, the decondensation process is not simply the reverse of the previously described condensation process. Importantly, the number of protruding DNA chains from the compact aggregates when the R ratio increases is less significant for the 16-s-16 series in comparison with the 12-s-12 series of gemini surfactants. This situation gives rise to such a diverse gradation in the decompaction process for both the 12-s-12 and 16-s-16 series. In such a way that, at the higher R values (R = 5), the situation is thoroughly different for both series (Fig. 8). That is, at high R ratios, the 12-s-12/DNA systems are fully decompacted (Fig. 8A and B), thus adopting a structure similar to the free DNA in solution (Fig. 5). However, for the 16-s-16/DNA systems, the complexes are not able to adopt fully decompacted structures, exhibiting a partially decompacted state (Fig. 8C and D). Fig. S8† shows an enlarged view for the 16-3-16/DNA and 16-6-16/DNA systems at the higher R ratio. In such images extended DNA chains together with compact globules structures are seen. Furthermore, from these images, it is noteworthy that the appearance of compact globules is much less frequent for the 16-6-16/DNA system than for the 16-3-16/DNA system, which reveals that the latter system is the worst one to be used as the delivery vehicle. Note that these results are in good agreement with DLS experiments, in which the presence of a significant percentage of compact structures is relevant for the 16-s-16 systems, while no condensed structures were seen for the 12-s-12 series at this stage of the interaction.
4. Discussion
The principal result stemming from the analysis of the data provided by the different experimental techniques considered in the present investigation is that, once the compaction of the system was accomplished, DNA decompaction was induced by gemini surfactants at high molar ratios. This behavior is particularly of interest, since even though the capacity of diverse gemini surfactants to compact DNA polymers was already recognized,16–18 such an ability to decompact DNA structures so far was not observed.Firstly, the viscosity results were examined. As is known, the viscosimetry is a well-established and simple methodology which is extremely sensitive to the length change of a biopolymer and it provides, perhaps, the least ambiguous criteria to identify the changes in extension of the DNA helix in solution upon ligand binding.14 The change in contour length (from L0 to L) has been related to the change in intrinsic viscosities (from η0 to η) of the free DNA and the DNA/gemini surfactant complexes by:42
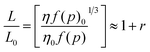 | (2) |
4.1. 12-s-12 effect on DNA structure
Fig. 3A shows that the CD spectrum of free DNA has a positive peak at approximately 279 nm and a negative peak at 249 nm which corresponds to B-DNA structure. These bands are caused by stacking interactions between the bases and the helical suprastructure of the polynucleotide that provides an asymmetric environment for the bases.44 In the same figure the CD changes induced by the 12-6-12 surfactant clearly demonstrates that the helical conformation is not maintained. Gradual addition of each of the two 12-s-12 gemini surfactants leads to remarkable perturbations on both positive and negative bands (Fig. 3A and C and S2A–C†). At low R ratios, Fig. 3A shows that the bands intensity are enhanced without appreciable shift in their position. These changes are indicative of a destabilization of the DNA helix and a disruption of staking contact of the bases, which could indicate that the gemini surfactant–DNA binding was via an intercalation mode.45 It is important to note that the extent of increase in the ellipticity is different for the two 12-s-12 gemini surfactants studied, the maximum being for the 12-3-12 gemini (see the inset of Fig. 2A). This result can be rationalized in light of the relative steric hindrance effect associated with the spacers' size on the surfactants. According to this steric effect, the intercalation into the DNA bases is restricted, most for 12-6-12 and least for 12-3-12. Therefore, the binding of 12-s-12 surfactant with shorter spacer length entails a larger perturbation on DNA strands and more extensive destabilization and destacking of the DNA