Functionalization of nano-graphenes by chimeric peptide engineering
Gayong Shima,
Jaiwoo Leeb,
Jinyoung Kima,
Hee-Jung Leec,
Young Bong Kimc and
Yu-Kyoung Oh*a
aCollege of Pharmacy, Research Institute of Pharmaceutical Sciences, Seoul National University, Seoul, Republic of Korea. E-mail: ohyk@snu.ac.kr
bDepartment of Molecular Medicine and Biopharmaceutical Sciences, Graduate School of Convergence Science, Seoul National University, Seoul, Republic of Korea
cDepartment of Bio-industrial Technologies, Konkuk University, Seoul, Republic of Korea
First published on 8th May 2015
Abstract
Here, we report the non-covalent functionalization of reduced graphene oxide (rGO) nanosheets using chimeric peptides engineered to have a biologically functional sequence, a spacer sequence, and an rGO-binding sequence. As a model peptide with biological activity, the cell-penetrating peptide buforin IIb (Bu) was used. A stretch of seven consecutive phenylalanine residues (7F) was used as the rGO-binding sequence. The various effects of tetraglycine (4G) and tetra-aspartate (4D) as spacers between the biologically active Bu and the rGO-binding 7F sequences were compared. All the chimeric peptides had α-helical structures at the carboxyl-terminal sequence, showing a structural similarity to the α-helical structure of Bu alone. Free chimeric peptides composed of 7F-Bu, 7F4G-Bu, or 7F4D-Bu in solution exhibited cell-penetrating abilities similar to that of Bu alone. However, following attachment onto rGO nanosheets, the compositions of the chimeric peptides affected the biological activity of Bu. Following modification, the 7F4D-Bu chimeric peptide yielded a higher cellular uptake of the rGO nanosheets than the other chimeric peptides. The levels of cellular uptake of the rGO nanosheets modified with the chimeric peptides were further evaluated by measuring the photothermal effect after near-infrared laser irradiation. The cells treated with 7F4D-Bu-modified rGO showed the greatest increase in temperature upon irradiation, with the temperature reaching 58.3 °C. The 7F4D-Bu-modified rGO also exhibited the highest photothermal cell-killing activity upon near-infrared laser irradiation. Our results demonstrate the utility of chimeric peptide engineering for simple and facile one-step non-covalent modification of rGO. The chimeric peptide composed of 7F4D can be further used to tether various functional peptides onto rGO nanosheets.
Introduction
Graphene-based nanosheets have received wide attention due to their potential applications in biomedical fields,1,2 including drug delivery,3 tissue engineering,4 and biosensors.5 They have also been modified with polymers,6 lipids,7 and functional peptides8 in order to increase their versatility for biomedical applications. To functionalize graphene-based nanosheets with biologically active peptides, covalent and non-covalent methods have been used. Graphene-based nanosheets have been covalently conjugated with enzyme-sensitive peptides such as a caspase-3-cleavable peptide9 and a protease-sensitive peptide10 for detection of enzymes. Although non-covalent modifications are simpler processes than are covalent modifications, the direct non-covalent anchoring of a peptide onto a graphene-based nanosheet can disrupt the three-dimensional conformation of the peptide, hence limiting its biological activity.11,12There is thus a need to develop a non-covalent modification technology in which peptides can bind graphene and at the same time retain their three-dimensional structures and hence functions. In this study, we aimed to engineer a chimeric peptide that consists of a biologically functional peptide sequence, a spacer sequence and a sequence that binds reduced graphene oxide (rGO) nanosheets in order to carry out a simple single-step non-covalent functionalization. For binding rGO, we tested a heptaphenylalanine sequence. Phenylalanine has been reported to adsorb onto carbon nanotubes13 and graphenes14 by π–π interaction. As a model peptide with biological activity, we used the buforin IIb (Bu) peptide. Bu peptide, a synthetic analogue of buforin II,15 is known to have cell-penetrating activity and enhance the cellular delivery of nanomaterials.16,17
Here, we report that the combination of rGO binding sequences and spacer sequences in engineered chimeric peptides containing Bu influenced the biological activity of the functional part of the peptide following attachment onto rGO nanosheets. By making the appropriate choice, we were able to carry out a simple non-covalent modification of rGO nanosheets while maintaining the biological activity of the functional peptide.
