Co-delivery of siRNA and paclitaxel into cancer cells by hyaluronic acid modified redox-sensitive disulfide-crosslinked PLGA–PEI nanoparticles†
Abstract
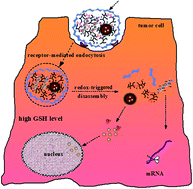
The main objective of our research was established in order to put forward a targeted anticancer co-delivery system for both micro-molecular chemotherapeutic drugs and small interfering RNA (siRNA). The drug delivery system can efficiently combine the advantages of the two therapeutic strategies with different mechanisms via their synergetic effects for cancer therapy. In this study, a cooperative nano-platform of docetaxel (DTX) and specific silencer select siRNA was developed by suppressing the cyclooxygenase-2 (COX-2) gene expression within one single nano-particle (NP) composed of poly(D,L-lactide-co-glycolide) (PLGA) bearing disulfide-linked reducible polyethyleneimine (PEIss) covered by hyaluronic acid (HA). PEIss was successfully synthesized by Michael addition of low molecular weight PEI (Mw = 1800 Da) with cystaminebisacrylamide (CBA). Docetaxel (DTX) was entrapped in the hydrophobic PLGA core, whereas negatively charged siRNA was adsorbed on the cationic shell of PEIss by electrostatic attraction. The redundant positive charge of PEIss allowed the introduction of HA onto the nanoparticles as active targeting groups. The whole complex was then delivered to both sensitive and resistant SGC-7901 gastric cancer cells that over-express CD44 receptors. This novel system was designated as HRPSP NPs (HA–PEIss–PLGA nanoparticles). HA–PEI–PLGA nanoparticles (HPP NPs) were also prepared to make a comparison. The electrophoresis confirmed the good stability of siRNA/HRPSP complex in the presence of PBS (pH 7.4), competitive heparin and RNase and remarkable sensitivity and reducibility to GSH. In in vitro cell transfection, siRNA/HRPSP complexes performed with higher transfection efficiency than siRNA/HPP and siRNA/RPSP and siRNA/PP complexes preferentially through caveolae-mediated endocytosis, which might be a desirable pathway to avoid the lysosomal degradation of delivered genes. Q-PCR and Western Blot experiments proved that COX-2 mRNA and COX-2 protein expressions were reduced to a statistically significant level in comparison with the control and the scrambled HRPSP/siRNA complex after treatment with the NPs/siRNA complex. In vivo, Dir/HRPSP/siRNA NPs complexes exhibited higher intensity at the tumor tissue compared to the non HA modified complexes. The biodistribution results demonstrated that DTX/HRPSP NPs could reduce DTX uptake particularly by the liver and lungs to limit the side effects while significantly increasing DTX accumulation in the tumors. Above these, HRPSP NPs were highly promising nano-carriers for the combinatorial delivery of siRNA and lipophilic anti-cancer drugs.

 Please wait while we load your content...
Please wait while we load your content...