helix. Subsequently, starting from a specific gemini surfactant concentration, the bands intensities are diminished significantly. This fact together with a small shift in the positive CD band to higher wavelength, indicates DNA compaction15,18 and partial denaturation of the double strand (Fig. 3B and S2B†).46,47 A direct observation of the above stated phenomenology can be obtained using the DLS technique. At low molar ratios, Fig. S3† shows a marked decrease in the average hydrodynamic diameter as the molar ratios increase. In the absence of surfactants, at the molar ratio R = 0, a high value of the hydrodynamic diameters with unimodal distribution is displayed which corresponds to free DNA in a buffer solution. Of particular interest is that a bimodal distribution appears, at almost the same R ratio (R = 0.03), in which the CD signals and the relative viscosity values were maximal, as was described above. This finding implies that there are two different populations of DNA with different weight-sized distributions, in accordance with previous studies.14,15 One of them can be assigned to DNA in elongated-coil conformation; the other, considering the fact that the average length of a condensed DNA segment is about 100 nm, can be ascribed to compact conformation.48 A closer examination of Table 1 reveals that, as the R ratio increases, the particle size corresponding to the elongated form decreases, while the corresponding one assigned to the compact forms is almost maintained. Then, at a specific R ratio of 0.25 for the 12-3-12/DNA and 0.60 for the 12-6-12/DNA system, the distribution turned out to be unimodal, exhibiting hydrodynamic diameters that can be easily identified with DNA compact structures. Interestingly, in good agreement with viscosity (Fig. 1A) and CD (Fig. 2A) results, Table 1 shows that the smaller values of the hydrodynamic diameters for both 12-s-12/DNA systems appear at an R ratio of 0.60. Finally, it is worth noting that with increasing the R ratios, a completely distinct behavior is revealed from the DLS experiments. That is, the diameters of this new unimodal distribution are size enhanced, highlighting the adoption hydrodynamic diameters even greater than that of the DNA free in solution. The features of DNA conformation described here correlated well with CD results. In such a way that at R > 0.5, the molar ellipticity of the positive and negative bands increased in their intensity and returned to show a similar form compared to that adopted for the DNA in the absence of any added surfactant (Fig. 3C and 2SC†).4.2. 16-s-16 effect on DNA structure
The intrinsic CD spectra of the 16-s-16/DNA systems, recorded at different values of the R ratio, show different behavior to those of the 12-s-12/DNA systems (Fig. 2B). At low R ratios, Fig. 3D shows that the binding of 16-6-16 to DNA leads to a shifting of the positive CD band (at 280 nm) to the longer wavelength (7 nm); this shift being accompanied by an increase in the intensity of the positive band. This behavior suggests that the complex 16-6-16-DNA goes from a non aggregated form to an aggregated form due to binding interaction, stabilizing the contact of the surfactant with nucleic acids.49 Again, it is important to mention here that the extent of increase in the ellipticity is maximum for the 16-3-16 gemini surfactant (see Fig. S2D†). On the other hand, the negative band became broader and showed a change in the intensity without any appreciable shift. The increase in the intensity observed for this band can be attributed to an increment in the stacking interaction between the DNA bases upon the binding of the surfactant.50,51 In a second stage of the interaction a different behavior can be observed, the positive band decreases in intensity and undergoes a blue shift, while the negative band decreases in its intensity (see Fig. 3E and S2E†). These changes can be attributed to DNA compaction induced by gemini surfactant interaction together with a disaggregation of the complex formed in the first stage of the binding.15,18,49 Concerning the DLS results, the behavior is quite similar to that observed for the 12-s-12 series. Thus, a bimodal distribution was registered starting from a R ratio of 0.03, the particle sizes of the elongated form being decreased with increasing the