Experimental details
Peptides
Four types of peptides were used in this study. The sequence for the buforin-derived peptide, Bu, was RAGLQFPVGRLLRRLLRRLLR. This peptide was modified with seven consecutive phenylalanine (7F) residues to form FFFFFFF-RAGLQFPVGRLLRRLLRRLLR (7F-Bu). To test the role of spacers, four glycine (4G) residues or four aspartic acid (4D) residues were placed between the 7F residues and the Bu sequence to form FFFFFFF-GGGG-RAGLQFPVGRLLRRLLRRLLR (7F4G-Bu), and FFFFFFF-DDDD-RAGLQFPVGRLLRRLLRRLLR (7F4D-Bu). The sequence of scrambled Bu as negative control was PRRGRQRGRFRLLLRRVLLLA (scBu). All peptides used in this study were purchased from Peptron Inc. (Daejeon, Republic of Korea). The secondary structures of the peptides were predicted by using the PEP-FOLD server18–20 (http://bioserv.rpbs.univ-paris-diderot.fr/services/PEP-FOLD). For fluorescence labeling of each peptide, 20 μmol of fluorescein 5(6)-isothiocyanate (FITC, Sigma-Aldrich, St. Louis, MO, USA) in 3 ml of anhydrous N,N-dimethylformamide Sigma-Aldrich) was added dropwise to a stirred solution of 20 μmol of each peptide and 40 μmol of N,N-diisopropylethylamine (Sigma-Aldrich) in 2 ml of anhydrous dimethyl sulfoxide (Sigma-Aldrich). The reaction mixture was stirred for 24 h at room temperature. The mixture was dialyzed against deionized water for 48 h. The resulting FITC-labeled peptides were lyophilized, and stored at 4 °C.Preparation of rGO nanosheets
rGO nanosheets were prepared by reducing graphene oxide (GO) nanosheets. GO nanosheets were prepared from graphite using Hummer's method with slight modification.6 Briefly, cold H2SO4 (23 ml) was combined with graphite powder (0.5 g, Sigma-Aldrich), KMnO4 (3 g, Sigma-Aldrich) and NaNO3 (0.5 g, Sigma-Aldrich), and stirred on ice. The resulting mixture was further stirred at 35 °C for 1 h. After addition of 46 ml of triple distilled water (TDW), the mixture was stirred at 90 °C for 1 h. The reaction was stopped with 140 ml of TDW and 10 ml of 30% H2O2. The reaction products were washed by repeated centrifugation, first with an aqueous 5% HCl solution, and then with TDW. Finally, the products were dispersed in TDW, sonicated, and centrifuged at 1600×g for 10 min. The supernatant containing GO nanosheets was collected and an extruder (Northern Lipid, British Columbia, Canada) was used to filter it through 0.2 μm polycarbonate membrane filters (Millipore Corp., Billerica, MA, USA). The resulting GO nanosheets were used for synthesis of rGO nanosheets. In brief, 2.0 ml of GO nanosheets (5 mg ml−1) in TDW was combined with 8.0 ml of TDW, 0.5 ml of ammonia solution (28 w/w% in water, Junsei Chemical, Tokyo, Japan), and 5.0 μl of hydrazine monohydrate (64 w/w% in water, Sigma-Aldrich). The resultant mixture was stirred at 80 °C for 10 min. After cooling down to room temperature, the excess hydrazine and ammonia were removed by dialysis (MWCO 100K; Spectrum Laboratories, Inc., Rancho Dominguez, CA, USA) against TDW. The obtained rGO nanosheets were stored at 4 °C until further use.Circular dichroism (CD) study
To determine the secondary structures of the Bu-derived peptides, CD spectra were obtained using a Chirascan-plus circular dichroism spectrometer (Applied Photophysics Ltd, Surrey, United Kingdom). The spectra were measured between the wavelengths of 190 and 260 nm in the presence of 50% (v/v) trifluoroethanol (Thermo Fisher Scientific Inc., Waltham, MA, USA) in phosphate-buffered saline (PBS). The α-helical content of the Bu-derived peptides was analyzed by using CDNN secondary structure analysis software (Applied Photophysics Ltd).Preparation of peptide-tethered rGO
To coat the surfaces of rGO with various buforin-derived peptides, rGO nanosheets in TDW (1 mg ml−1) were mixed with the same volume of Bu (50 μM), 7F-Bu (50 μM), 7F4G-Bu (50 μM), or 7F4D-Bu (50 μM). Unloaded peptides were removed by gel filtration through a Sephadex G-25M column (GE Healthcare, Piscataway, NJ, USA), yielding peptide-rGO nanosheets. The extent of peptide loading onto rGO or GO was determined by measuring the quenching of FITC-labeled peptides at a wavelength of 525 nm after adsorption onto nano-graphenes. The fluorescence intensity was measured using a fluorescence microplate reader (Gemini XS; Molecular Device, Sunnyvale, CA, USA). To test serum stability, 0.1 mg of peptide-tethered rGO was incubated in 0.8 ml of 100% mouse serum for 6 h. The stability in serum was measured by checking the formation of aggregates or precipitates of rGO.Atomic force microscopy (AFM)
AFM was used to measure topography, thickness, and lateral sizes of peptide-tethered rGO nanosheets. Precut silicon wafer sections (5 mm × 5 mm) were rinsed with ethanol for 30 min with sonication. Then the sections were silanized in a solution by mixing 3-aminopropyltriethoxysilane (Sigma-Aldrich) with TDW (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9, v/v) and one drop of hydrochloric acid (Sigma-Aldrich) for 30 min, and washed with TDW. The silanized wafer sections were immersed in solutions of rGO (10 μg ml−1) for 10 min and rinsed with TDW. Next, the wafer sections were dipped into buforin-derived peptide solutions (0.5 μM) for 10 min, and baked in a 70 °C oven for 20 min. Topography, thickness, and lateral sizes were measured using AFM (XE-100; Park Systems, Suwon, Republic of Korea) in non-contact mode.
