Assessment of the therapeutic potential of hesperidin and proteomic resolution of diabetes-mediated neuronal fluctuations expediting Alzheimer’s disease
Sapna Khowal†
a,
Malik M. A. Mustufa†a,
Naveen K. Chaudharya,
Samar Husain Naqvib,
Suhel Parvezc,
Swatantra K. Jaind and
Saima Wajid*a
aDepartment of Biotechnology, Faculty of Science, Hamdard University (Jamia Hamdard), New Delhi – 110 062, India. E-mail: swajid@jamiahamdard.ac.in; wajid_saima@hotmail.com; Tel: +91-9899823898
bMolecular Diagnostics, Genetix Biotech Asia, New Delhi – 110 015, India
cDepartment of Medical Elementology and Toxicology, Faculty of Science, Hamdard University (Jamia Hamdard), New Delhi – 110 062, India
dDepartment of Biochemistry, Hamdard Institute of Medical Sciences and Research, Hamdard University (Jamia Hamdard), New Delhi – 110 062, India
First published on 6th May 2015
Abstract
Alzheimer’s disease (AD) has been proposed as type III diabetes mellitus (DM). Prognosis and early stage diagnosis of AD is essentially required in diabetic patients to avoid extensive irreversible neuronal damage. Also, simple medication regimes including therapeutics for maintaining glucose levels and simultaneous resistance to neuronal damage are quintessential. In the present study, secretome and hippocampus proteome modulations were investigated for serum-based markers that have correlations with DM-mediated neurological alterations which extend to AD. Concurrently, the therapeutic effect of hesperidin on DM and DM-mediated neurodegeneration was investigated. Twenty one male Wistar rats were separated into three groups, namely, healthy control, diabetic (65 mg kg−1 STZ i.p., single) and diabetic administered with hesperidin (STZ i.p. + 50 mg kg−1 hesperidin orally, for four weeks). Secretome and hippocampus proteome profiling was accomplished by two dimensional electrophoresis, and proteins showing differential expression were characterized by MALDI-TOF MS PMF and validated by relative expression analysis. APO A-IV and secretory AGK were found to have prognostic and/or diagnostic potential in detecting the early stage of DM-associated AD. A novel protein, ‘WajidSaima_Diabetes protein’ (WSDP), was found to have a probable role in neural homeostasis. The therapeutic potential of hesperidin in DM and DM-mediated neuronal fluctuations has successfully been determined through proteomic resolution. Our study emphasizes the DM-mediated neuronal fluctuations that expedite AD.
Introduction
Diabetes mellitus (DM) is a frequently reported ailment.1–5 The global incidence of DM in the year 2000 was 171 million and by 2030 the total number of cases has been estimated to increase up to 366 million.6 In India, the prevalence of DM was 31.7 million (2000) and by 2030 the total number of cases has been estimated to increase up to 79.4 million.7 In accordance with an Indian study on diabetes, conducted by the Indian Council of Medical Research (ICMR-INDIAB, 2011), approximately 62.4 million people in India were suffering with DM and within two decades, the number of sufferers is expected to rise to over 100 million.8 Diabetes mellitus is a complex ailment comprising of heterogeneous disorders arising due to insulin deficiency, impaired insulin action or both, resulting in hyperglycemia and glucose intolerance.9In the medication regime of DM, chemical drugs are available which specifically target hyperglycemia, causing a lowering of the blood glucose levels to an extent that normal or near to normal blood glucose levels are achieved in DM patients.10,11 Antidiabetic drugs have multiple ill effects on key organs, to wit, heart, brain, kidneys, eyes, liver, etc.10,11 These pernicious and irreversible side effects increase health complexities in diabetes patients, leading to an incorporation of medicine for secondary diseases in the existent treatment regime. Current antidiabetic drugs are inefficient and have multiple ill effects.12 Development of simple medication systems, including therapeutics for maintaining glucose levels in a normal range and simultaneous resistance of secondary damages to organs, is quintessential. Hesperidin (HP), an abundant flavonone glycoside found in citrus fruits, is involved in plant defense mechanisms.13,14 HP reduces cholesterol,15 blood pressure16 and bone density loss,17 and carries significant anti-inflammatory and analgesic ramifications.18,19 HP exhibits anticancer activity20 and can penetrate the blood–brain barrier.21 HP possesses antioxidant and neuroprotective attributes on the brain tissue against the hyperglycemia-mediated diabetic oxidative damage in STZ-DM rat models.14 HP oral administrations significantly diminish the elevated levels of oxidative stress and neurotoxicity biomarkers induced during STZ-mediated diabetes.14 Conjointly, depleted activities of both enzymatic and non-enzymatic antioxidants get restored.14 Antidiabetic attributes of HP mediated by its antihyperglycemic and antidyslipidemic efficacies have been reported in STZ-DM rodent models.14,22,23 But its potential in controlling diabetes associated neuronal complications needs exploration.
Neurological impairments in DM is a sign of ongoing neurodegenerative pathologies.24 Several prospective studies have perceived DM as a perilous factor for the decline of cognitive functions and the eventuation of Alzheimer’s disease (AD).25 Researchers have evaluated the threat of developing AD to be approximately doubled in DM.26 Type 1 diabetic patients are at an increased risk of damaged cognitive functions. Additionally, type 2 diabetic patients have an increased risk of developing AD in their middle and later lives.27 Conjointly, a pre-diabetic condition called borderline diabetes has been found to aggrandize the threat of developing dementia and AD.28 The risk of developing Alzheimer’s disease (AD) in DM is so strong that it has been categorized as type III diabetes mellitus (DM).29 Both AD and DM have interlinked pathologies.30,31
Deposition of amyloid occurs in target organs, namely, Aβ and tau in AD brains, and amylin in the pancreatic islets of type-2 diabetes.32 Moreover, insulin deficiency in STZ-mediated DM mice results in hyperphosphorylation of tau proteins.33–35 Also, in AD, insulin signalling and glucose metabolism pathways become flawed in the brain.36–38 The molecular links existent between them need to be investigated further. An increased load of insoluble Aβ (amyloid beta) plaques, soluble Aβ oligomers, and neurofibrillary tangles are existent in AD brains.31,39–43 Aβ, a peptide containing 39–43 amino acids, comes into existence after the sequential cleavage of amyloid precursor protein (APP) by beta and gamma secretases; the cellular functional operations conducted by APP in a healthy brain are still enigmatic.31,44–46 The neurofibrillary tangles constituting hyper-phosphorylated ‘tau’ proteins, are damaging to neural tissues.47 Detection of such proteinaceous plaques and tangles in the neural tissue is a late stage diagnostic strategy for AD. By the time plaques and tangles are visible, significant irreversible neural damage has already been done.24,48 Development of serum-based methods may be beneficial in the prognosis and early stage diagnosis of AD in diabetic patients.
Streptozotocin (STZ) is a glucosamine–nitrosourea compound.14 STZ is being used to treat cancers of the Islets of Langerhans (U.S. Food and Drug Administration).13,49–52 The colossal pertinence of STZ is found in animal models of diabetes for experimental investigations.53 Intraperitoneal (i.p.) administration of STZ is used for the induction of DM in rodent models, by virtue of its toxic attributes for insulin-secreting beta cells.14 Intracerebroventricular (i.c.v.) administration of STZ causes neurological pathologies that are similar to those observed in sporadic AD, which is useful for developing animal models to conduct studies on the early pathophysiological alterations prevalent in AD.54–57 In the present study, the therapeutic effect of HP in DM and DM-associated neurological complications was investigated. Concurrently, simultaneous modulations in the secretome and hippocampus proteome of the STZ-DM rat model were investigated for serum-based biomarkers that have correlations with neurological alterations extending into AD. The objectives comprised of (i) the evaluation of the protective role of HP in STZ-DM by blood glucose monitoring, (ii) the evaluation of the protective role of HP in STZ-DM-mediated neuronal damage by proteomic techniques and validation by relative gene expression using Real Time PCR, and (iii) to find secretome based biomarkers corresponding to STZ-DM neuronal fluctuations advancing into AD.
Methods
Ethics Statement
The experimental plan was approved by the Institutional Animal Ethics Committee (Hamdard University). The study was conducted in accordance with the guidelines given by the Committee for the Purpose of Control and Supervision of Experiments on Animals (Ministry of Environment & Forests, India).Treatment schedule
Twenty one male Wistar rats weighing between 270 and 300 g were procured from the Central Animal House Facility of Jamia Hamdard (Hamdard University). Animals were acclimatized for a week before starting the treatment. They were kept in an environment with a temperature of 25 ± 2 °C, relative humidity at 45–55% and at a photoperiod of 12 h light/dark cycles. A standard pellet rodent diet and water were provided to the animals ad libitum. One month diabetic animal models were prepared according to Parvez et. al. (our group).14 Twenty one animals were divided into three groups with seven animals per group, as follows:(i) Group I (control): non-diabetic. Animals were administered orally with normal saline.
(ii) Group II (toxicant): diabetic. Animals were administered intraperitoneally with a single injection of STZ (65 mg kg−1 b. w.−1).
(iii) Group III (toxicant + protectant): Diabetic animals were administered with HP. Animals were administered intraperitoneally with a single injection of STZ (65 mg kg−1 b. w.−1). After accomplishing a hyperglycemic state, animals were administered orally with HP (50 mg kg−1 b. w.−1) daily for four weeks.
STZ was dissolved in sterile milliQ water and HP (50 mg kg−1 b. w.−1) was dissolved in 0.5% w/v sodium carboxymethyl cellulose (CMC) solution. The administered doses of STZ and HP were based on previously published reports.14,58,59 A hyperglycemic state was accomplished three days after STZ i.p. administration.14 Hence, the establishment of diabetes was assured by a blood glucose assessment 3 days after STZ administration, and the animals that maintained blood glucose levels significantly higher than the controls were considered diabetic. The treatment schedule culminated on the 31st day after STZ administration.