surfactant concentrations. Then, these bimodal distributions were converted into a unimodal, at about R ∼ 0.5. Therefore, the different CD behavior of the 16-s-16 series compared to 12-s-12, in the R range of 0.03–0.5, cannot be distinguished by using the DLS technique; consequently, this fact will be discussed later, on the basis of AFM results. Finally, another diverse behavior is noted at high R ratios for both negative and positive CD bands which are increased strongly in their intensity (see Fig. 3F and S2F†). These changes are accompanied by a blue shift of the maximum negative band (∼7 nm). It is worth noting that, contrary to what occurred with the 12-s-12 series, the final form of the 16-s-16/DNA CD spectrum is remarkably different to the characteristic of the free B-DNA in solution. In such a way that the intensity of both bands are strongly augmented and the position of the broad negative band is displaced to the blue. The hypochromic shift along with the enhancement in negative intensity can be attributed to changes in local base pairing geometry and a great increment in stacking interaction of the bases on the surfactant–DNA interaction.50,51 Besides, the strong increase in the positive band signify strong alteration in duplex helicity upon 16-6-16 binding to DNA. Furthermore, Fig. 2B shows that the extent of increase in the ellipticity is somewhat different for the two 16-s-16 surfactants, the 16-3-16 compound being the maximum; thus, this implies that binding of 16-3-16 to DNA causes a larger perturbation and greater destabilization of the DNA helix.52 With respect to the DLS results at these high R ratios, the observed trend is somewhat similar to that observed in a previous work for the CTAB/DNA system.15 In such a way that the hydrodynamic diameter first increases, probably by forming greater size compact structures, that is, favoring globules aggregation. Then, at a R ratio of 4.0 for the 16-3-16/DNA system and of 2.5 for the 16-6-16/DNA system, a new bimodal distribution appear, which presumably denotes the coexistence of globules and more extended DNA chains. Note that contrary to what occurred with the CTAB/DNA system,15 the percentage of the smaller species is higher than the population corresponding to the extended DNA helix. Additionally, looking at the values of hydrodynamic size and the percentage of the populations collected in Table 1, it can be concluded that the shorter the spacer length of the 16-s-16 surfactant is, the shorter and less frequent the elongated forms become. A possible explanation would be that in the case of 16-6-16 gemini surfactant the distance between head groups induced by the spacer size is more similar to that of the monomer CTAB.33 Besides, as was mentioned in the Introduction section, the effect of the spacer on the aggregation properties of the gemini surfactants is significant.29–33 Consequently, taking into account these facts, we can reasonable suppose that the 16-6-16 aggregates structures in the 16-6-16/DNA system could be like that of the CTAB/DNA, and thus, the elongated DNA forms are similar in sizes. Note that the hydrodynamic diameter of the longer species, at R = 5.0, was 613 nm for the CTAB/DNA system,15 and for the 16-6-16/DNA system studied here it was 615 nm (see Table 1).On the basis of the previous discussion, it seems clear that the CD and DLS data confirm the features of DNA–gemini surfactants interaction revealed by the viscosity technique. That is, the existence of different DNA conformational changes induced by the gemini surfactants: compaction for the lower R ratio, and decompaction (or partial decompaction) for the higher molar ratios. However, some important points still remain to be clarified: (i) the motive by which maxima, at the low R ratios, observed by the distinct experimental techniques are more prominent for the surfactants with short spacer length; (ii) the stages by which the DNA compaction/decompaction phenomena occur induced by the gemini surfactants; (iii) the reasons for which the degree of decompaction is different depending on the surfactants tail's length. Therefore, in order to gain more direct and precise information on the DNA conformational change, the AFM ultrasensitive technique was employed in conjunction with zeta potential experiments.
