Four related mixed-ligand nickel(II) complexes: effect of steric encumbrance on the structure, DNA/BSA binding, DNA cleavage and cytotoxicity†
Chun-Yan Gao‡
ab,
Zhong-Ying Ma‡c,
Yong-Po Zhangb,
Si-Tong Liad,
Wen Guad,
Xin Liuad,
Jin-Lei Tian*ad,
Jing-Yuan Xu*c,
Jin-Zhong Zhaob and
Shi-Ping Yana
aCollege of Chemistry, Collaborative Innovation Center of Chemical Science and Engineering (Tianjin), Nankai University, Tianjin 300071, P. R. China. E-mail: tiant@nankai.edu.cn; xujingyuan@tmu.edu.cn
bCollege of Arts and Sciences, Shanxi Agricultural University, Taigu, Shanxi 030801, P. R. China
cCollege of Pharmacy, Tianjin Medical University, Tianjin 300070, P. R. China
dKey Laboratory of Advanced Energy Materials Chemistry (MOE), Tianjin 300071, P. R.China
First published on 20th March 2015
Abstract
Four closely related mononuclear nickel(II) complexes [Ni(L)(diimine)Cl](ClO4) (1–4), where L is a tridentate polypyridyl ligand of 4-methyl-N,N-bis(pyridin-2-ylmethyl)aniline and diimine is 2,2′-bipyridine (bpy, 1), 1,10-phenanthroline (phen, 2), dipyrido[3,2-d:20,30-f]quinoxaline (dpq, 3) or dipyrido[3,2-a:20,30-c]phenazine (dppz, 4), have been synthesized and characterized using various physico-chemical techniques. All Ni centers adopt a distorted octahedral geometry with N5Cl donor sets. From 1 to 4, the dihedral angles between the benzene ring of L and the plane of the diimine gradually decline (52.5–6.8°), leading to increasing steric encumbrance. The interactions of the complexes with CT-DNA and BSA have been explored using absorption and emission spectral methods. These complexes display binding propensity to CT-DNA in the order: 4 (dppz) > 3 (dpq) > 2 (phen) > 1 (bpy), and the quenching mechanisms of BSA by all the complexes are static procedures. In the absence of any external agents, only 1 (bpy) and 4 (dppz) exhibit apparent DNA cleavage activity, while with the addition of GSH or on the irradiation with UV-A light of 365 nm, the DNA cleavage abilities of the complexes are obviously enhanced, which vary as 1 > 2 > 3 > 4 (GSH) and 4 > 3 > 2 > 1 (UV-A). In addition, the in vitro cytotoxicity of the complexes on tumor cells lines (MCF-7, HepG-2 and SGC-7901) have been examined by MTT and the morphological assessment obtained using Hoechst 33342 staining reveals that 4 induces apoptosis against HepG-2.
1. Introduction
Metallointercalators play an important role in nucleic acid chemistry due to their wide applications, such as DNA foot printing, sequence specific binding and reactions, as new structural probes and therapeutic agents.1–7 Hence, large numbers of metallointercalators having planar N-donor heterocyclic bases have been widely used as chemical or photochemical reagents in nucleic acid chemistry in recent decades. For example, Barton and co-workers1,8–12 have been making significant contributions to polypyridyl complexes of 3d–5d transition metals for the chemistry of metallointercalators and DNA charge transport. Also, numerous other important contributions to chemical nuclease activities of phenanthroline complexes have been reported by Sigman and co-workers.6,13–17 The 2,2′-bipyridine (bpy) and 1,10-phenanthroline (phen) have been extensively used as chelating ligands in both analytical and preparative coordination chemistry for a long time.18 Over the last decades, systematic studies of other α-diimine derivatives such as dipyrido[3,2-d:20,30-f]quinoxaline (dpq) and dipyrido[3,2-a:20,30-c]phenazine (dppz) have been undertaken.19–27 A key feature of these six-membered fused N-heterocyclic rings is their π-electron deficiency, which make them excellent π-acceptor ligands and suitable as intercalation agents and photophysical probes for investigating DNA binding and cleavage properties.Transition metal complexes with their various coordination modes, redox behavior, feasible substitution kinetic pathways and highly sensitive diagnostic agents have been synthesized and developed rapidly for chemotherapeutic applications.28 Among these DNA-targeted artificial metallonucleases, ruthenium, copper and cobalt complexes are numerous in the literature.29–31 Though nickel is an essential element related to life process and an active component in various types of enzymes, studies on the interactions of nickel(II) complexes with DNA are comparably less.32–35 In recent years, reports on the role of nickel in bioinorganic chemistry have been rapidly expanding, The interaction of nickel(II) complexes with DNA mainly depends on the structure of the ligand exhibiting intercalative nature, consequently, design of novel varieties of ligands as new chemical nucleases is an area of current research interest.