Blood glucose monitoring
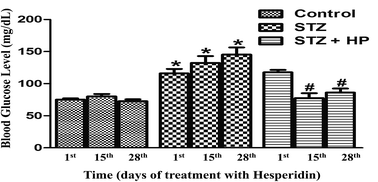
Fasting blood glucose levels were evaluated on the 4th, 18th and 31st day after STZ administration, which respectively corresponds to the 1st, 15th and 28th day after HP treatment. This further corresponds to the conditions of no HP treatment, two and four weeks of HP treatment respectively. The results were expressed as mean ± standard error (SE). All data were analyzed using analysis of variance (ANOVA) followed by Tukey’s test; values of p < 0.05 were considered significant. All the statistical analyses were performed using Graph pad Prism 5 software (Graph Pad Software Inc.).Proteomic analysis
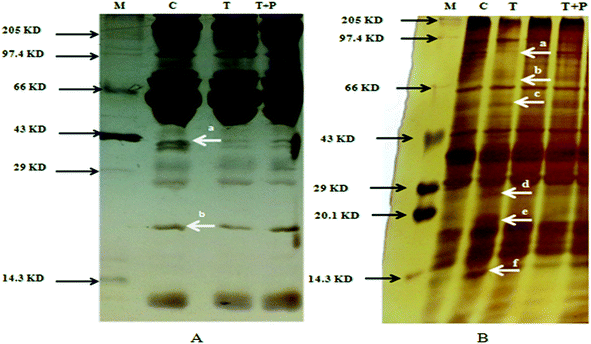
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 V h.61 After isoelectric focusing, the strips were equilibrated in equilibration buffer I for 15 min, followed by equilibration buffer II for another 15 min. The composition of equilibration buffer I was 6 M urea, 75 mM Tris pH 8.8, 29.3% glycerol (v/v), 2% SDS (w/v), 0.002% bromophenol blue (w/v) and 2% DTT (w/v).61 Equilibration buffer II comprised of 6 M urea, 75 mM Tris pH 8.8, 29.3% glycerol (v/v), 2% SDS (w/v), 0.002% bromophenol blue (w/v) and 135 mM IAA.61 Equilibrated IPG strips were placed on 12% homogenous SDS polyacrylamide gel (14 × 16 cm) and sealed with low melting point (1%) agarose for two-dimensional gel electrophoresis at a constant current (180 V) in a running buffer (Tris–glycine-SDS, pH 8.3).61 The two-dimensional gels were silver stained using standard protocols.61 The spot patterns obtained were analyzed using PD Quest 2D analysis software (Bio-Rad) and the spots of interest were characterized by MALDI TOF MS peptide mass fingerprint analysis.61,64
000 V h.61 After isoelectric focusing, the strips were equilibrated in equilibration buffer I for 15 min, followed by equilibration buffer II for another 15 min. The composition of equilibration buffer I was 6 M urea, 75 mM Tris pH 8.8, 29.3% glycerol (v/v), 2% SDS (w/v), 0.002% bromophenol blue (w/v) and 2% DTT (w/v).61 Equilibration buffer II comprised of 6 M urea, 75 mM Tris pH 8.8, 29.3% glycerol (v/v), 2% SDS (w/v), 0.002% bromophenol blue (w/v) and 135 mM IAA.61 Equilibrated IPG strips were placed on 12% homogenous SDS polyacrylamide gel (14 × 16 cm) and sealed with low melting point (1%) agarose for two-dimensional gel electrophoresis at a constant current (180 V) in a running buffer (Tris–glycine-SDS, pH 8.3).61 The two-dimensional gels were silver stained using standard protocols.61 The spot patterns obtained were analyzed using PD Quest 2D analysis software (Bio-Rad) and the spots of interest were characterized by MALDI TOF MS peptide mass fingerprint analysis.61,64![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio and the resulting 2 μl was spotted onto the MALDI plate. After air drying, the sample was analyzed on the MALDI TOF/TOF Ultraflex III instrument and further analysis was done with Flex Analysis Software Version 3.2 for obtaining the peptide mass fingerprint. The masses obtained in the peptide mass fingerprint were submitted to the Mascot server for a peptide mass fingerprint search in the “NCBInr” database for protein characterization.61 The fixed modification(s) and variable modification(s) considered during analysis, including the residue specificity of trypsin (the protein digesting enzyme), respectively comprised of carbamidomethylation at cysteine and oxidation at methionine. The peptide mass tolerance for precursor ions varied for each spot as ±150 ppm (SSP 6708), ±500 ppm (SSP 7418), ±100 ppm (SSP 7122), and ±70 ppm (SSP 2107); no contaminants were excluded from the PMF data. The significance threshold score/expectation value for accepting individual spectra was selected as p < 0.05. The protein score was −10 × Log(P), where P is the probability that the observed match is a random event. Protein scores greater than 59 were selected to be significant (p < 0.05). The number of missed and/or non-specific cleavages permitted during analysis lied between 0–2.
1 ratio and the resulting 2 μl was spotted onto the MALDI plate. After air drying, the sample was analyzed on the MALDI TOF/TOF Ultraflex III instrument and further analysis was done with Flex Analysis Software Version 3.2 for obtaining the peptide mass fingerprint. The masses obtained in the peptide mass fingerprint were submitted to the Mascot server for a peptide mass fingerprint search in the “NCBInr” database for protein characterization.61 The fixed modification(s) and variable modification(s) considered during analysis, including the residue specificity of trypsin (the protein digesting enzyme), respectively comprised of carbamidomethylation at cysteine and oxidation at methionine. The peptide mass tolerance for precursor ions varied for each spot as ±150 ppm (SSP 6708), ±500 ppm (SSP 7418), ±100 ppm (SSP 7122), and ±70 ppm (SSP 2107); no contaminants were excluded from the PMF data. The significance threshold score/expectation value for accepting individual spectra was selected as p < 0.05. The protein score was −10 × Log(P), where P is the probability that the observed match is a random event. Protein scores greater than 59 were selected to be significant (p < 0.05). The number of missed and/or non-specific cleavages permitted during analysis lied between 0–2.Validation by Real Time PCR
Dissected hippocampus tissues were stored at −70 °C in the RNA later (Ambion) solution. Total RNA was isolated using a mini SureSpin total RNA isolation kit (Fermentas) including on-column DNase treatment. The isolated samples were quantified on a NanoDrop 2000c Spectrophotometer (Thermo Fisher Scientific Incorporation). The genomic DNA contamination was checked by PCR, with RNA samples and β-actin primers (targeted for 1234 bp amplicon) in the reaction composition of 50 ng RNA, 1X Taq buffer, 1U Taq polymerase, 1.5 mM MgCl2, 0.4 mM dNTPs, 0.4 μM β-actin Forward primer and 0.4 μM β-actin Reverse primer. The thermal cycling conditions performed on a Thermo Scientific Arktik thermal cycler (Thermo Fisher Scientific Incorporation) were (i) 96 °C for 5 min (initial denaturation), (ii) 35 cycles at 96 °C for 1 min, 58 °C for 1 min, and 72 °C for 1 min, and (iii) a final extension (72 °C for 10 min). The amplification products were analyzed by horizontal gel electrophoresis on a 1.5% agarose 1X TAE buffer system, followed by visualization on a UV gel documentation system (AlphaImager HP System, Protein Simple). For cDNA synthesis, 0.5 μg of normalized RNA were reverse transcribed using a RevertAid first strand cDNA synthesis kit (Fermentas). The integrity of the cDNA was verified by PCR with β-actin specific primers resulting in 207 bp amplicons. The reaction