9, v/v) and one drop of hydrochloric acid (Sigma-Aldrich) for 30 min, and washed with TDW. The silanized wafer sections were immersed in solutions of rGO (10 μg ml−1) for 10 min and rinsed with TDW. Next, the wafer sections were dipped into buforin-derived peptide solutions (0.5 μM) for 10 min, and baked in a 70 °C oven for 20 min. Topography, thickness, and lateral sizes were measured using AFM (XE-100; Park Systems, Suwon, Republic of Korea) in non-contact mode.
Size and zeta potential measurement
The lateral sizes of the peptide-rGO nanosheets were measured using dynamic light scattering. The rGO and peptide-rGO nanosheets were, in separate experiments, diluted with TDW and placed in an ELS-Z (Photal, Osaka, Japan). The hydrodynamic diameters of the nanosheets were determined by He–Ne laser (10 mW) light scattering, and the zeta potential values of rGO and peptide-rGO nanosheets were determined using an ELS-8000 instrument (Photal, Osaka, Japan). The samples were diluted with TDW and zeta potential values were determined by laser Doppler microelectrophoresis at an angle of 22°.Cell-penetrating ability of Bu-derived peptide
The ability of Bu-derived peptides to penetrate cells of a BT-20 breast cancer cell line (American Type Culture Collection, Rockville, MD, USA) was tested using flow cytometry. BT-20 cells were seeded onto cover glasses at a density of 8 × 104 cells per well in 24-well plates. In the next day, the cells were treated with FITC-labeled Bu, 7F-Bu, 7F4G-Bu, or 7F4D-Bu, each at a concentration of 10 μM. After 15 min incubation at 37 °C, the cells were harvested and washed thrice with cold PBS containing 2% fetal bovine serum. Then the cells were analyzed by using BD FACSCalibur flow cytometry and Cell Quest Pro software (BD Bioscience, San Jose, CA, USA).Cellular uptake test of peptide-rGO nanosheets
The cellular uptake of rGO with or without the various peptides was tested in BT-20 cells using confocal microscopy and flow cytometry. To visualize the cellular uptake, rGO was labeled with 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[poly (ethyleneglycol) 5000-Alexa Fluor®680 (DSPE-PEG5000-Alexa Fluor®680), as described previously.6 BT-20 cells were seeded onto cover glasses at a density of 8 × 104 cells per well in 24-well plates. The next day, the cells were treated with DSPE-PEG5000-Alexa Fluor®680-labeled plain rGO or peptide-rGO nanosheets at a concentration of 10 μM of each peptide and 80 μg ml−1 of the rGO nanosheets. After 15 minutes of incubation, the cells were washed and fixed with 4% paraformaldehyde in PBS for 15 min, and stained with 4′,6-diamidino-2-phenylindole dihydrochloride (Sigma-Aldrich). The fluorescence intensity of cellular Alexa Fluor®680 was observed using a confocal laser scanning microscope (LSM 5 Exciter; Carl Zeiss, Inc., Jena, Germany). For flow cytometry, the cells were harvested and washed thrice with cold PBS containing 2% fetal bovine serum. Then the cells were analyzed by BD FACSCalibur flow cytometry using Cell Quest Pro software (BD Bioscience).Laser irradiation of cells treated with peptide-rGO nanosheets
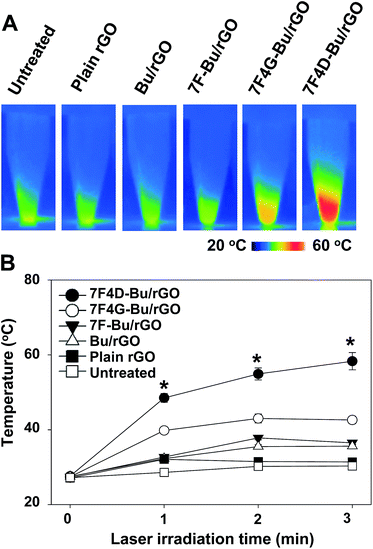