Concerning the first question: a hypothesis by which the partial intercalation of few gemini surfactants could promote DNA–DNA interactions was already done in order to explain this anomalous behavior. In spite of both the viscosity and CD techniques supporting this mode of interaction, it is still not clear if the gemini surfactant intercalation promotes the interaction between other DNA helix or within the base pairs of the same helix. In light of AFM experiments we observed that this interaction conduces to the formation of small globules and large-sized aggregates, the latter being larger in size in the case of surfactants with short spacer length (see Fig. 6). Note that, as shown in Fig. 4, at R ratio of 0.03, the DNA–surfactant complexes are negatively charged, so, the mica had to be functionalized with APTES. As to the zeta potential trend, which is observed in Fig. 4, the reason by which these aggregates are larger in size can easily be surmised. That is, taking into account the height of the maximums, R ∼ 0.03, it seems clear that the zeta potential values were higher for the complexes with spacer length s = 3; consequently, the charge shift from negative to positive was greater. Hence, this finding shows that the gemini surfactants with short spacer length possess a larger number of surfactants per DNA aggregates; this circumstance contributes to the larger size of the complexes with spacers s = 3 compared to the surfactants with spacers s = 6. On the basis of both the AFM and the zeta potential results, we can reasonably conclude that the initial step in DNA condensation event induced by gemini surfactants is the stabilization in the contacts of DNA strands–strands, along the length of the strands in the aggregates. This phenomenon is accompanied by a bending and a decrease in the persistent length of the DNA molecules which leads to the formation and stabilization of intramolecular loops. The appearance of flower-like intermediate structures are consistent with the model for DNA condensation proposed by Montigny.53 According to this model, these intermediates are the results from the binding of single cationic molecules and multiple free DNA loops.
The next recognizable step in DNA condensation is revealed by the appearance of DNA compact globules emerging from DNA strands which contain, in some cases, flower-like structures (see Fig. S5†); these structures being considered as a type of DNA strand–strand stabilization.54 According to these structures, the intramolecular condensation is feasible when the bound ligand reaches a critical concentration. Finally, the resultant complex can collapse from the addition of more cationic gemini surfactants, giving rise to a stable compact particle.53 Hence, the condensation of DNA induced by the cationic gemini surfactants studied here occurs in three stages: (i) the binding of single cationic surfactant molecules to DNA which produce a bending of the macromolecule and the formation of different loops and flower-like structures; (ii) the addition of a higher quantity of cationic surfactants leads to intramolecular DNA condensation with the formation of compact globules emerging from DNA strands; (iii) further addition of surfactants produces the collapse of DNA loops onto cation cores resulting in stable compact particles.
Once the stages of DNA condensation have been identified, the differences observed by CD technique, in the range 0.03–0.5, on the behavior of 12-s-12/DNA and 16-s-16/DNA systems will be considered again. Since AFM images reveal the formation of ordered aggregates at R = 0.03, it seems clear that to achieve the collapse of these structures into a nucleation center, the breakage of some of these aggregates is needed. Therefore, taking into account that the aggregates formed were larger in size and more in order for the 16-s-16/DNA, compared to the 12-s-12/DNA systems, the disaggregation and breakage phenomena should be more perceivable for the 16-s-16/DNA (Fig. S5†). This gives support to the significant shift in the CD maximum observed for the 16-s-16/DNA system. Nonetheless, a comment about the values of zeta potential when the compaction has been reached seems pertinent. Note that, after passing the first maximum (R = 0.03), the zeta potential becomes more negative until approximately the R values at which the compaction is accomplished (R ∼ 0.6 for the 12-s-12 series and R ∼ 0.4 for the 16-s-16 series). This behavior is, in some sense, counterintuitive due to such an increase in the surfactants concentrations that should be produced, to some extent, the neutralization of the DNA charges. However, a valuable explanation could be made taking into account the intermediates of the DNA compaction process observed by the AFM technique. That is, in spite of being in the first stage of the compaction process (R ∼ 0–0.03) the surfactant charges are more exposed in the aggregates, in the second stage the charges of surfactants could be shielded with the collapse of DNA structures forming globules on the cationic surfactant core.
We next examine the R values when the complexes become neutrals. Since, starting from zero point charge, the AFM measurements would be done in the absence of APTES to ensure the correct adsorption of the DNA–surfactant complexes, the localization of this point was of particular importance. The neutralization occurs at R values of about 0.7–1.0 for the 16-s-16/DNA systems, and only for the 12-3-12/DNA gemini surfactant, it was accomplished at a molar ratio of about R ∼ 1. On the other hand, the fact that the 12-s-12/DNA complexes displayed much lower values of the zeta potential compared to the 16-s-16/DNA systems, it is noteworthy once the compaction was fully accomplished. In fact, this behavior could be directly related to the fact that the degree of decompaction is better for the surfactants with shorter alkyl chains.
