Versatility of divinylsulfone supports permits the tuning of CALB properties during its immobilization
Abstract
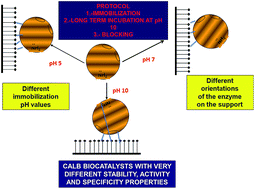
The lipase B from C. antarctica (CALB) has been immobilized on divinylsulfone (DVS) activated agarose beads under different conditions (pH 5–10). In the presence of 0.3% Triton X-100, the immobilization rate was rapid at pH 10 and the slowest one was at pH 5. Incubation at pH 10 for 72 h of the immobilized enzymes before blocking of the support with ethylenediamine permitted improvement of the enzyme stability. Enzyme features (activity, stability, specificity versus different substrates, effect of the pH on enzyme properties) were quite different on the different CALB preparations, suggesting the different orientation of the enzyme. The alkaline incubation produced an increase in enzyme activity with some substrates, and some of the DVS-CALB preparations exhibited a higher specific activity than the octyl-preparations. The indirect fluorescence spectrum of the different immobilized preparations confirmed that different structures of the CALB molecules were generated after immobilization.

 Please wait while we load your content...
Please wait while we load your content...