mixture constituted of 1 μl of cDNA, 1X Taq buffer, 1U Taq polymerase, 1.5 mM MgCl2, 0.4 mM dNTPs, 0.4 μM β-actin Forward primer and 0.4 μM β-actin Reverse primer. The thermal cycling conditions were (i) 95 °C for 5 min (initial denaturation) (ii) 35 cycles at 95 °C for 1 min, 58 °C for 1 min, 72 °C for 1 min (iii) final extension (72 °C for 10 min). Quantitative PCR was performed on the Rotor-Gene Q real-time PCR cycler (Qiagen) using SYBR Green dye. All reactions were performed in duplicates. The total reaction volume of 25 μL contained 12.5 μL of Maxima SYBR Green qPCR Master Mix (Fermentas), 0.4 μM of each primer (Table 3), 1 μl of diluted cDNA, and 9.5 μl of nuclease free water. The thermal cycling conditions were (i) 95 °C for 10 min (initial denaturation), (ii) 45 cycles at 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 60 s, followed by (iii) melt curve analysis at a temperature range of 72 °C to 95 °C. The raw data were analyzed with the Rotor-Gene Q series software 1.7 (Qiagen). The Ct values were obtained using a constant threshold value for all the genes examined. The relative gene expression was quantified with a comparative ΔCt(2−ΔΔCT) method.65 The control group was used as the calibrator. HPRT1 served as the normalizer gene.66Results
Monitoring of blood glucose levels
Prior to the 1st HP dose, group I showed a normal fasting range (70–100 mg dL−1); groups II and III had average blood glucose levels >70–100 mg dL−1 and <126 mg dL−1, implying establishment of hyperglycemia. On the 15th day of HP administration, group II showed an average blood glucose level of 132 mg dL−1 (>126 mg dL−1), significantly higher than group I, indicating the development of severe hyperglycemia and DM. On the contrary, group III ranged in normal limits, indicative of the role of HP in controlling hyperglycemia and preventing STZ-DM in group III animals. On the 28th day of HP administration, group II displayed an average glucose level of 145 mg dL−1 (>126 mg dL−1), a gradually overwhelming DM condition. Group I and group III possessed normal fasting blood glucose levels, indicating a persistent ameliorative attribute of HP for STZ-DM in group III animals (Fig. 1).Proteomic analysis
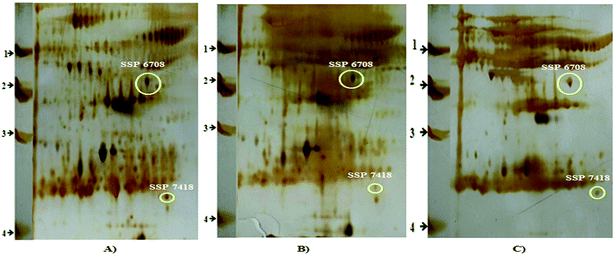
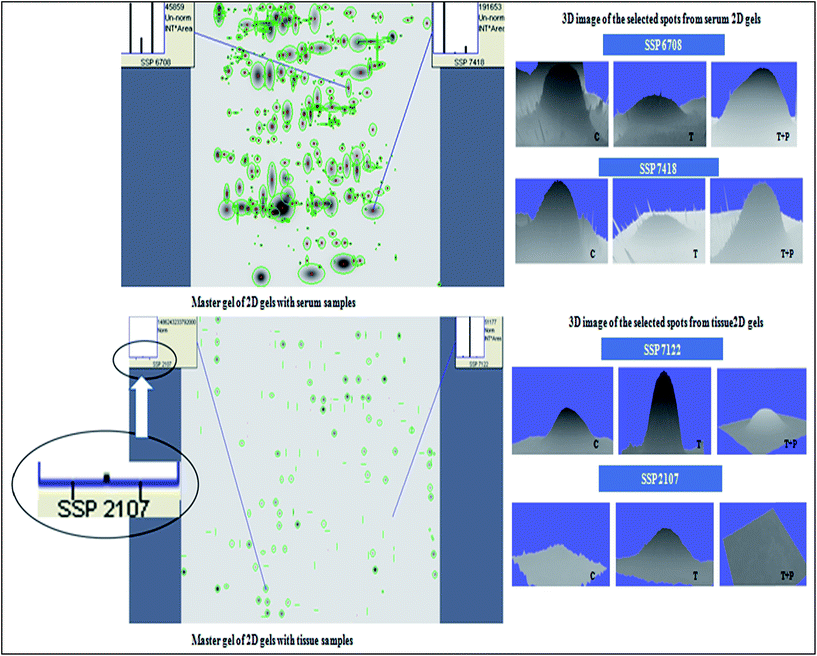
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 640.84 INT*area (control); 13
640.84 INT*area (control); 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 050.26 INT*area (STZ); 45
050.26 INT*area (STZ); 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 859.16 INT*area (STZ + HP). SSP 7418: 191
859.16 INT*area (STZ + HP). SSP 7418: 191![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 653.44 INT*area (control); 1572.06 INT*area (STZ); 22
653.44 INT*area (control); 1572.06 INT*area (STZ); 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 656.49 INT*area (STZ + HP). SSP 7122: 896.37 INT*area (control); 26
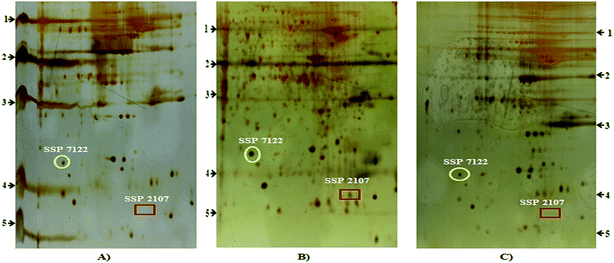
656.49 INT*area (STZ + HP). SSP 7122: 896.37 INT*area (control); 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 638.75 INT*area (STZ); 7.04 INT*Area (STZ + HP). SSP 2107: 0.00 INT*area (control); 158.33 INT*area (STZ); 0.00 INT*area (STZ + HP).
638.75 INT*area (STZ); 7.04 INT*Area (STZ + HP). SSP 2107: 0.00 INT*area (control); 158.33 INT*area (STZ); 0.00 INT*area (STZ + HP).
| SSP no. | Spot intensity | Proteins (Rattus norvegicus) | Accession no. | Score | Queries matched | Expected | Theoretical molecular weight (kD) | Experimental molecular weight (kD) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Group I (C) | Group II (T) | Group III (T + P) | ||||||||
| 6708 | ↑ | ↓ | ↑ | Apolipoprotein A-IV precursor | 114008 | 177 | 16 | 8 × 10−14 | 44.429 | <66 and >43 |
| 7418 | ↑ | ↓ | ↑ | Acylglycerol kinase, mitochondrial | NP_001120969.1 | 64 | 12 | 0.015 | 47.195 | <29 and >20.1 |
| 7122 | ↓ | ↑ | ↓ | Hypothetical protein | XP_001067747.1 | 61 | 4 | 0.033 | 6.869 | <29 and > 20.1 |
| 2107 | − | + | − | Tubulin beta 2 A or T beta-15 | P04691 | 60 | 11 | 0.04 | 50.361 | <20.1 and >14.3 |
| S. no. | Observed | Mr(expt) | Mr(calc) | ppm | Start–end | Miss | Peptide |
|---|---|---|---|---|---|---|---|
| SPOT: SSP 6708 | |||||||
| 1 | 1018.6734 | 1017.6662 | 1017.5607 | 104 | 156–163 | 0 | R.QLTPYIQR.M |
| 2 | 1261.8066 | 1260.7993 | 1260.6463 | 121 | 295–304 | 0 | K.QLDQQVEVFR.R |
| 3 | 1287.8163 | 1286.8090 | 1286.6466 | 126 | 210–220 | 0 | K.ATIDQNLEDLR.S |
| 4 | 1417.9395 | 1416.9323 | 1416.7474 | 131 | 295–305 | 1 | K.QLDQQVEVFRR.A |
| 5 | 1510.0647 | 1509.0574 | 1508.8715 | 123 | 80–93 | 0 | K.LVPFAVQLSGHLTK.E |
| 6 | 1546.9603 | 1545.9531 | 1545.7432 | 136 | 234–246 | 0 | K.LNHQMEGLAFQMK.K |
| 7 | 1733.0628 | 1732.0555 | 1731.8726 | 106 | 190–204 | 1 | K.FNQNMEGLKGQLTPR.A |
| 8 | 1843.1594 | 1842.1521 | 1841.9483 | 111 | 205–220 | 1 | R.ANELKATIDQNLEDLR.S |
| 9 | 2025.3182 | 2024.3109 | 2024.1055 | 101 | 80–97 | 1 | K.LVPFAVQLSGHLTKETER.V |
| 10 | 2061.2857 | 2060.2784 | 2060.0538 | 109 | 288–304 | 1 | K.SLEDLNKQLDQQVEVFR.R |
| 11 | 2148.3301 | 2147.3229 | 2147.0755 | 115 | 306–324 | 1 | R.AVEPLGDKFNMALVQQMEK.F |
| 12 | 2379.4164 | 2378.4091 | 2378.1503 | 109 | 325–345 | 1 | K.FRQQLGSDSGDVESHLSFLEK.N |
| 13 | 2812.7311 | 2811.7238 | 2811.3973 | 116 | 131–154 | 0 | K.LQEHLRPYATDLQAQINAQTQDMK.R |
| 14 | 2968.8711 | 2967.8638 | 2967.4984 | 123 | 131–155 | 1 | K.LQEHLRPYATDLQAQINAQTQDMKR.Q |
| 15 | 3309.1745 | 3308.1673 | 3307.6936 | 143 | 362–391 | 0 | K.GSPDQPLALPLPEQVQEQVQEQVQPKPLES.- |
| 16 | 3437.3078 | 3436.3005 | 3435.7885 | 149 | 361–391 | 1 | K.KGSPDQPLALPLPEQVQEQVQEQVQPKPLES.- |
| SPOT: SSP 7418 | |||||||
| 1 | 584.3458 | 583.3385 | 583.2867 | 88.9 | 10–13 | 0 | R.NHWK.K |
| 2 | 637.3290 | 636.3217 | 636.3483 | −41.71 | 78–82 | 0 | R.TLFEK.N |
| 3 | 713.4107 | 712.4034 | 712.4820 | −110.29 | 277–281 | 2 | R.RILRR.L |
| 4 | 1575.3419 | 1574.3346 | 1574.8515 | −328.25 | 304–317 | 0 | K.DVQLSTIELSITTR.N |
| 5 | 1709.0763 | 1708.0690 | 1708.8872 | −478.76 | 1–13 | 2 | -.MTTFFKTLRNHWK.K |
| 6 | 2211.2213 | 2210.2140 | 2211.0895 | −395.94 | 197–215 | 2 | K.GEKEQPVYAMTGLRWGSFR.D |
| 7 | 2384.0670 | 2383.0597 | 2383.1743 | −48.07 | 200–220 | 2 | K.EQPVYAMTGLRWGSFRDAGVK.V + oxidation (M) |
| 8 | 2399.1303 | 2398.1230 | 2398.2930 | −70.88 | 78–99 | 1 | R.TLFEKNAAPILHLSGMDVTVVK.T + oxidation (M) |
| 9 | 2566.4061 | 2565.3989 | 2566.3438 | −368.21 | 40–62 | 2 | R.RAACQEAQVFGNQLIPPNAQVKK.A |
| 10 | 2872.5448 | 2871.5375 | 2871.4034 | 46.7 | 327–351 | 2 | K.EDFMNICIEPDTVSKGDFIIIGSKK.V + oxidation (M) |
| 11 | 3582.0028 | 3580.9955 | 3582.7109 | −478.82 | 370–401 | 1 | R.CTLSLPEGTEGSFSIDSEEYEAMPVEVKLLPR.K |
| 12 | 3598.0102 | 3597.0029 | 3598.7059 | −473.21 | 370–401 | 1 | R.CTLSLPEGTEGSFSIDSEEYEAMPVEVKLLPR.K + oxidation (M) |