In order to understand the different degrees of decompaction of 12-s-12/DNA and 16-s-16/DNA systems, the morphology of surfactant aggregates, free in solution, was explored by the AFM technique. Fig. S9 and S10† show micellar aggregates of 12-3-12 and 16-6-16 gemini surfactants, in the free state. Note that the apparent diameter of these structures gives, generally, uncertainties because of the image convolution with the tip diameter.55,56 However, the height of DNA molecules can be determined without taking into account this convolution effect. That is, the average height of these aggregates was 4.5 nm for the 12-3-12 surfactants, while aggregates of about 12 nm and 3 nm were observed for the 16-6-16 surfactants. Consequently, these figures show that with the increase of the alkyl chain length, the size of the aggregates in solution increases. This is probably due to the inter-molecular hydrophobic interactions between the surfactant molecules which are optimized as the alkyl chains increase in length. In the same figure, images of 12-3-12/DNA and 16-6-16/DNA systems in a decompacted state can be observed. Comparing the z values measured for the surfactant aggregates, with the height of DNA–surfactant complexes in this intermediate decompaction state, certain similarities were revealed. It is remarkable that from the analysis of the height of the surfactants aggregates bound to DNA structures, only the smaller size surfactants aggregates, of about 4 nm, still remained attached to the DNA chains. Taking into account this fact and in consideration of the fact that the zeta potential of the 12-s-12/DNA complexes was lower than that of the 16-s-16/DNA complexes, we can reasonably conclude that more quantity of small aggregates of gemini surfactant were able to be inserted in the 12-s-12/DNA systems; therefore, the positive charges of the 12-s-12 surfactants aggregates became more hidden. Hence, the effect of alkyl chain length, on the degree of DNA decompaction is, probably, directly related to the size of the surfactants aggregates. That is, the aggregates of smaller sizes such as that of the 12-3-12 surfactant favor DNA decompaction. Furthermore, the distinct gradation observed in the decompaction process for 12-s-12 and 16-s-16 surfactants can also be explained by attention to the diverse nature of the intermediates formed. In such a way that contrary to what occurs with the 12-s-12 systems, the intermediates that appear in the DNA decompaction process of 16-s-16/DNA systems are somewhat different to those observed in the corresponding DNA compaction. Similarly, as happened in the case of the analogous monoquaternary surfactant CTAB,15 in the first stage of decompaction process linear DNA chains protruding from 16-s-16/DNA compact globules, are seen. In a second stage of decompaction, “pearling” structures, in which spherical small globules are generated on a single polymer chain, are visualized. In a previous work we postulate that the decompaction of CTAB–DNA complexes could be attributed to the existence of a micelles phase transition from spherical to rod-like micelles, the latter unable to fit well with the polymer and thus, decompaction was favored.15 In this study we have looked more carefully at the effect of spacer length on the decompaction phenomena of the analogous 16-s-16 gemini surfactants. The fact that, the breakage of globules is favored following the trend: CTAB/DNA > 16-6-16/DNA > 16-3-16/DNA, denotes that not only steric hinderance reasons are playing an important role in the decompaction phenomena. According to Atkin et al., the spacer group of the gemini surfactants controls the separation between the head groups that may be greater or lesser than the mean separation of the corresponding monomer.33 For gemini surfactants of lower spacer groups, the head group are in close proximity and thus, the smaller head group areas could lead to aggregates of lower curvature and a higher aggregation number than those of the corresponding monomer. In contrast, for gemini surfactants with longer spacers, the distance between the head groups resemble that of the monomer, and consequently, the structure of aggregates could result more similar. Therefore, a counterbalance between the effect of size and the charge of aggregates is, probably taking place in the case of the 16-3-16 gemini surfactants. That is, the more closed packing of the head groups of 16-3-16 compared to that found for both, 16-6-16 and the free CTAB micelles, could indicate that the 16-3-16 aggregates would act as a “laterally extended highly charged counterion” for the polymer strands, thus hindering DNA decompaction.