Exploration of the mixed-ligands spans main areas of inorganic chemistry, recently, effect of steric encumbrance between the mixed-ligands on the structure and properties have been reported deeply by the A. R. Chakravarty and co-workers,36–38 and the result indicated that the steric encumbrance has positive effects on the photoinduced DNA-cleavage activity of the complexes. With an aim to explore the steric effect further, in this work, we have chosen a tridentate polypyridyl ligand L derived from 2-picolyl chloride and 4-methylaniline (L = 4-methyl-N,N-bis(pyridin-2-ylmethyl)aniline) (Scheme 1). The steric interaction between L and diimine (bpy, phen, dpq and dppz) ligand leads to significant differences in their physicochemical properties, which are mainly dependent on the dihedral angle between the average plane of the diimine and the benzene ring. The selected ligands allow us to make direct comparisons on structures and biological activities of the four Ni(II) complexes, on the whole, the steric effect and intercalative nature of the diimine ligand could be the two critical factors. Interestingly, the DNA cleavage efficiencies of the four complexes exhibit remarkably different with change of external conditions, such as no additive agents, with the presence of GSH (glutathione) or on the photoirradiation at 365 nm. Recently, several nickel(II) complexes have been tested for inhibition of cancer cell proliferation and found several potential applications in medicine.39–42 Also, the cytotoxicity and preliminary apoptotic experiments of these Ni complexes have been measured and analyzed in this work.
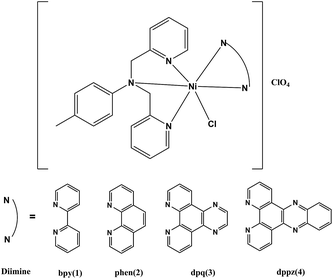 | ||
| Scheme 1 Schematic structures of complexes [Ni(L)(diimine)Cl](ClO4) (1–4) (L = 4-methyl-N,N-bis(pyridin-2-ylmethyl)aniline; diimine = bpy, (1); phen, (2); dpq, (3); dppz, (4)). | ||
2. Experimental
CAUTION: perchlorate salts of metal complexes are potentially explosive and therefore should be prepared in small quantities.2.1 Materials and measurements
All reagents and chemicals were purchased from commercial sources and used as received. Plasmid pBR322 DNA, agarose, ethidium bromide (EB), bovine serum albumin (BSA) and calf thymus (CT-DNA) were obtained from Sigma. Stock solutions of Ni(II) complexes (1.0 × 10−4 M in 1% DMF/H2O) were stored at 4 °C and prepared to required concentrations for all experiments. Tris–HCl and phosphate buffer solution were prepared using deionized sonicated triple-distilled water. Human breast adenocarcinoma cell line (MCF-7), human liver hepatocellular carcinoma cell line (HepG-2) and human gastric cancer cell line (SGC-7901) were obtained from the American Type Culture Collection (Rockville, MD, USA). Fetal bovine serum (FBS) was obtained from Hyclone. DMEM medium was obtained from Gibco. 3-(4,5-Dimathylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and Hoechst 33342 were purchased from Sigma. Elemental analyses for C, H and N were obtained on a Perkin-Elmer analyzer model 240. Infrared spectra were recorded as KBr pellets using a Perkin-Elmer FT-IR spectrometer in the range 4000–400 cm−1. Electronic spectra were measured on a JASCO V-570 spectrophotometer. Fluorescence spectral data were obtained on a MPF-4 fluorescence spectrophotometer at room temperature. The Gel Imaging and documentation DigiDoc-It System were assessed using Labworks Imaging and Analysis Software (UVI, England). The MTT assay was determined by measuring the absorbance of each well at 570 nm using a Bio-Rad 680 microplate reader (Bio-Rad, USA). The morphology of cells after treatment with complexes was obtained on the EPI fluorescence microscopy (Nikon eclipse E800).2.2 Synthesis of ligand L and corresponding Ni(II) complexes