To test the photothermal effect of peptide-rGO nanosheets upon laser irradiation, the cells were treated with various peptide-rGO nanosheets, and irradiated with a near infrared (NIR) laser. BT-20 cells were seeded onto 12-well plates at a density of 1 × 105 cells per well. The following day, cells were treated with plain rGO or peptide-rGO nanosheets at a concentration of 10 μM of peptide and 80 μg ml−1 of nanosheets. After a 15 min incubation, the cells were washed twice with cold PBS and re-suspended in complete RPMI-1640 media. The cell suspensions were irradiated with an 808 nm-wavelength continuous-wave NIR diode laser (BWT Beijing LTD, Beijing, China) with an output power of 1.2 W. The temperature and thermal images of the graphene-based suspensions during laser irradiation were recorded using an infrared thermal imaging system every 60 seconds (FLIR T420, FLIR Systems Inc., Danderyd, Sweden). Immediately after irradiation, the cells were diluted 5-fold using complete RPMI-1640 media and transferred to 96-well plates for the cell viability assays.Quantitative cell viability assay following laser irradiation
The viability of the cells treated with peptide-rGO and laser irradiation was quantified using fluorescent staining of live/dead cells (Invitrogen Corp., Carlsbad, CA, USA) and Cell Counting Kit-8™ (CCK8, Dojindo Molecular Technologies, Inc., Rockville, MD, USA). Briefly, the cells were washed twice with PBS, and added with 200 μl of a staining solution (2 μM of calcein and 2 μM of ethidium homodimer-1) for live and dead cell identification. After 10 min, the cells were washed twice with PBS and observed under a fluorescence microscope (Leica DM IL; Leica, Wetzla, Germany). For the CCK8 assay, 20 μl of CCK-8 (water-soluble tetrazolium salt) solution was added to each well for 30 min, and absorbance was measured at 450 nm using a microplate reader (Sunrise-Basic TECAN, Männedorf, Switzerland). The cell viability in each group was expressed as a percentage of that in control cells.Statistics
ANOVA was used for statistical evaluation of experimental data, using the Student–Newman–Keuls test as a post-hoc test. All statistical analyses were carried out using SigmaStat software (version 3.5, Systat Software, Richmond, CA, USA), and a p-value < 0.05 was considered significant.Results and discussion
Rationale of peptide-tethered rGO nanosheets
Various Bu-derived peptides were coated onto the surfaces of rGO. The predicted adsorption patterns of Bu-derived peptides on rGO are shown in Fig. 1. To provide the functional peptide with conformational flexibility, an oligoglycine was often used as a spacer between anchoring and active peptide domains.21,22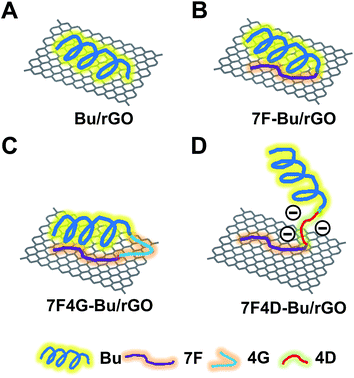 | ||
| Fig. 1 Diagrams of the peptide-tethered rGO. Schematic illustrations of Bu/rGO (A), 7F-Bu/rGO (B), 7F4G-Bu/rGO (C), and 7F4D-Bu/rGO (D) are shown. | ||
In this study, a stretch of 7F residues was used as a moiety to non-covalently anchor the Bu peptide to rGO nanosheets (Fig. 1D). This simple engineering of the peptide sequence avoids the multi-step covalent modification process for anchoring of functional peptides. Various approaches have been investigated to anchor peptides onto graphene-based nanosheets.9,23 A covalent anchorage of a peptide was done by conjugating protease-cleavable peptide to the surfaces of GO nanosheets.9 In another study, interleukin-13 receptor-binding peptide was covalently linked to a pegylated mesoporous silica-coated graphene nanosheet.23
Noncovalent anchoring has been mainly done using lipids or peptide moieties. A PEG lipid conjugate of RGD was used to anchor the hydrophobic lipid moiety to rGO.7 Recently, the nonapeptide HNWYHWWPH was used to attach elastin-like polypeptides consisting of 250 amino acids to graphene nanosheets.8 However, there have been few studies on rGO-binding peptides that anchor relatively small functional peptides such as buforin without affecting their biological properties.