5. Conclusions
Different aspects of DNA conformational changes induced by 12-s-12 and 16-s-16 gemini surfactants whose structures vary, both in the size of the chain as in the length of the spacers, have been analyzed using spectroscopic, viscometric and structural approaches. All of these methods concur with the fact that distinctive conformational changes are displayed as a function of the Cm-s-m/CDNA ratio. At low R ratios, the formation of DNA–surfactants aggregates precedes DNA compaction. Then, starting from a R ratio of about 0.6 for the 12-s-12/DNA system, and of R ∼ 0.4 for the corresponding 16-s-16/DNA, decompaction is beginning to occur. In such a way that at an appropriate surfactant concentration, in the absence of any additional chemical agents, the surfactant is able to switch DNA conformation from compact globular structures to extended DNA chains. The extent of the decompaction process depends on the size of the chain length and also on the surfactant spacer's length. Importantly, this process is fully accomplished for dodecyl gemini surfactants and is only partial for the 16-s-16 series. This behavior is particularly interesting, since in spite of the ability of gemini surfactants to induce DNA compaction, such an ability was already recognized, but the DNA decompaction up to now had not been observed. These findings open a new strategy for the design of non-toxic carriers, based on the gemini surfactants, for drug delivery and gene therapy applications. Due to the greater efficiency in the DNA compaction/decompaction, to the best of our knowledge, the most effective gemini surfactants for medical applications are those belonging to the 12-s-12 series and those of the 16-s-16 series with large-sized spacers. We hope that these physicochemical and structural studies of DNA conformational changes induced by gemini surfactants not only will increase the understanding of reversible DNA compaction, but also it can be applied, as a perspective of future, to improve gene delivery application in cells.Acknowledgements
This work was financed by the Consejería de Educación y Ciencia of the Junta de Andalucía (FQM-03623). We thank to Dr C. Cerrillos (CITIUS, University of Seville) for her assistance in obtaining the AFM images.References
- C. Bombelli, L. Giansanti, P. Luciani and G. Mancini, Curr. Med. Chem., 2009, 16(13), 171–183 CrossRef CAS.
- M. C. Garnett, Crit. Rev. Ther. Drug Carrier Syst., 1999, 16(2), 147–207 CrossRef CAS.
- C. Tros de Ilarduya, Y. Sun and N. Düzgünes, Eur. J. Pharm. Sci., 2010, 40(3), 159–170 CrossRef CAS PubMed.
- M. E. Dowty, P. Williams, G. F. Zhang, J. E. Hagstrom and J. A. Wolff, Proc. Natl. Acad. Sci. U. S. A., 1995, 92(10), 4572–4576 CrossRef CAS.
- Z. Lin, C. Wang, X. Feng, M. Liu, J. Li and C. Bai, Nucleic Acids Res., 1998, 26(13), 3228–3234 CrossRef CAS PubMed.
- D. A. Erie, G. Yang, H. C. Schultz and C. Bustamante, Science., 1994, 266(5190), 1562–1566 CAS.
- K. Besteman, K. van Eijk and S. G. Lemay, Nat. Phys., 2007, 3, 641–644 CrossRef CAS.
- F. Jiménez-Ángeles and M. A. Lozada Cassou, J. Phys. Chem. B, 2004, 108, 7286–7296 CrossRef.
- J. C. Girod, W. C. J. Johson, S. K. Huntington and M. F. Maestre, Biochemistry, 1983, 12, 5092–5096 CrossRef.
- R. Huey and S. C. Mohr, Biopolymers, 1981, 20, 2533–2552 CrossRef CAS PubMed.
- V. A. Bloomfield, Biopolymers, 1991, 31, 1471–1481 CrossRef CAS PubMed.