| SPOT: SSP 7122 | |||||||
| 1 | 1179.6898 | 1178.6825 | 1178.5947 | 74.5 | 56–64 | 0 | R.WLWCAFLLA.- |
| 2 | 2097.1117 | 2096.1044 | 2095.9116 | 92.0 | 3–21 | 0 | R.DPSPHSPGSLEASELCEER.V |
| 3 | 2210.1958 | 2209.1886 | 2209.1199 | 31.1 | 33–55 | 1 | R.KPRGSALSDSGEAVAGAHLSGSR.W |
| 4 | 2384.1013 | 2383.0941 | 2383.0532 | 17.1 | 1–21 | 1 | -.MRDPSPHSPGSLEASELCEER.V |
| SPOT: SSP 2107 | |||||||
| 1 | 808.3505 | 807.3432 | 807.3399 | 4.16 | 157–162 | 0 | R.EEYPDR.I |
| 2 | 1077.5709 | 1076.5636 | 1076.5250 | 35.8 | 155–162 | 1 | K.IREEYPDR.I |
| 3 | 1615.9212 | 1614.9139 | 1614.8287 | 52.8 | 63–77 | 0 | R.AILVDLEPGTMDSVR.S |
| 4 | 1631.9194 | 1630.9121 | 1630.8236 | 54.3 | 63–77 | 0 | R.AILVDLEPGTMDSVR.S + oxidation (M) |
| 5 | 1707.8544 | 1706.8471 | 1706.8549 | −4.60 | 283–297 | 0 | R.ALTVPELTQQMFDSK.N |
| 6 | 1871.0236 | 1870.0163 | 1869.9373 | 42.2 | 47–62 | 1 | R.INVYYNEAAGNKYVPR.A |
| 7 | 1959.0514 | 1958.0441 | 1957.9745 | 35.5 | 104–121 | 0 | K.GHYTEGAELVDSVLDVVR.K |
| 8 | 2087.1149 | 2086.1076 | 2086.0695 | 18.3 | 104–122 | 1 | K.GHYTEGAELVDSVLDVVRK.E |
| 9 | 2110.1325 | 2109.1253 | 2109.0571 | 32.3 | 1–19 | 1 | -.MREIVHIQAGQCGNQIGAK.F |
| 10 | 2141.1186 | 2140.1114 | 2139.9969 | 53.5 | 157–174 | 1 | R.EEYPDRIMNTFSVMPSPK.V |
| 11 | 3102.5552 | 3101.5479 | 3101.4003 | 47.6 | 20–46 | 0 | K.FWEVISDEHGIDPTGSYHGDSDLQLER.I |
| Genes | Forward primer | Reverse primer | Amplicon size | Amplicon sequence |
|---|---|---|---|---|
| HPRT1 | GTCAAGCAGTACAGCCCCAAAATG | AAATCCAACAAAGTCTGGCCTGTA | 96 bp (cDNA) | GTCAAGCAGTACAGCCCCAAAATGGTTAAGGTTGCAAGCTTGCTGGTGAAAAGGACCTCTCGAAGTGTTGGATACAGGCCAGACTTTGTTGGATTT |
| Apo A-IV | GCCATCACCGGGACCCAGGC | TCCAGTGGCATTGAGCTGTT | 147 bp (cDNA) | GCCATCACCGGGACCCAGGCTGAGGTCACTTCCGACCAGGTGGCCAATGTGATGTGGGACTACTTCACCCAGCTAAGCAACAATGCCAAGGAGGCTGTGGAACAACTGCAGAAGACAGATGTCACTCAACAGCTCAATGCCACTGGA |
| AGK | CACTCTTCAGGAGTGGCCTC | GGGGAGAAGGCATCCTGTGG | 168 bp (cDNA) | CACTCTTCAGGAGTGGCCTCAGACCCATCAGGCCTCCATCTCTTACACGGGCCCTACAGAGAGACCTCCCATTGGGCCTGAAGATGCTGCTCCCCGGCCTTCTCTGTACAGGAGAATATTACGTAGGCTTGCCTCATTCTGGGCACAGCCACAGGATG CCTTCTCCCC |
| HYPO | TCGCCTGGGAGCCTGGAGGC | GCCCAAACACCCCATGCCTG | 210 bp (cDNA) | TCGCCTGGGAGCCTGGAGGCGTCGGAGCTGTGCGAGGAGCGCGTACCCATACGCACTGGGGCGCGGGGTGCAAGAAAGCCACGCGGGTCAGCGTTATCAGACAGCGGAGAAGCCGTCGCGGGAGCGCACTTATCGGGGTCTCGCTGGCTCTGGTGCGCCTTCCTACTTGCCTGATCGGAGTGCGAGCCAGCAGGCATGGGGTGTTTGGGC |
| TUBB2A | CGGCGCTAAGTTTTGGGAGG | TATTTGTTGCCAGCAGCTTC | 129 bp (cDNA) | CGGCGCTAAGTTTTGGGAGGTGATAAGCGATGAGCATGGCATCGACCCCACTGGCAGTTACCATGGCGACAGTGACTTGCAGCTGGAGAGAATCAATGTGTACTACAATGAAGCTGCTGGCAACAAATA |
| β-Actin | CACCCGCGAGTACAACCTTC | CCCATACCCACCATCACACC | 1,234 bp (gDNA) | CACCCGCGAGTACAACCTTCTTGCAGCTCCTCCGTCGCCGGTCCACACCCGCCACCAGGTAAGCAGGGACGTCGGGCCCAGCGGGCCCCAACTTTACCTTGGCCACTACCTCGCTGCAGGATCGTGAGGAACACTCAGAAGGGACACCGTAGAGGGGTGGAGCGTGGTACCGGGCCGCGGAGCGGACACTGGCAAAGCTTAACTTTCCGCCTAGGGTGTAGAGTGTTTGCAGTCGTATTCCCGCGGTGTAGACACTCGTGGGCACGCTCCTGCTTGGTGCGCGGGGCTTGGGGACACACTAGAGTCGCGGTGTGGGCATTTGGAGAGCCGGTGCGGCTTGCGGGTGTTAAGCCGCATCTGTCCACCTTGAGGGGACACAGTATTGGGAGTCAGGCGTTACAATCACGCTTTGATGGCCTATGGGTCTTTGTCCAAACCGGTTTTGCCCATTCGGCTTGGCGGGCGCGGCGGGGCCGGCTCGGCCGGGTGGGGGCTGGGATGCCATTGCGCGTGCGCGCTCTATCACTGGGCATTGGGGCGCGTGCGCGCTGGGGAGGGAACTCTTCCTCTCCCCCTCTTCCGAGTTAAGAGTTGCGCGTGCGTATTGAGACTAGGAGCGCGGCCGCCCCGGGTTGGGCGAGGGCGGGGCCGTTGCCCGGAAGGGGCGGGGTCGTAGCGGCTAGGGCGCCTGCTCGCGCTTCCTGCTGGGTGTGGTCGCCTCCCGCGCGCGCACTAGCCGCCCGTCGCCTCAGTGTAGGCGGGGCCTGTGCCCGTTTGGGGAGGGGGCGGAGGCCTGGCTTCCTGCCGTGGGTCCGCCTCCGGGCCAGCGTTTGCCTTTTATGGTAATAATGCGGCTGTCCTGCGCTTCCTTTGTCCCCTGAGCTTGGGCGCGCGCCCCCTGGCGGCTCGAGGCCGCGGCTTGCCGGAAGTGGGCAGGGCGGCAGCGGCTGCTCTTGGCGGCTCCGCGGTGACCATAGCCCTCTTTTGTGCCTTGATAGTTCGCCATGGATGACGATATCGCTGCGCTCGTCGTCGACAACGGCTCCGGCATGTGCAAGGCCGGCTTCGCGGGCGACGATGCTCCCCGGGCCGTCTTCCCCTCCATCGTGGGCCGCCCTAGGCACCAGGTAAGTGACCCTTTACTTTGGGAGTGGCAGCCCTAGGGTTTTCTTGGGGGTCGATGCCAGTGCTGAGAACGTTGTTCTCCTCCGCAGGGTGTGATGGTGGGTATGGG |
| β-Actin | CACCCGCGAGTACAACCTTC | CCCATACCCACCATCACACC | 207 bp (cDNA), | CACCCGCGAGTACAACCTTCTTGCAGCTCCTCCGTCGCCGGTCCACACCCGCCACCAGTTCGCCATGGATGACGATATCGCTGCGCTCGTCGTCGACAACGGCTCCGGCATGTGCAAGGCCGGCTTCGCGGGCGACGATGCTCCCCGGGCCGTCTTCCCCTCCATCGTGGGCCGCCCTAGGCACCAGGGTGTGATGGTGGGTATGGG |
Validation of differential expression proteins
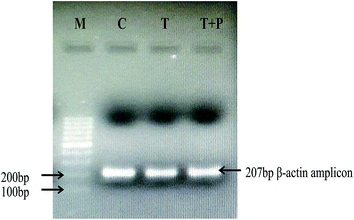
The PCR reactions with RNA samples and β-actin specific primers produced no amplification, hence verifying the absence of genomic DNA in the isolated total RNA. The cDNA samples were verified by amplifying 207 bp β actin amplicons (Fig. 6). Real time PCR was performed for the relative expression analysis of the AGK, APO A-IV, TUBB2A and HYPO genes, in the hippocampus tissue of STZ-DM rats and HP treated STZ-DM rats in comparison with that of the control healthy rats as calibrator samples. Fig. 7 shows the amplification plots and melting curves generated during real time PCR. As shown in Fig. 8, the AGK and APO A-IV genes were down-regulated in STZ-DM rats. The expression increased for both these genes in the HP treated STZ-DM rats compared to STZ-DM rats. The expression of the HYPO gene was up-regulated in STZ-DM rats, while its expression was reduced in the HP treated STZ-DM rats. The relative expression patterns for the AGK, APO A-IV and HYPO genes were consistent with the 2DE proteomic results, while the expression pattern of TUBB2A showed a different trend. The expression of TUBB2A in STZ-DM rats was down-regulated, while its expression was found to be increased in HP treated STZ-DM rats.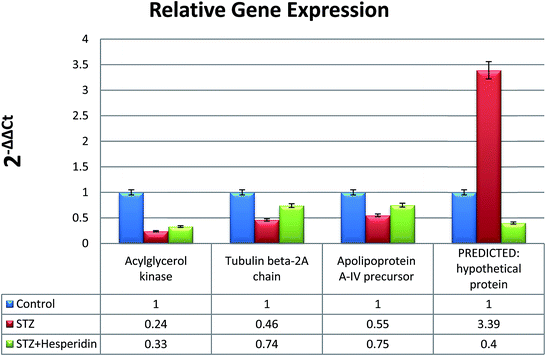 | ||
| Fig. 8 Differential expression of the AGK, APO A-IV, TUBB2A and HYPO genes in the control, toxicant (STZ) and toxicant + protectant (STZ + hesperidin) groups. | ||
Discussion
The hippocampus, a region in the brain, is responsible for learning and supports memory, and is affected by oxidative damages.71 Hyperglycemia promotes oxidative stress and the resulting damage increases complications.14 In the hippocampus proteome, Tubulin β 2A corresponding protein spot was present only in STZ-DM rats, while it was absent in the HP treated group and the control group. In our study, the experimental molecular weight of the TUBB2A chain was much lower than its theoretical weight. Such a reduction in the size may be due to fragmentation or cleavage of the full length chain.67 Significantly changed expression of TUBB2A chains has been linked to both type 2 diabetes mellitus and Alzheimer’s disease.72 The protein spot was lower than 20 kD and may indicate higher rates of degradation in STZ-DM rats compared to HP treated and control groups. Regulated degradation pathways exist for tubulin via cofactor E-like or parkin.73 The degradation of beta tubulin has been implicated in the pathogenesis of Alzheimer’s disease. Cleavage of tubulins and actins has been