The driving force behind the adsorption of the 7F residue peptide by rGO can be attributed to the hydrophobic interaction between aromatic rings of phenylalanine and the planar surface of rGO. Indeed, the parallel orientation of aromatic rings and planar sheet of graphene was reported to contribute to non-covalent π–π interactions of the aromatic phenylalanine side chain with the graphene nanosheets.14 Moreover, it has been reported that aromatic side chains of peptides can maximize the π–π interactions with graphene surfaces.24
Biophysical characterization and cell-penetrating ability of buforin-derived peptides
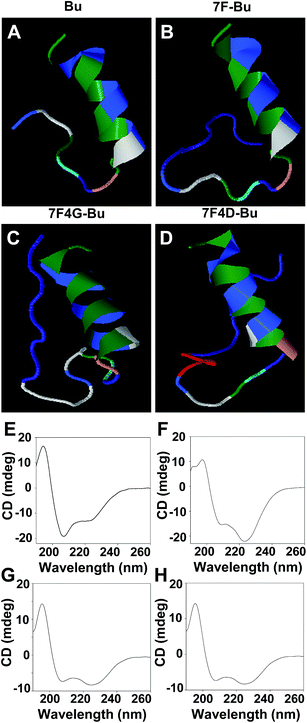
The modification of the Bu peptide with 7F residues did not affect the α-helical structure and cell-penetrating ability of the Bu peptide. Secondary structure predictions and CD spectra indicate that the typical α-helix structure of the buforin motif in the Bu peptide (Fig. 2A) and the other three peptides, 7F-Bu (Fig. 2B), 7F4G-Bu (Fig. 2C), and 7F4D-Bu (Fig. 2D), were similar (Fig. 2).We observed CD peaks at 208 nm and 222 nm for Bu peptide (Fig. 2E), the 7F-Bu (Fig. 2F), 7F4G-Bu (Fig. 2G), and 7F4D-Bu (Fig. 2H) peptides. It has been reported that two peaks at 208 nm and 222 nm at CD spectra indicate the presence of α-helical structure.25,26 The previous study supports that Bu-derived peptides maintain distinct α-helical structure of Bu. Note that these CD spectra were obtained in the presence of 50% trifluoroethanol, and this condition is known to mimic the lipid membrane.25
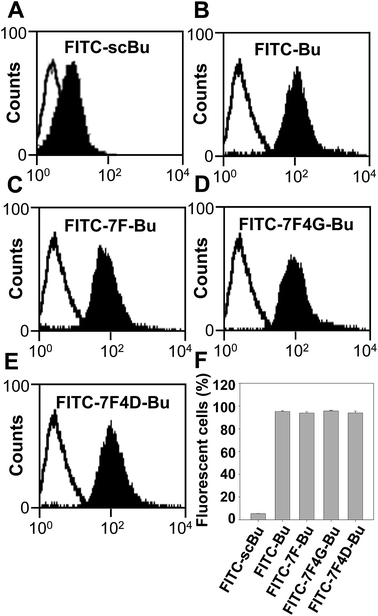
In addition, the modifications of the Bu peptide sequence in this study did not affect the ability of the resulting peptides to penetrate BT-20 cells: the fluorescence intensities of the cells treated with FITC-Bu peptide (Fig. 3B and F) did not significantly differ from those of the cells treated with FITC-7F-Bu (Fig. 3C and F), FITC-7F4G-Bu (Fig. 3D and F), and FITC-7F4D-Bu (Fig. 3E and F) peptides.
The driving force behind the adsorption of the 7F residue peptide by rGO can be attributed to the hydrophobic interaction between aromatic rings of phenylalanine and the planar surface of rGO. Indeed, the parallel orientation of aromatic rings and planar sheet of graphene was reported to contribute to non-covalent π–π interactions of the aromatic phenylalanine side chain with the graphene nanosheets.14 Moreover, it has been reported that aromatic side chains of peptides can maximize the π–π interactions with graphene surfaces.24
Characterization of peptide-tethered rGO nanosheets
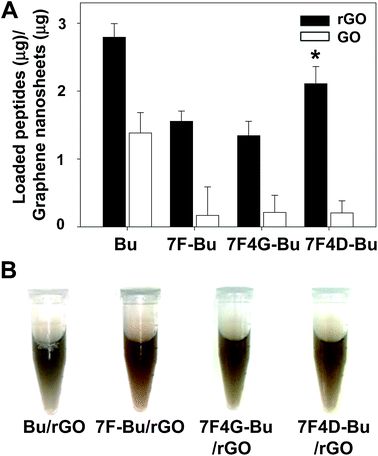
Among various Bu-derived peptides, 7F4D-Bu revealed the highest loading efficiency on rGO nanosheets (Fig. 4A). The higher loading efficiency of 7F4D-Bu than other Bu-derived peptides could be due to the uplifted configuration which allows the occupation of smaller spaces on rGO nanosheets. Regardless of peptide sequences, rGO showed higher loading of peptides than GO nanosheets. The lower loading of peptides on GO nanosheets might be contributed by the limited planar surfaces of GO nanosheets as compared to rGO nanosheets. Previously, it has been reported that the planar surfaces of GO nanosheets were not enough to facilitate stacking interaction with hydrophobic peptides.7,27Moreover, coating the rGO surfaces with Bu or Bu-derived peptides provided the stability of nano-graphene dispersion in serum. Regardless of the peptide types, peptide-coated nano-graphenes did not show aggregation or precipitation up to 6 h of incubation in mouse serum (Fig. 4B).
