α-Mangostin induces G1 cell cycle arrest in HCT116 cells through p38MAPK-p16INK4a pathway†
Abstract
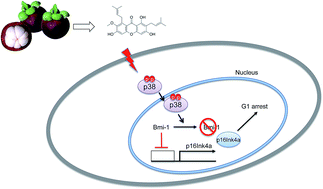
α-Mangostin (α-MG), one of the active substances in Garcinia mangostana, has been shown to exhibit anti-cancer effects in various cancer cell types. α-MG treatment induces G1 arrest in cancer cell models through the induction of cyclin-dependent kinase inhibitors (CDKIs) and the subsequent loss of CDK activity. However, outside its role in the p53-p21CIP1 axis, the precise molecular mechanisms underlying the effect of α-MG on cell cycle arrest remain unclear. In this study, we observed that α-MG inhibits the proliferation of HCT116 cells in a dose-dependent manner. Interestingly, although the loss of p53 rescued the α-MG effect on cell cycle arrest, in agreement with previous reports, p21Cip1 expression was only marginally delayed in the absence of p53 after α-MG treatment. Instead, we found that the activation of p38 mitogen activated protein kinase (MAPK) and the subsequent downregulation of Bmi-1 also contributed to the induction of p16Ink4a, which is responsible for G1 arrest upon α-MG treatment. These findings indicate that α-MG exerts cytostatic effects on colon cancer cells by inducing G1 arrest via the p38MAPK-p16INK4a axis.

 Please wait while we load your content...
Please wait while we load your content...