reported to cause axonal degeneration.74 The reduction in expression of the full length beta tubulin chain exists in AD.75 Transgenic AD model rats expressing Swedish mutant human APP proteins have down-regulated tubulin beta chain A in the hippocampus proteome.76 APP proteins are involved in familial AD.74,75,77 The down-regulation of tubulin beta 2A is associated with DM.72 Also, low levels of tubulin beta have been reported in the hippocampus of AD patient.72 Diabetes patients are at increased risk for developing Alzheimer’s disease.14 Diabetes patients have decreased risk of Parkinson’s disease.78 The oxidative stress built in diabetes is responsible for AD pathogenesis.79–83 The cellular oxidative stress is involved in neuronal degeneration in living animals81 and a strong relationship exists between oxidative stress and the build up of amyloid plaques in neural tissues.82,84–88 DM-mediated oxidative stress is associated with an increased risk of developing AD.89–91 DM and AD share insulin deficiency and have linked pathologies.31,92–94 Thus, in the present study, an increased levels of degraded hippocampal TUBB2A chain in combination with reduced mRNA levels within STZ-DM rats, may indicate that DM neuronal fluctuations have ultimately been expedited as AD, and hence may reflect the establishment of AD in STZ-DM rats. The experimental i.c.v. STZ administration has been known to produce early pathophysiological changes of AD.14–16,57 The i.c.v. administration of STZ interferes with brain energy metabolic pathways, reducing the usage of glucose in the brain and the functioning of glycolytic key enzymes, ultimately lowering the concentration of ATP and creatinine phosphate,95–97 causing neuron death. On the contrary, i.p. STZ administration induces dose-dependent DM53 by its explicit toxicity towards the beta cells of the pancreas. STZ enters beta cells by the GLUT2 glucose transporter and causes DNA alkylation, leading to cell death. Additionally, STZ promotes oxidative stress leading to diabetes associated neuronal degenerative ambiguities.14,98 In the present study, proteomic analysis mediated detection of AD related changes was carried out in a DM rat model developed by Parvez et al. (our group).14 Hence, further studies may be performed to analyze the potential of this designed rat model to serve as the experimental model for studying the pathologies of DM-mediated AD.The hippocampus protein, corresponding to the HYPO (XP_001067747.1) lacks functional recognition. The availability of HYPO in the NCBI protein database was generated by the reliability of computational scrutiny of the genomic DNA of Rattus norvegicus using protein prediction tools. The only procurable information (NCBI) presents HYPO as a stretch of 64 aa. We have named this hypothetical protein as ‘WajidSaima_Diabetes protein’ (WSDP), signifying that the protein was experimentally first reported in DM conditions in the STZ-DM rat model. This protein was present in all three groups reflecting its involvement in neuronal homeostasis. The expression of WSDP was enhanced in DM and associated AD.
The two secretomic or serum proteins, APO A-IV and sAGK, showed disparate levels in STZ-DM rats. APO A-IV has an association with triacylglycerol-rich lipoproteins and plays a role in lipid absorption, transport, and triacylglycerol-rich lipoprotein cellular internalization via many different mechanisms.99 APO A-IV improves glucose homeostasis by augmenting insulin secretion.100 The APO A-IV containing pre-β-migrating particles are present in cerebrospinal fluid.31,120 The APO A-IV 2 phenotype has been associated with an increased susceptibility for the development of AD.31,120 APO A-I down-regulation is associated with poorly controlled DM.101 APOA-IV has been linked to type 2 DM (The GeneCards Human Gene Database; http://www.genecards.org/cgi-bin/listdiseasecards.pl?type=full#). APO A-IV plays an important role in brain metabolism and genetic reduction of APO A-IV inflates extracellular amyloid-β peptide (Aβ), provoking neuronal loss in the brain, accelerating spatial learning deficits and increased mortality.102 Polymorphism in APO A-IV genes serves as a risk factor for depression.103 APO A-IV interacts with neurotoxic oligomeric Aβ42.104 Studies by Kronenberg and coworkers have associated low APO A-IV with coronary artery disorders.105 The diminished serum levels of APO A-IV reported in our study were obtained in STZ-DM rats. Also, its reduced expression was observed in the hippocampus of the STZ-DM rats. These observations propose a key role of APO A-IV in DM and associated AD. Apolipoproteins play a well-established role in the transport and metabolism of lipids within the central nervous system and are critical for healthy brain functions.106 Apolipoproteins are present in human cerebrospinal fluid.106 The expression of brain apolipoproteins is significantly altered in several brain disorders, for example, late-onset AD is linked to apolipoprotein E (apoE).107 APOA4 Gln360His polymorphism has an association with a risk of coronary artery calcium progression in type 1 diabetes patients.108
The mAGK, a multiple substrate lipid kinase, phosphorylates both mono and diacylglycerol to form lysophosphatidic acid (LPA) and phosphatidic acid (PA), respectively.109 The LPAs activate diverse groups of G-protein-coupled receptors that are widely expressed to regulate decisive cellular functions.110 They have been implicated in development, regulation of the cardiovascular, immune and nervous systems, inflammation, arteriosclerosis and cancer.110 The increased levels of mAGK in the vitreous fluid of the eye have been reported in retinopathy in a DM rat model and serve as a clinical biomarker for retinopathy in DM patients.111 The mAGK has cellular (mitochondrial) locations with 47.195 kD molecular weight (UniProt). In the present work, a protein similar to mAGK was isolated from the secretome of all three groups. Its experimental molecular weight ranged between 29 kD and 20.1 kD, significantly lower than the theoretical molecular weight of mAGK. These observations may signify the presence of a less bulky AGK isoform in the serum, and hence it may be named the ‘secretory AGK’ (sAGK). In Rattus norvegicus, the gene for AGK is located on chromosome 4 and comprises of 16 exons (Ensembl Genome Browser). The isoform sAGK might have originated by alternate splicing from the full length AGK precursor RNA, resulting in the shorter reading frame. sAGK may have a role in maintaining lipid homeostasis. On the contrary, sAGK shows different expression profiles, STZ-DM rats exhibited lower expression than the controls, and furthermore, the low expression level was also simulated in HP administered STZ-DM rats.