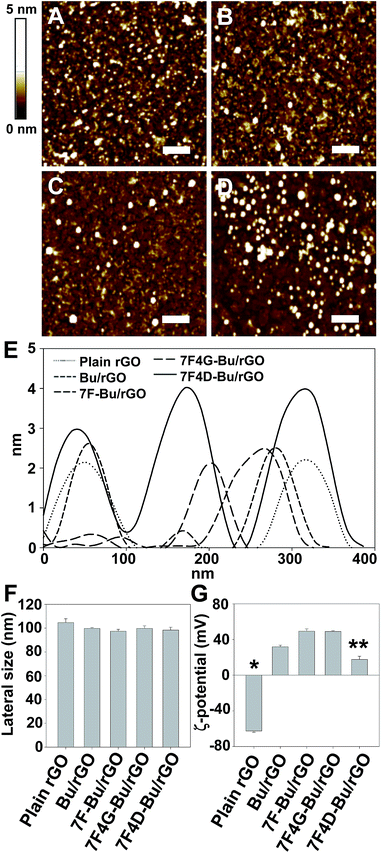
The adsorption of buforin-derived peptides on the rGO nanosheet did not affect the sizes of the nanosheets, but did affect their thickness and zeta potentials. The AFM imaging showed a uniform topography of rGO nanosheets after peptide modification (Fig. 5A–D). The thickness of most peptide-tethered rGOs including rGO nanosheets was approximately ∼2 nm (Fig. 5E). The 7F4D-Bu-rGO showed slight increase in thickness from ∼2 nm to 4 nm, which supports a role of 4D as a charge repulsion spacer.
The lateral size of the rGO nanosheet remained unaffected by the binding of buforin-derived peptides (Fig. 5F). In contrast, the surface charge of peptide-tethered rGO nanosheets depended on the type of peptide (Fig. 5G). Whereas plain rGO is negatively charged, the adsorption of Bu increased the zeta potentials of the rGOs to positive values due to the positive charge of Bu motif (Fig. 5G). Among the various peptide-tethered rGO nanosheets, 7F4D-Bu-rGO showed the lowest zeta potential values. The lowest zeta potential can be explained by the presence of negatively charged 4D spacer on the surfaces of rGO nanosheets.
Cellular uptake of peptide-tethered rGO nanosheets
Coating the surface of rGO nanosheets with buforin-derived peptides did affect the cellular delivery of such nanosheets that were also loaded with DSPE-PEG5000-Alexa Fluor®680. This lipid was loaded onto rGO nanosheets in order to visualize the cellular uptake of the nanosheets. For the cellular uptake study, rGO was loaded with this lipid and each type of peptide. Compared to untreated cells, the cells treated with plain rGO showed negligible increase in fluorescence of the DSPE-PEG5000-Alexa Fluor®680 on the rGO nanosheets (Fig. 6B and G). In contrast, surface coating of rGO with buforin-derived peptides did enhance the cellular delivery of the fluorescent lipid-loaded rGO nanosheets. The greatest increase in the cellular delivery of rGO was observed for 7F4D-Bu/rGO (Fig. 6F and G), followed by 7F4G-Bu/rGO (Fig. 6E and G), 7F-Bu/rGO (Fig. 6D and G), and then Bu/rGO (Fig. 6C and G), which displayed the smallest increase.We observed that the biological activity of an rGO-anchored functional peptide was better retained by the use of 4D as the spacer (Fig. 6F) than when 4G was used as the spacer (Fig. 6E). The enhanced biological properties of 7F4D-Bu that we observed could be due to the negative surface charges of rGO that have carboxyl groups at the edges of the nanosheets.28 Charge repulsion between the anionic 4D spacer and the negatively charged rGO could cause the tethered functional Bu peptide to become oriented away from the rGO nanosheet, which in turn would allow it to exert its biological function.