- L. Guan-Ying, G. Rui-Lin, J. Liang-Nian and C. Hui, DNA condensation induced by metal complexes, Coord. Chem. Rev., 2014, 281, 100–113 CrossRef PubMed.
- E. Grueso, P. Perez-Tejeda, R. Prado-Gotor and C. Cerrillos, J. Phys. Chem. C, 2014, 118(8), 4416–4428 CAS.
- S. Marchetti and G. Onori, J. Phys. Chem. B, 2005, 109, 3676–3680 CrossRef CAS PubMed.
- E. Grueso, C. Cerrillos, J. Hidalgo and P. Lopez-Cornejo, Langmuir, 2012, 28(30), 10968–10979 CrossRef CAS PubMed.
- Z. Ting, X. Guiying, A. Mingqi, Y. Yanlian and W. Chen, Colloids Surf., A, 2012, 414, 33–40 CrossRef PubMed.
- J. P. García, E. Marrón, V. I. Martín, M. L. Moyá and P. Lopez-Cornejo, Colloids Surf., B, 2014, 118, 90–100 CrossRef PubMed.
- E. Grueso, E. Kuliszewska, R. Prado-Gotor, P. Perez-Tejeda and E. Roldan, Phys. Chem. Chem. Phys., 2013, 15, 20064–20074 RSC.
- R. Zana, Adv. Colloid Interface Sci., 2002, 97(1–3), 205–253 CrossRef CAS.
- A. Bhadani and S. Singh, Langmuir, 2009, 25, 11703–11712 CrossRef CAS PubMed.
- S. D. Wettig, R. E. Verrall and M. Foldvari, Curr. Gene Ther., 2008, 8(1), 9–23 CrossRef CAS.
- Y. He, Y. Shang, S. Shao, H. Liu and Y. Hua, J. Colloid Interface Sci., 2011, 358(2), 513–520 CrossRef CAS PubMed.
- L. Karlsson, M. C. van Eijk and O. Söderman, J. Colloid Interface Sci., 2002, 252(2), 290–296 CrossRef CAS PubMed.
- I. Badea, R. Verrall, M. Baca-Estrada, S. Tikoo, A. Rosenberg, P. Kumar and M. Foldvari, J. Gene Med., 2005, 7(9), 1200–1214 CrossRef CAS PubMed.
- R. Dias, M. Rosa, A. Canelas Paisa, M. Miguel and B. Lindman, J. Chin. Chem. Soc., 2004, 51, 447–469 CAS.
- A. Gonzalez-Perez and R. S. Diaz, Front. Biosci., Elite Ed., 2009, E1, 228–241 CAS.
- A. Gonzalez-Perez, Curr. Top. Med. Chem., 2014, 14, 766–773 CrossRef CAS.
- H. Fabre, N. Kamenka, A. Khan, G. Lindblom, B. Lindman and G. J. T. Tiddy, J. Phys. Chem., 1980, 84, 3428–3433 CrossRef CAS.
- Y. Han and Y. Wang, Phys. Chem. Chem. Phys., 2011, 13, 1939–1956 RSC.
- R. Oda, I. Huc and S. J. Candau, Chem. Commun., 1997, 21, 2105–2106 RSC.
- R. Oda, L. Bourdieu and M. Schmutz, J. Phys. Chem., 1997, 101(31), 5913–5916 CrossRef CAS.
- R. Oda, I. Huc, J.-C. Homo, B. Beinrich, M. Schmutz and S. Candau, Langmuir, 1999, 15, 2384–2390 CrossRef CAS.
- R. Atkin, V. S. J. Craig, E. J. Wanless and S. Biggs, J. Phys. Chem. B, 2003, 107, 2978–2985 CrossRef CAS.
- F. Secco, M. Venturini, T. Biver, F. Sanchez, R. Prado-Gotor and E. Grueso, J. Phys. Chem. B, 2010, 114, 4686–4691 CrossRef CAS PubMed.