AD is the most prevalent form of dementia, constituting more than 60–80% of reported cases,112,113 and affecting more than 27 million persons worldwide. By the year 2050, the number of patients is expected to reach 86 million.114 AD involves a loss of synapses and neurons and a reduction in brain volume114 causing deterioration to cognitive functions, tarnishing personal-social lifestyle and eventually death.39,43,115 Early and definitive diagnosis of Alzheimer’s disease (AD) can lead to a better and more targeted treatment and/or prevention for diabetes patients.89,114 For the prognostic and diagnostic biomarkers of AD, the serum-based methods represent more non-invasive, inexpensive and acceptable sources for repeated measurements than cerebrospinal fluid.89 The identification of peripheral biomarkers would enable presymptomatic detection of AD and would be valuable for monitoring the efficacy of disease interventions during clinical trials.114 We suggest that a reduction in APO A-IV and sAGK may serve as a diagnostic marker for DM and DM-associated AD and in addition, it has therapeutic potential. Further studies on sAGK may uncover involved lipid homeostatic pathways and the pathological alterations causing DM. Conjointly, the role of sAGK in DM-associated neurodegeneration and AD may be investigated for insight. WSDP seems to have an important role in DM-mediated AD and possesses high potentiality as a therapeutic target.
Antidiabetic drugs are targeted towards controlling glucose levels in the normal range and are ultimately aimed at preventing hyperglycemia.116 The oxidative stress generated during DM is not taken into consideration by such drugs.117 HP, along with controlling the normal glucose levels, has been recognized to overcome the oxidative stress developed in the STZ-DM rat model.14 These dual properties of HP are important in preventing or managing DM and associated neurological complications.14 HP may be useful in developing drugs with dual antidiabetic and AD treating attributes.31 Prolonged STZ induced hyperglycemia enhances blood–brain barrier permeability, exposing neural tissue to the potentially pernicious agent.118 HP crosses the blood–brain barrier119 and restores oxidative damage.14 In the present study, a daily oral dose of HP (50 mg kg−1 b. w.−1) to STZ-DM rats efficiently maintained glucose levels in the normal range, while untreated STZ-DM rats suffered with progressively increasing hyperglycemia and diabetes. Along with maintaining blood glucose levels in the normal range, HP treatment was found to restore normal secretome and hippocampal proteome profiles. Interestingly, this restorative effect was more pronounced in the hippocampal proteome.
Conclusions
Diabetes mellitus possesses an appreciably significant association with AD. Persistent hyperglycemia and oxidative stress in diabetes leads to numerous changes in the secretome and proteomic profiles of hippocampus tissues. Hesperidin is a strong candidate as a potential therapeutic drug for controlling and treating diabetes mellitus, as well as preventing and treating diabetes mellitus-associated Alzheimer’s disease. Hesperidin is capable of controlling hyperglycemia. Conjointly its administration can restore normal proteomic expression profiles in the diabetic conditions. Early detection of diabetes mellitus-associated Alzheimer’s disease is a pre-requisite for preventing irreversible cognitive loss and neural impairments. APO A-IV and secretory AGK may serve as diagnostic markers for early stages detection of diabetes mellitus-associated Alzheimer’s disease. The predicted Hypothetical protein (XP_001067747.1) earlier unrecognized, was found to have an important role in maintaining neural homeostasis and was named ‘WSDP’, signifying that the protein was experimentally first reported in the diabetic conditions. Its experimental molecular weight lies between 29 kD and 20.1 kD. The serum proteins (APO A-IV and secretory AGK) and hippocampus proteins (WSDP and Tubulin beta 2A) scrutinized in this work may serve as therapeutic targets for the development of efficient treatment regimes for diabetes mellitus and associated Alzheimer’s disease. The current study established the capability of a regular oral dose of hesperidin in diminishing oxidative stress and associated proteomic alterations in the hippocampus and serum of a STZ-induced diabetes rat model. Our results strongly suggest that hesperidin may play an important role in restoring the neurologically harmful complications induced by diabetes-mediated oxidative stress.Conflicts of interest
None.Abbreviations
| aa | Amino acids |
| AGK | Acylglycerol kinase |
| mAGK | Mitochondrial acylglycerol kinase |
| sAGK | Secretory AGK |
| AD | Alzheimer’s disease |
| Aβ | Amyloid β |
| APO A-IV | Apolipoprotein A-IV |
| b.w. | Body weight |
| CNS | Central nervous system |
| DM | Diabetes mellitus |
| WSDP | WajidSaima_Diabetes protein |
| DTT | Dithiothreitol |
| HCCA | α-Cyano-4-hydroxycinnamic acid |
| HP | Hesperidin |
| HYPO | Hypothetical protein accession Id XP_001067747.1 |
| i.c.v | Intracerebroventricular |
| i.p. | Intraperitoneal |
| kD | Kilo Dalton |
| kg | Kilogram |
| mg | Milligram |
| mins | minutes |
| PMSF | Phenylmethanesulfonyl fluoride |
| RT | Room temperature |
| SE | Standard error |
| SSP | Standard spot parameter |
| STZ | Streptozotocin |
| STZ-DM | Streptozotocin mediated diabetes mellitus |
| Tris | Tris(hydroxymethyl)aminomethane |
| TUBB2A | Tubulin beta-2A chain |
Acknowledgements
We sincerely acknowledge the financial support from the University Grant Commission (UGC-SAP) to Department of Biotechnology, Jamia Hamdard during the course of this study.References
- American Diabetes Association, Diabetes Care, 2009, 32, S62–S67 Search PubMed.
- K. L. Chaichana, M. J. McGirt, G. F. Woodworth, G. Datoo, R. J. Tamargo, J. Weingart, A. Olivi, H. Brem and A. Quinones-Hinojosa, Neurol. Res., 2010, 32, 442–448 CrossRef PubMed.
- S. M. Seyed Saadat, E. Bidabadi, S. N. Seyed Saadat, M. Mashouf, F. Salamat and S. Yousefzadeh, Child’s Nerv. Syst., 2012, 28, 1773–1777 CrossRef PubMed.
- W. H. Pan, L. B. Cedres, K. Liu, A. Dyer, J. A. Schoenberger, R. B. Shekelle, R. Stamler, D. Smith, P. Collette and J. Stamler, Am. J. Epidemiol., 1986, 123, 504–516 CAS.
- B. Fuentes, M. A. Ortega-Casarrubios, B. Sanjose, J. Castillo, R. Leira, J. Serena, J. Vivancos, A. Davalos, A. Gil-Nunez, J. Egido and E. Diez-Tejedor, Stroke, 2010, 41, 2362–2365 CrossRef CAS PubMed.
- D. R. Whiting, L. Guariguata, C. Weil and J. Shaw, Diabetes Res. Clin. Pract., 2011, 94, 311–321 CrossRef PubMed.
- S. A. Kaveeshwar and J. Cornwall, Australas. Med. J., 2014, 7, 45–48 CrossRef.
- R. M. Anjana, R. Pradeepa, M. Deepa, M. Datta, V. Sudha, R. Unnikrishnan, A. Bhansali, S. R. Joshi, P. P. Joshi, C. S. Yajnik, V. K. Dhandhania, L. M. Nath, A. K. Das, P. V. Rao, S. V. Madhu, D. K. Shukla, T. Kaur, M. Priya, E. Nirmal, S. J. Parvathi, S. Subhashini, R. Subashini, M. K. Ali and V. Mohan, Diabetologia, 2011, 54, 3022–3027 CrossRef CAS PubMed.
- H. Umegaki, Adv. Exp. Med. Biol., 2012, 724, 258–265 CAS.
- Z. C. Chen, S. L. Zhang, L. Yan, M. C. Wu, L. H. Chen and L. N. Ji, Zhonghua Yixue Zazhi, 2011, 91, 229–233 Search PubMed.
- S. Singh, P. P. Singh, A. G. Singh, M. H. Murad and W. Sanchez, Am. J. Gastroenterol., 2013, 108, 881–891 CrossRef CAS PubMed ; quiz 892.
- C. L. T. Chang, Y. Lin, A. P. Bartolome, Y.-C. Chen, S.-C. Chiu and W.-C. Yang, J. Evidence-Based Complementary Altern. Med., 2013, 2013, 33 Search PubMed.
- M. M. A. Mustufa, S. Chandra and S. Wajid, Int. J. Pharma Bio Sci., 2014, 5, B1033–B1042 Search PubMed.
- M. Ashafaq, L. Varshney, M. H. A. Khan, M. Salman, M. Naseem, S. Wajid and S. Parvez, BioMed Res. Int., 2014, 2014, 9 Search PubMed.
- M. Monforte, A. Trovato, S. Kirjavainen, A. Forestieri, E. Galati and C. R. Lo, Farmaco, 1995, 50, 595–599 CAS.
- K. Ohtsuki, A. Abe, H. Mitsuzumi, M. Kondo, K. Uemura, Y. Iwasaki and Y. Kondo, J. Nutr. Sci. Vitaminol., 2003, 49, 447–450 CrossRef CAS.
- H. Chiba, M. Uehara, J. Wu, X. Wang, R. Masuyama, K. Suzuki, K. Kanazawa and Y. Ishimi, J. Nutr., 2003, 133, 1892–1897 CAS.
- J. A. D. S. Emim, A. B. Oliveira and A. J. Lapa, J. Pharm. Pharmacol., 1994, 46, 118–122 CrossRef CAS PubMed.
- E. Galati, M. Monforte, S. Kirjavainen, A. Forestieri, A. Trovato and M. Tripodo, Farmaco, 1994, 40, 709–712 CAS.
- H. A. Al-Ashaal and S. T. El-Sheltawy, Pharm. Biol., 2011, 49, 276–282 CrossRef CAS PubMed.
- K. A. Youdim, M. S. Dobbie, G. Kuhnle, A. R. Proteggente, N. J. Abbott and C. Rice-Evans, J. Neurochem., 2003, 85, 180–192 CrossRef CAS.
- A. M. Mahmoud, M. B. Ashour, A. Abdel-Moneim and O. M. Ahmed, J. Diabetes Complications, 2012, 26, 483–490 CrossRef PubMed.