Photothermal effect of peptide-tethered rGO nanosheets
In line with the different levels of cellular uptake of the different peptide-tethered rGOs, the photothermal effect also varied depending on the type of buforin-derived peptide bound on the rGO nanosheets. Those cells treated with buforin-derived peptides containing a spacer peptide between 7F and Bu showed the greatest increases in temperature when irradiated with an NIR laser and the 4D spacer yielded a greater increase than did the 4G spacer. The temperature of the NIR-irradiated cells treated with 7F4D-Bu/rGO reached as high as 58.3 ± 2.3 °C (Fig. 7A and B). In the case of the cells treated with 7F4G-Bu/rGO, the temperature reached 42.6 ± 1.0 °C. The NIR-induced temperature increase of the cells treated with 7F-Bu/rGO as well as the increase for cells treated with Bu/rGO was significantly less, with the temperature remaining below 40 °C.Photoresponsive cell-killing effect of 7F4D-Bu/rGO nanosheets
The substantial increase of temperature upon NIR irradiation could exert cell-killing effects in cancer cells treated with 7F4D-Bu/rGO nanosheets. The viability of the cells treated with various buforin-derived peptide-tethered rGOs was visualized by fluorescence staining of live cells with calcein and dead cells with ethidium homodimer-1. The live/dead cell staining images showed that the cells treated with 7F4D-Bu/rGO were significantly killed upon NIR irradiation (Fig. 8F). Quantitation of cell viability revealed that only 33.2 ± 0.4% of the cells treated with 7F4D-Bu/rGO remained viable upon NIR irradiation (Fig. 8G). The other peptides, in contrast, did not yield any significant decrease of cell viability after NIR irradiation.Our results with the use of the 7F4D motif for tethering of Bu onto rGO suggest that this motif can be applied to the delivery of other functional peptides including tumor targeted ligands for anticancer therapy, and to the peptide-based biosensors for biomarker detection and diagnosis.
Since peptide-modified rGO nanosheets can be used for detection of enzymes,10,29 antibodies,30 and cells,31 the 7F4D motif can be used to engineer such diagnostic peptides without cumbersome chemical modification. Moreover, the use of oligopeptides of seven residues as an anchoring moiety can decrease the costs of the peptide engineering.
Conclusions
In this study, we provided evidence that the 7F4D motif can be used for anchoring the biologically functional peptide Bu on rGO without hampering its biological function. The use of 4D contributed to minimizing the undesirable interaction of Bu with rGO, resulting in the substantially enhanced cell-penetrating ability of Bu, as compared to native Bu, or the peptide containing the 4G spacer. These results suggest the potential for extensive applications of the 7F4D motif for non-covalent tethering of functional peptides onto rGO for protein delivery as well as for a broad spectrum of peptide-based biosensors on rGO nanosheets.Acknowledgements
This work was supported by research grants from the Ministry of Science, ICT and Future Planning (NRF-2014K2A2A4001156; NRF-2015R1A2A1A01005674), and from the Technology Innovation Program (Grant no. 10050648, Development of Advanced Technologies of Raw Materials for HIV Diagnosis) funded by the Ministry of Trade, Industry & Energy, Republic of Korea.Notes and references
- D. Bitounis, H. Ali-Boucetta, B. H. Hong, D. H. Min and K. Kostarelos, Adv. Mater., 2013, 25, 2258–2268 CrossRef CAS PubMed.
- S. Goenka, V. Sant and S. Sant, J. Controlled Release, 2014, 173, 75–88 CrossRef CAS PubMed.
- G. Shim, J. Y. Kim, J. Han, S. W. Chung, S. Lee, Y. Byun and Y.-K. Oh, J. Controlled Release, 2014, 189, 80–89 CrossRef CAS PubMed.
- Y. Wang, W. C. Lee, K. K. Manga, P. K. Ang, J. Lu, Y. P. Liu, C. T. Lim and K. P. Loh, Adv. Mater., 2012, 24, 4285–4290 CrossRef CAS PubMed.
- W. Miao, G. Shim, S. Lee and Y.-K. Oh, Biomaterials, 2014, 35, 4058–4065 CrossRef CAS PubMed.
- W. Miao, G. Shim, C. M. Kang, S. Lee, Y. S. Choe, H.-G. Choi and Y.-K. Oh, Biomaterials, 2013, 34, 9638–9647 CrossRef CAS PubMed.
- J. T. Robinson, S. M. Tabakman, Y. Liang, H. Wang, H. Sanchez Casalongue, D. Vinh and H. Dai, J. Am. Chem. Soc., 2011, 133, 6825–6831 CrossRef CAS PubMed.
- E. Wang, M. S. Desai and S.-W. Lee, Nano Lett., 2013, 13, 2826–2830 CrossRef CAS PubMed.