The dpq and dppz ligands were prepared according to the previous reported procedures.44,45
2.3 X-ray crystallography
Diffraction data were collected at 113(2) or 293(2) K for 1–4 with a Bruker Smart 1000 CCD diffractometer using Mo-Kα radiation (λ = 0.71073 Å) with the ω–2θ scan technique. The structures were solved by direct methods (SHELXS-97) and refined with full-matrix least-squares technique on F2 using the SHELXL-97.46,47 The hydrogen atoms were added theoretically, and riding on the concerned atoms and refined with fixed thermal factors. The details of crystallographic data and structure refinement parameters are summarized in Table 1, and selected bond angles and distances are listed in Table 2 (ESI†).| Complex | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Empirical formula | C30H31Cl2N5NiO5 | C31H29Cl2N5NiO5 | C34H31Cl2N7NiO5 | C38H32Cl2N7NiO5 |
| Mr | 671.20 | 681.19 | 747.25 | 796.32 |
| T/K | 293(2) | 113(2) | 293(2) | 113(2) |
| λ/Å | 0.71073 | 0.71073 | 0.71073 | 0.71073 |
| Crystal system | Monoclinic | Monoclinic | Triclinic | Monoclinic |
| Space group | C2/c | P2(1)/c | P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
P2(1)/c |
| a/Å | 22.221(4) | 7.6563(3) | 7.5420(15) | 7.5063(15) |
| b/Å | 15.276(3) | 35.2380(17) | 11.202(2) | 22.913(5) |
| c/Å | 17.848(4) | 12.7241(8) | 19.311(4) | 20.750(4) |
| α/° | 90 | 90 | 97.04(3) | 90 |
| β/° | 103.16(3) | 110.532(4) | 90.51(3) | 92.90(3) |
| γ/° | 90 | 90 | 90.79(3) | 90 |
| V/A3 | 5899(2) | 3214.8(3) | 1619.0(5) | 3564.2(12) |
| Z | 8 | 4 | 2 | 4 |
| D/g cm−3 | 1.502 | 1.403 | 1.525 | 1.484 |
| F (000) | 2752 | 1400 | 764 | 1644 |
| θ range for data collection/° | 3.03 to 25.01 | 2.43 to 25.01 | 3.19 to 25.01 | 2.84 to 25.00 |
| Limiting indices, hk1 | −26 ≤ h ≤ 26 | −9 ≤ h ≤ 5 | −8 ≤ h ≤ 8 | −8 ≤ h ≤ 8 |
| −18 ≤ k ≤ 18 | −41 ≤ k ≤ 34 | −13 ≤ k ≤ 13 | −27 ≤ k ≤ 27 | |
| −21 ≤ l ≤ 20 | −15 ≤ l ≤ 15 | −22 ≤ l ≤ 22 | −24 ≤ l ≤ 24 | |
| Reflections collected | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 912 912 |
12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 479 479 |
9248 | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 371 371 |
| Independent refletions (R(int)) | 5194 [R(int) = 0.0638] | 5668 [R(int) = 0.0343] | 5689 [R(int) = 0.0477] | 6271 [R(int) = 0.0487] |
| Goodness-of-fit on F2 | 1.108 | 1.059 | 1.053 | 1.029 |
| R1/wR2 [I > 2s(I)] | R1 = 0.0543 | R1 = 0.0663 | R1 = 0.0718 | R1 = 0.0549 |
| wR2 = 0.1158 | wR2 = 0.1887 | wR2 = 0.1564 | wR2 = 0.1391 | |
| R1/wR2 (all data) | R1 = 0.0705 | R1 = 0.0893 | R1 = 0.1162 | R1 = 0.0702 |
| wR2 = 0.1234 | wR2 = 0.2054 | wR2 = 0.1770 | wR2 = 0.1512 | |
| Largest diff. peak/e A−3 | 0.496 and −0.330 | 0.907 and −0.458 | 0.720 and −0.446 | 1.130 and −0.627 |
| Complex 1 | |||||
|---|---|---|---|---|---|
| Ni(1)–N(5) | 2.056(3) | Ni(1)–N(3) | 2.076(3) | Ni(1)–N(4) | 2.262(3) |
| Ni(1)–N(1) | 2.068(3) | Ni(1)–N(2) | 2.100(3) | Ni(1)–Cl(1) | 2.3759(10) |
| N(5)–Ni(1)–N(1) | 96.07(12) | N(3)–Ni(1)–N(2) | 94.49(12) | N(5)–Ni(1)–Cl(1) | 96.34(8) |
| N(5)–Ni(1)–N(3) | 90.64(12) | N(5)–Ni(1)–N(4) | 80.25(10) | N(1)–Ni(1)–Cl(1) | 89.99(8) |