- A. M. Mahmoud, O. M. Ahmed, A. Abdel-Moneim and M. B. Ashour, Int. J. Bioassays, 2013, 2, 756–761 Search PubMed.
- R. Tarawneh and D. M. Holtzman, CNS Neurol. Disord.: Drug Targets, 2009, 8, 144–159 CAS.
- Z. Arvanitakis, R. S. Wilson, J. L. Bienias, D. A. Evans and D. A. Bennett, Arch. Neurol., 2004, 61, 661–666 CrossRef PubMed.
- L. Gasparini, W. J. Netzer, P. Greengard and H. Xu, Trends Pharmacol. Sci., 2002, 23, 288–293 CrossRef CAS.
- A. M. Tolppanen, P. Lavikainen, A. Solomon, M. Kivipelto, M. Uusitupa, H. Soininen and S. Hartikainen, Diabetes Care, 2013, 36, 2015–2019 CrossRef PubMed.
- W. Xu, C. Qiu, B. Winblad and L. Fratiglioni, Diabetes, 2007, 56, 211–216 CrossRef CAS PubMed.
- S. M. de la Monte and J. R. Wands, J. Diabetes Sci. Technol., 2008, 2, 1101–1113 CrossRef PubMed.
- K. Gauthaman, P. Natesan Pushparaj, M. Rajeshkumar, K. Narasimhan, M. Al-Qahtani, N. Sang Cheung and J. Manikandan, CNS Neurol. Disord.: Drug Targets, 2014, 13, 247–258 CAS.
- K. Akter, E. A. Lanza, S. A. Martin, N. Myronyuk, M. Rua and R. B. Raffa, Br. J. Clin. Pharmacol., 2011, 71, 365–376 CrossRef CAS PubMed.
- J. Götz, L. Ittner and Y.-A. Lim, Cell. Mol. Life Sci., 2009, 66, 1321–1325 CrossRef PubMed.
- E. Planel, Y. Tatebayashi, T. Miyasaka, L. Liu, L. Wang, M. Herman, W. H. Yu, J. A. Luchsinger, B. Wadzinski, K. E. Duff and A. Takashima, J. Neurosci., 2007, 27, 13635–13648 CrossRef CAS PubMed.
- B. J. Clodfelder-Miller, A. A. Zmijewska, G. V. W. Johnson and R. S. Jope, Diabetes, 2006, 55, 3320–3325 CrossRef CAS PubMed.
- M. Hong and V. M.-Y. Lee, J. Biol. Chem., 1997, 272, 19547–19553 CrossRef CAS PubMed.
- E. J. Rivera, A. Goldin, N. Fulmer, R. Tavares, J. R. Wands and S. M. de la Monte, J. Alzheimer’s Dis., 2005, 8, 247–268 CAS.
- S. Craft, E. Peskind, M. W. Schwartz, G. D. Schellenberg, M. Raskind and D. Porte, Neurology, 1998, 50, 164–168 CrossRef CAS.
- L. Mosconi, Eur. J. Nucl. Med. Mol. Imaging, 2005, 32, 486–510 CrossRef CAS PubMed.
- A. F. Maiorini, M. J. Gaunt, T. M. Jacobsen, A. E. McKay, L. D. Waldman and R. B. Raffa, J. Clin. Pharm. Ther., 2002, 27, 169–183 CrossRef CAS.
- D. M. Walsh, I. Klyubin, J. V. Fadeeva, W. K. Cullen, R. Anwyl, M. S. Wolfe, M. J. Rowan and D. J. Selkoe, Nature, 2002, 416, 535–539 CrossRef CAS PubMed.
- S. Hong, B. L. Ostaszewski, T. Yang, T. T. O’Malley, M. Jin, K. Yanagisawa, S. Li, T. Bartels and D. J. Selkoe, Neuron, 2014, 82, 308–319 CrossRef CAS PubMed.
- D. Iacono, S. M. Resnick, R. O’Brien, A. B. Zonderman, Y. An, O. Pletnikova, G. Rudow, B. Crain and J. C. Troncoso, J. Neuropathol. Exp. Neurol., 2014, 73, 295–304 CrossRef CAS PubMed 210.1097/NEN.0000000000000052.
- S. Grimm, A. Hoehn, K. J. Davies and T. Grune, Free Radical Res., 2011, 45, 73–88 CrossRef CAS PubMed.
- P. D. Gorevic, F. Goni, B. Pons-Estel, F. Alvarez, N. S. Peress and B. Frangione, J. Neuropathol. Exp. Neurol., 1986, 45, 647–664 CrossRef CAS PubMed.
- I. Grundke-Iqbal, K. Iqbal, Y.-C. Tung, M. Quinlan, H. M. Wisniewski and L. I. Binder, Proc. Natl. Acad. Sci. U. S. A., 1986, 83, 4913–4917 CrossRef CAS.
- D. G. Smith, R. Cappai and K. J. Barnham, Biochim. Biophys. Acta, Biomembr., 2007, 1768, 1976–1990 CrossRef CAS PubMed.
- K. S. KoSIK, C. L. Joachim and D. J. Selkoe, Proc. Natl. Acad. Sci. U. S. A., 1986, 83, 4044–4048 CrossRef CAS.
- G. B. Irvine, O. M. El-Agnaf, G. M. Shankar and D. M. Walsh, Mol. Med., 2008, 14, 451–464 CAS.
- S. Chandra, S. Khowal and S. Wajid, Int. J. Pharma Bio Sci., 2014, 5, B274–B292 Search PubMed.
- G. Brosky and J. Logothetopoulos, Diabetes, 1969, 18, 606–611 CrossRef CAS.
- M. Ito, Y. Kondo, A. Nakatani and A. Naruse, Biol. Pharm. Bull., 1999, 22, 988–989 CAS.
- R. B. Weiss, Cancer Treat. Rep., 1982, 66, 427–438 CAS.
- M. A. Abeeleh, Z. B. Ismail, K. R. Alzaben, S. A. Abu-Halaweh, M. K. Al-Essa, J. Abuabeeleh and M. M. Alsmady, Eur. J. Sci. Res., 2009, 32, 398–402 Search PubMed.
- M. Šalković-Petrišić, Period. Biol., 2008, 110, 17–25 Search PubMed.
- M. Salkovic-Petrisic and S. Hoyer, Central insulin resistance as a trigger for sporadic Alzheimer-like pathology: an experimental approach, Springer, 2007 Search PubMed.
- E. Grünblatt, M. Salkovic-Petrisic, J. Osmanovic, P. Riederer and S. Hoyer, J. Neurochem., 2007, 101, 757–770 CrossRef PubMed.
- R. Deshmukh, V. Sharma, S. Mehan, N. Sharma and K. Bedi, Eur. J. Pharmacol., 2009, 620, 49–56 CrossRef CAS PubMed.
- M. Torres-Piedra, R. Ortiz-Andrade, R. Villalobos-Molina, N. Singh, J. L. Medina-Franco, S. P. Webster, M. Binnie, G. Navarrete-Vazquez and S. Estrada-Soto, Eur. J. Med. Chem., 2010, 45, 2606–2612 CrossRef CAS PubMed.
- A. M. Mahmoud, M. B. Ashour, A. Abdel-Moneim and O. M. Ahmed, J. Diabetes Complications, 2012, 26, 483–490 CrossRef PubMed.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS.
- N. Chaudhary, S. Bhatnagar, S. Malik, D. P. Katare and S. K. Jain, Chem.-Biol. Interact., 2013, 204, 125–134 CrossRef CAS PubMed.
- U. K. Laemmli, Nature, 1970, 227, 680–685 CrossRef CAS PubMed.
- A. Shevchenko, O. N. Jensen, A. V. Podtelejnikov, F. Sagliocco, M. Wilm, O. Vorm, P. Mortensen, H. Boucherie and M. Mann, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 14440–14445 CrossRef CAS.
- O. Yanes, J. Villanueva, E. Querol and F. X. Aviles, Nat. Protoc., 2007, 2, 119–130 CrossRef CAS PubMed.
- K. J. Livak and T. D. Schmittgen, Methods, 2001, 25, 402–408 CrossRef CAS PubMed.
- B. E. Bonefeld, B. Elfving and G. Wegener, Synapse, 2008, 62, 302–309 CrossRef CAS PubMed.
- M. D. Person, J. Shen, A. Traner, S. C. Hensley, H.-H. Lo, J. L. Abbruzzese and D. Li, J. Biomol. Tech., 2006, 17, 145–156 Search PubMed.
- B. Schulenberg, T. N. Goodman, R. Aggeler, R. A. Capaldi and W. F. Patton, Electrophoresis, 2004, 25, 2526–2532 CrossRef CAS PubMed.
- C. Hart, B. Schulenberg, T. H. Steinberg, W. Y. Leung and W. F. Patton, Electrophoresis, 2003, 24, 588–598 CrossRef CAS PubMed.
- K. Hiller, M. Schobert, C. Hundertmark, D. Jahn and R. Münch, Nucleic Acids Res., 2003, 31, 3862–3865 CrossRef CAS PubMed.
- J. R. Manns, R. O. Hopkins and L. R. Squire, Neuron, 2003, 38, 127–133 CrossRef CAS.
- Z. Mirza, M. A. Kamal, A. M. Buzenadah, M. H. Al-Qahtani and S. Karim, CNS Neurol. Disord.: Drug Targets, 2014, 13, 501–516 CAS.
- V. F. Lundin, M. R. Leroux and P. C. Stirling, Trends Biochem. Sci., 2010, 35, 288–297 CrossRef CAS PubMed.
- J. D. Sokolowski, K. K. Gamage, D. S. Heffron, A. C. LeBlanc, C. D. Deppmann and J. W. Mandell, Acta Neuropathol. Commun., 2014, 2, 16 CrossRef PubMed.