- H. Wang, Q. Zhang, X. Chu, T. Chen, J. Ge and R. Yu, Angew. Chem., Int. Ed. Engl., 2011, 50, 7065–7069 CrossRef CAS PubMed.
- S.-Y. Kwak, J.-K. Yang, S.-J. Jeon, H.-I. Kim, J. Yim, H. Kang, S. Kyeong, Y.-S. Lee and J.-H. Kim, Adv. Funct. Mater., 2014, 24, 5119–5128 CrossRef CAS PubMed.
- T. Alava, J. A. Mann, C. C. Théodore, J. J. Benitez, W. R. Dichtel, J. M. Parpia and H. G. Craighead, Anal. Chem., 2013, 85, 2754–2759 CrossRef CAS PubMed.
- J. A. Mann and W. R. Dichtel, ACS Nano, 2013, 7, 7193–7199 CrossRef CAS PubMed.
- L. Piao, Q. Liu, Y. Li and C. Wang, J. Nanosci. Nanotechnol., 2009, 9, 1394–1399 CrossRef CAS PubMed.
- C. Rajesh, C. Majumder, H. Mizuseki and Y. Kawazoe, J. Chem. Phys., 2009, 130, 124911 CrossRef PubMed.
- M. E. Bustillo, A. L. Fischer, M. A. LaBouyer, J. A. Klaips, A. C. Webb and D. E. Elmore, Biochim. Biophys. Acta, 2014, 1838, 2228–2233 CrossRef CAS PubMed.
- S. A. Jang, H. Kim, J. Y. Lee, J. R. Shin, D. J. Kim, J. H. Cho and S. C. Kim, Peptides, 2012, 34, 283–289 CrossRef CAS PubMed.
- W. P. Verdurmen and R. Brock, Trends Pharmacol. Sci., 2011, 32, 116–124 CrossRef CAS PubMed.
- P. Thévenet, Y. Shen, J. Maupetit, F. Guyon, P. Derreumaux and P. Tufféry, Nucleic Acids Res., 2012, 40, W288–W293 CrossRef PubMed.
- J. Maupetit, P. Derreumaux and P. Tuffery, Nucleic Acids Res., 2009, 37, W498–W503 CrossRef CAS PubMed.
- J. Maupetit, P. Derreumaux and P. Tufféry, J. Comput. Chem., 2010, 31, 726–738 CAS.
- Y. Cui, S. N. Kim, S. E. Jones, L. L. Wissler, R. R. Naik and M. C. McAlpine, Nano Lett., 2010, 10, 4559–4565 CrossRef CAS PubMed.
- M. S. Mannoor, H. Tao, J. D. Clayton, A. Sengupta, D. L. Kaplan, R. R. Naik, N. Verma, F. G. Omenetto and M. C. McAlpine, Nat. Commun., 2012, 3, 763 CrossRef PubMed.
- Y. Wang, K. Wang, J. Zhao, X. Liu, J. Bu, X. Yan and R. Huang, J. Am. Chem. Soc., 2013, 135, 4799–4804 CrossRef CAS PubMed.
- S. N. Kim, Z. Kuang, J. M. Slocik, S. E. Jones, Y. Cui, B. L. Farmer, M. C. McAlpine and R. R. Naik, J. Am. Chem. Soc., 2011, 133, 14480–14483 CrossRef CAS PubMed.
- G. Hao, Y.-H. Shi, Y.-L. Tang and G.-W. Le, J. Microbiol., 2013, 51, 200–206 CrossRef CAS PubMed.
- D. Ghosh, P. K. Singh, S. Sahay, N. N. Jha, R. S. Jacob, S. Sen, A. Kumar, R. Riek and S. K. Maji, Sci. Rep., 2015, 5, 9288 CrossRef PubMed.
- O. C. Compton and S. T. Nguyen, Small, 2010, 6, 711–723 CrossRef CAS PubMed.
- H. Huang, P. Chen, X. Zhang, Y. Lu and W. Zhan, Small, 2013, 9, 1397–1404 CrossRef CAS PubMed.
- S. K. Bhunia and N. R. Jana, ACS Appl. Mater. Interfaces, 2011, 3, 3335–3341 CAS.
- Y.-M. Wu, Y. Cen, L.-J. Huang, R.-Q. Yu and X. Chu, Chem. Commun., 2014, 50, 4759–4762 RSC.
- Z. Wang, P. Huang, A. Bhirde, A. Jin, Y. Ma, G. Niu, N. Neamati and X. Chen, Chem. Commun., 2012, 48, 9768–9770 RSC.
| This journal is © The Royal Society of Chemistry 2015 |