- G. Felsenfeld and S. Z. Hirschman, J. Mol. Biol., 1965, 13, 409–419 CrossRef.
- E. Kuliszewska and L. Brecker, J. Surfactants Deterg., 2014, 17, 951–957 CrossRef CAS PubMed.
- B. P. Pozniak and E. Kuliszewska, Int. J. Mass Spectrom., 2013, 348, 29–38 CrossRef CAS PubMed.
- R. Zana, M. Benrraou and R. Rueff, Langmuir, 1991, 7, 1072–1075 CrossRef CAS.
- N. C. Garbett, P. A. Ragazzon and J. B. Chaires, Nat. Protoc., 2007, 2(12), 3166–3172 CrossRef CAS PubMed.
- N. Berova, K. Nakanishi and R. W. Woody, Circular Dichroism: Principles and Applications, Wiley-VCH, New York, 2nd. edn, 2000, p. 741 Search PubMed.
- N. Miyazawa, T. Sakaue, K. Yoshikawa and R. Zana, J. Phys. Chem., 2005, 122, 044902–044904 CrossRef PubMed.
- G. Cohen and H. Eisenberg, Biopolymers, 1969, 8, 45–55 CrossRef CAS.
- M. R. Beccia, T. Biver, A. Pardini, J. Spinelli, F. Secco, M. Venturini, N. Busto, P. Lopez-Cornejo, V. I. Martin Herrera and R. Prado Gotor, Chem. Asian J., 2012, 7(8), 1803–1810 CrossRef CAS PubMed.
- S. Neidle, Nucleic Acid Structure and Recognition, Oxford University Press, New York, 2002 Search PubMed.
- M. Monnot, O. Mauffret, E. Lescot and S. Fermandjian, Eur. J. Biochem., 1992, 204(3), 1035–1039 CrossRef CAS PubMed.
- M. F. Maestre and J. C. Wang, Biopolymers, 1971, 10(6), 1021–1030 CrossRef CAS PubMed.
- B. Alberts, D. Bray, J. Lewis, M. Raff, K. Roberts and J. D. Watson, Molecular Biology of the Cell, Galland, New York, 4th. edn. 1994 Search PubMed.
- X. L. Wang, X. H. Zhang, M. W. Cao, H. Z. Zheng, B. Xiao and Y. Wang, J. Phys. Chem. B, 2009, 113, 2328–2332 CrossRef CAS PubMed.
- A. Gopala Krishna, D. Vijay Kumar, B. M. Khan, S. K. Rawal and K. N. Ganesh, Biochim. Biophys. Acta, 1998, 1381, 104–112 CrossRef.
- L. F. Tietze, B. Krewer, F. Major and I. Schuberth, J. Am. Chem. Soc., 2009, 131, 13031–13036 CrossRef CAS PubMed.
- D. Meierhans, M. Sieber and R. K. Allemann, Nucleic Acids Res., 1997, 25, 4537–4544 CrossRef CAS PubMed.
- D. Sarkar, P. Das, S. Basak and N. Chattopadhyay, J. Phys. Chem. B, 2008, 112, 9243–9249 CrossRef CAS PubMed.
- W. J. Montigny, C. R. Houchens, S. Illenye, J. Gilbert, E. Coonrod, Y. C. Chang and N. H. Heintz, Nucleic Acids Res., 2001, 29, 1982–1988 CrossRef CAS PubMed.
- Y. Fang and J. H. Hoh, J. Am. Chem. Soc., 1998, 120, 8903–8909 CrossRef CAS.
- H. G. Hansma, R. L. Sinsheimer, M. Q. Li and P. Q. Hansma, Nucleic Acids Res., 1992, 20, 3585–3590 CrossRef CAS PubMed.
- J. Pelta, F. Livolant and J. L. Sikorav, J. Biol. Chem., 1996, 271(10), 5656–5662 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra03944d |
| This journal is © The Royal Society of Chemistry 2015 |