| N(1)–Ni(1)–N(3) | 171.61(12) | N(1)–Ni(1)–N(4) | 99.38(11) | N(3)–Ni(1)–Cl(1) | 94.27(8) |
| N(5)–Ni(1)–N(2) | 173.62(12) | N(3)–Ni(1)–N(4) | 76.77(10) | N(2)–Ni(1)–Cl(1) | 87.05(8) |
| N(1)–Ni(1)–N(2) | 78.49(13) | N(3)–Ni(1)–N(4) | 97.23(10) | N(4)–Ni(1)–Cl(1) | 170.30(8) |
| Complex 2 | |||||
|---|---|---|---|---|---|
| Ni(1)–N(5) | 2.065(4) | Ni(1)–N(2) | 2.088(4) | Ni(1)–N(4) | 2.259(4) |
| Ni(1)–N(3) | 2.086(4) | Ni(1)–N(1) | 2.101(4) | Ni(1)–Cl(1) | 2.3839(14) |
| N(5)–Ni(1)–N(3) | 92.68(17) | N(2)–Ni(1)–N(1) | 79.00(17) | N(5)–Ni(1)–Cl(1) | 97.34(12) |
| N(5)–Ni(1)–N(2) | 168.64(17) | N(5)–Ni(1)–N(4) | 79.98(15) | N(3)–Ni(1)–Cl(1) | 96.05(13) |
| N(3)–Ni(1)–N(2) | 93.51(18) | N(3)–Ni(1)–N(4) | 77.00(16) | N(2)–Ni(1)–Cl(1) | 91.47(13) |
| N(5)–Ni(1)–N(1) | 93.60(16) | N(2)–Ni(1)–N(4) | 92.16(16) | N(1)–Ni(1)–Cl(1) | 91.21(11) |
| N(3)–Ni(1)–N(1) | 169.72(16) | N(1)–Ni(1)–N(4) | 96.11(15) | N(4)–Ni(1)–Cl(1) | 172.34(11) |
| Complex 3 | |||||
|---|---|---|---|---|---|
| Ni(1)–N(5) | 2.069(5) | Ni(1)–N(7) | 2.078(4) | Ni(1)–N(6) | 2.255(4) |
| Ni(1)–N(1) | 2.081(4) | Ni(1)–N(2) | 2.106(4) | Ni(1)–Cl(1) | 2.3685(16) |
| N(5)–Ni(1)–N(1) | 166.22(17) | N(7)–Ni(1)–N(2) | 170.53(16) | N(5)–Ni(1)–Cl(1) | 95.92(13) |
| N(5)–Ni(1)–N(7) | 94.23(17) | N(5)–Ni(1)–N(6) | 79.73(16) | N(1)–Ni(1)–Cl(1) | 93.73(13) |
| N(1)–Ni(1)–N(7) | 94.83(17) | N(1)–Ni(1)–N(6) | 92.02(16) | N(7)–Ni(1)–Cl(1) | 94.40(12) |
| N(5)–Ni(1)–N(2) | 91.72(17) | N(7)–Ni(1)–N(6) | 78.03(16) | N(2)–Ni(1)–Cl(1) | 92.31(12) |
| N(1)–Ni(1)–N(2) | 78.05(17) | N(2)–Ni(1)–N(6) | 95.83(15) | N(6)–Ni(1)–Cl(1) | 170.86(11) |
| Complex 4 | |||||
|---|---|---|---|---|---|
| Ni(1)–N(7) | 2.064(3) | Ni(1)–N(5) | 2.093(3) | Ni(1)–N(6) | 2.284(3) |
| Ni(1)–N(1) | 2.086(3) | Ni(1)–N(2) | 2.106(3) | Ni(1)–Cl(1) | 2.3607(12) |
| N(7)–Ni(1)–N(1) | 168.96(12) | N(5)–Ni(1)–N(2) | 168.07(12) | N(7)–Ni(1)–Cl(1) | 95.19(10) |
| N(7)–Ni(1)–N(5) | 93.52(13) | N(7)–Ni(1)–N(6) | 79.63(12) | N(1)–Ni(1)–Cl(1) | 92.52(10) |
| N(1)–Ni(1)–N(5) | 93.79(12) | N(1)–Ni(1)–N(6) | 93.86(12) | N(5)–Ni(1)–Cl(1) | 94.10(9) |
| N(7)–Ni(1)–N(2) | 92.92(12) | N(5)–Ni(1)–N(6) | 77.81(12) | N(2)–Ni(1)–Cl(1) | 95.30(9) |
| N(1)–Ni(1)–N(2) | 78.48(12) | N(2)–Ni(1)–N(6) | 93.51(11) | N(6)–Ni(1)–Cl(1) | 170.01(8) |
2.4 UV-visible studies
The electronic absorption spectra of 1–4 were recorded at 25 °C on a JASCO V-570 spectrophotometer over the spectral range of 200–1100 nm, in 1 cm cuvettes. Samples containing 50 μL 10−3 M Ni(II) complexes in acetonitrile and 2 mL H2O were prepared.2.5 DNA-binding and cleavage experiments
The UV absorbance at 260 nm and 280 nm of the CT-DNA solution in 5 mM Tris–HCl/50 mM NaCl buffer (pH = 7.2) gives a ratio of 1.8–1.9, indicating that the DNA was sufficiently free of protein.48 The concentration of CT-DNA was determined from its absorption intensity at 260 nm with a molar extinction coefficient of 6600 M−1 cm−1.49 The absorption spectra of complexes 1–4 binding to DNA were performed by increasing amounts of CT-DNA to the complexes in Tris–HCl buffer (pH = 7.2).The relative binding of complexes to CT-DNA was studied with an EB-bound CT-DNA solution in 5 mM Tris–HCl/50 mM NaCl buffer (pH = 7.2). The fluorescence spectra were recorded at room temperature with excitation at 510 nm and emission at about 602 nm. The experiments were carried out by titrating complexes into EB–DNA solution containing 2.4 × 10−6 M EB and 4.8 × 10−5 M CT-DNA.