- R. Sultana, D. Boyd-Kimball, J. Cai, W. M. Pierce, J. B. Klein, M. Merchant and D. A. Butterfield, J. Alzheimer’s Dis., 2007, 11, 153–164 CAS.
- R. A. Sowell, J. B. Owen and D. A. Butterfield, Ageing Res. Rev., 2009, 8, 1–17 CrossRef CAS PubMed.
- A. Goate, M.-C. Chartier-Harlin, M. Mullan, J. Brown, F. Crawford, L. Fidani, L. Giuffra, A. Haynes, N. Irving, L. James, R. Mant, P. Newton, K. Rooke, P. Roques, C. Talbot, M. Pericak-Vance, A. Roses, R. Williamson, M. Rossor, M. Owen and J. Hardy, Nature, 1991, 349, 704–706 CrossRef CAS PubMed.
- L. Lu, D.-L. Fu, H.-Q. Li, A.-J. Liu, J.-H. Li and G.-Q. Zheng, PLoS One, 2014, 9, e85781 Search PubMed.
- S. M. de la Monte, A. Neusner, J. Chu and M. Lawton, J. Alzheimer’s Dis., 2009, 17, 519–529 CAS.
- A. Gella and N. Durany, Cell Adhes. Migrat., 2009, 3, 88–93 CrossRef.
- H. Xie, S. Hou, J. Jiang, M. Sekutowicz, J. Kelly and B. J. Bacskai, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 7904–7909 CrossRef CAS PubMed.
- Y. Zhao and B. Zhao, Oxid. Med. Cell. Longevity, 2013, 2013, 10 Search PubMed.
- E. R. Mayeda, R. A. Whitmer and K. Yaffe, Clin. Geriatr. Med., 2015, 31, 101–115 CrossRef PubMed.
- M. Padurariu, A. Ciobica, R. Lefter, I. L. Serban, C. Stefanescu and R. Chirita, Psychiatr. Danubina, 2013, 25, 401–409 CAS.
- P. H. Axelsen, H. Komatsu and I. V. Murray, Physiology, 2011, 26, 54–69 CrossRef CAS PubMed.
- D. Pratico, K. Uryu, S. Leight, J. Q. Trojanoswki and V. M. Lee, J. Neurosci., 2001, 21, 4183–4187 CAS.
- S. Varadarajan, S. Yatin, M. Aksenova and D. A. Butterfield, J. Struct. Biol., 2000, 130, 184–208 CrossRef CAS PubMed.
- E. Tamagno, M. Guglielmotto, D. Monteleone and M. Tabaton, Neurotoxic. Res., 2012, 22, 208–219 CrossRef CAS PubMed.
- K. Inoue, H. Tsuchiya, T. Takayama, H. Akatsu, Y. Hashizume, T. Yamamoto, N. Matsukawa and T. Toyo’oka, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2015, 974, 24–34 CrossRef CAS PubMed.
- N. T. Vagelatos and G. D. Eslick, Epidemiol. Rev., 2013, 35, 152 CrossRef PubMed.
- M. Malek-Ahmadi, T. Beach, A. Obradov, L. Sue, C. Belden, K. Davis, D. G. Walker, L. Lue, A. Adem and M. N. Sabbagh, Curr. Alzheimer Res., 2013, 10, 654–659 CrossRef CAS.
- B. Kim, C. Backus, S. Oh and E. L. Feldman, J. Alzheimer’s Dis., 2013, 34, 727–739 CAS.
- C. Long-Smith, S. Manning, P. McClean, M. Coakley, D. O’Halloran, C. Holscher and C. O’Neill, NeuroMol. Med., 2013, 15, 102–114 CrossRef CAS PubMed.
- Y. Liu, F. Liu, I. Grundke-Iqbal, K. Iqbal and C.-X. Gong, J. Pathol., 2011, 225, 54–62 CrossRef CAS PubMed.
- R. Duelli, H. Schröck, W. Kuschinsky and S. Hoyer, Int. J. Dev. Neurosci., 1994, 12, 737–743 CrossRef CAS.
- K. Plaschke and S. Hoyer, Int. J. Dev. Neurosci., 1993, 11, 477–483 CrossRef CAS.
- H. Lannert and S. Hoyer, Behav. Neurosci., 1998, 112, 1199 CrossRef CAS.
- S. Lenzen, Diabetologia, 2008, 51, 216–226 CrossRef CAS PubMed.
- Z. Sun, F. K. Welty, G. G. Dolnikowski, A. H. Lichtenstein and E. J. Schaefer, Am. J. Clin. Nutr., 2001, 74, 308–314 CAS.
- F. Wang, A. B. Kohan, T. L. Kindel, K. L. Corbin, C. S. Nunemaker, S. Obici, S. C. Woods, W. S. Davidson and P. Tso, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 9641–9646 CrossRef CAS PubMed.
- H. S. Bhonsle, A. M. Korwar, A. D. Chougale, S. S. Kote, N. L. Dhande, K. M. Shelgikar and M. J. Kulkarni, Mol. Med. Rep., 2013, 7, 495–498 CAS.
- Y. Cui, M. Huang, Y. He, S. Zhang and Y. Luo, Am. J. Pathol., 2011, 178, 1298–1308 CrossRef CAS PubMed.
- V. K. Ota, E. S. Chen, T. F. Ejchel, T. K. Furuya, D. R. Mazzotti, M. S. Cendoroglo, L. R. Ramos, L. Q. Araujo, R. R. Burbano and A. Smith Mde, J. Invest. Med., 2011, 59, 966–970 CAS.
- M. Rahman,
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Proteomics studies of Alzheimer's Aβ-oligomers to identify interactions with other proteins in human serum and cerebrospinal fluid,
Proteomics studies of Alzheimer's Aβ-oligomers to identify interactions with other proteins in human serum and cerebrospinal fluid,![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Epsilon Archive for Student Projects,
Epsilon Archive for Student Projects,![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SLU University Library, Uppsala, Second cycle, A2E. SLU, Dept. of Molecular Biology (until 131231), 2012 Search PubMed.
SLU University Library, Uppsala, Second cycle, A2E. SLU, Dept. of Molecular Biology (until 131231), 2012 Search PubMed. - F. Kronenberg, M. Stühlinger, E. Trenkwalder, F. Geethanjali, O. Pachinger, A. von Eckardstein and H. Dieplinger, J. Am. Coll. Cardiol., 2000, 36, 751–757 CrossRef CAS.
- K. Akter, E. A. Lanza, S. A. Martin, N. Myronyuk, M. Rua and R. B. Raffa, Br. J. Clin. Pharmacol., 2011, 71, 365–376 CrossRef CAS PubMed.
- J. Poirier, J. Miron, C. Picard, P. Gormley, L. Théroux, J. Breitner and D. Dea, Neurobiol. Aging, 2014, 35(Suppl. 2), S3–S10 CrossRef CAS PubMed.
- A. Kretowski, J. E. Hokanson, K. McFann, G. L. Kinney, J. K. Snell-Bergeon, D. M. Maahs, R. P. Wadwa, R. H. Eckel, L. G. Ogden, S. K. Garg, J. Li, S. Cheng, H. A. Erlich and M. Rewers, Diabetologia, 2006, 49, 1946–1954 CrossRef CAS PubMed.
- S. Spiegel and S. Milstien, J. Biol. Chem., 2007, 282, 2125–2129 CrossRef CAS PubMed.
- D. Meyer zu Heringdorf and K. H. Jakobs, Biochim. Biophys. Acta, Biomembr., 2007, 1768, 923–940 CrossRef CAS PubMed.
- A. M. A. El-Asrar, G. Mohammad, M. I. Nawaz, M. M. Siddiquei, D. Kangave and G. Opdenakker, Acta Diabetol. Lat., 2013, 50, 363–371 CrossRef PubMed.
- A. Alzheimer, R. A. Stelzmann, H. N. Schnitzlein and F. R. Murtagh, Clin. Anat., 1995, 8, 429–431 CrossRef CAS PubMed.
- W. Thies and L. Bleiler, Alzheimer’s Dementia, 2013, 9, 208–245 CrossRef PubMed.
- J. D. Doecke, S. M. Laws, N. G. Faux, W. Wilson, S. C. Burnham, C.-P. Lam, A. Mondal, J. Bedo, A. I. Bush and B. Brown, Arch. Neurol., 2012, 69, 1318–1325 CrossRef PubMed.
- A. P. Association, The Diagnostic and Statistical Manual of Mental Disorders: DSM 5, bookpointUS, 2013 Search PubMed.
- M. Hamaty, Cleveland Clin. J. Med., 2011, 78, 332–342 CrossRef PubMed.
- R. Kulkarni, J. Acharya, S. Ghaskadbi and P. Goel, PLoS One, 2014, 9, e100897 Search PubMed.
- J. D. Huber, R. L. VanGilder and K. A. Houser, Streptozotocin-induced diabetes progressively increases blood–brain barrier permeability in specific brain regions in rats, 2006 Search PubMed.
- J. Nones, T. E. Spohr and F. Gomes, Neurochem. Res., 2011, 36, 1776–1784 CrossRef CAS PubMed.
- N. Demeester, G. Castro, C. Desrumaux, C. De Geitere, J. C. Fruchart, P. Santens, E. Mulleners, S. Engelborghs, P. P. De Deyn, J. Vandekerckhove, M. Rosseneu and C. Labeur, J. Lipid Res., 2000, 41, 963–974 CAS.
Footnote |
| † Sapna Khowal and Malik M. A. Mustufa have contributed equally. |
| This journal is © The Royal Society of Chemistry 2015 |