The DNA cleavage experiments were done by agarose gel electrophoresis, which was performed by incubation at 37 °C as follows: pBR322 DNA (0.1 μg μL−1) in 50 mM Tris–HCl/18 mM NaCl buffer (pH = 7.2) was treated with 1–4. The samples were incubated for 3 h, and then loading buffer (0.25% bromophenol blue, 45% glycerol and 2 mM EDTA) was added. Then the samples were electrophoresed for 2 h at 120 V on 0.9% agarose gel using Tris–boric acid–EDTA buffer. After electrophoresis, bands were visualized by UV light and photographed. The extent of cleavage of the SC DNA was determined by measuring the intensities of the bands using the Gel Documentation System.50
Cleavage mechanistic investigation of pBR322 DNA was carried out in the presence of standard radical scavengers and reaction inhibitors. These reactions were carried out by adding standard radical scavengers of NaN3, KI, D2O, SOD, catalase and EDTA to pBR322 DNA prior to the addition of complex. Cleavage experiment was initiated by the addition of complex and quenched with 2 μL of loading buffer. Further analysis was carried out by the above standard method.
2.6 Protein binding studies
The protein binding study was performed by tryptophan fluorescence quenching experiments using bovine serum albumin stock solution (BSA, 1.5 mM) in 10 mM phosphate buffer (pH = 7.0). A concentrated stock solution of the compounds was prepared as used for the DNA binding experiments, except that the phosphate buffer was used instead of a Tris–HCl buffer for all of the experiments. The fluorescence spectra were recorded at room temperature with excitation wavelength of BSA at 280 nm and the emission at 342 nm by keeping the concentration of BSA constant (29.4 μM) while varying the complex concentration from 0 to 8.89 μM.2.7 Cell culture
Human breast cancer cell line (MCF-7), human liver hepatocellular carcinoma cell line (HepG-2) and human gastric cancer cell line (SGC-7901) obtained from the American Type Culture Collection (Rockville, MD, USA) were grown in DMEM medium which were supplemented with 100 units mL−1 penicillin, 100 μg mL−1 streptomycin and 10% FBS. Cells were maintained in a humidified atmosphere containing 95% air and 5% CO2 at 37 °C. The cells were harvested and plated for subsequent drug treatments when they reached 80% confluence.2.8 MTT assay
Cell viability was examined by MTT assay which is a colorimetric assay based on the conversion of the yellow tetrazolium salt MTT to purple formazan crystals by metabolically active cells.51 Varied kinds of Cells (5 × 103 per well) were plated in 96-well microplates and three replica wells were used for controls. Graded amounts of complexes 1–4 at various concentrations of 0, 12.5, 25.0, 50.0 and 100.0 μM in 10 μL of FBS-free culture medium were added to the wells when the cells reached 80% confluence and the plates were incubated in a 5% CO2 humidified atmosphere for 48 h, respectively. After the drug treatments, 20 μL of 5 mg mL−1 MTT in phosphate buffered saline (PBS, pH 7.4) was added to each well for an additional 4 h. Then the medium was removed and 100 μL of DMSO was added to dissolve the MTT formazan precipitate. The absorbance of samples was measured at 570 nm with an enzyme-linked immunosorbent assay (ELISA) reader. Cytotoxicity effect was revealed as the percentage of treated cells relative to untreated cells at A570 nm. The value of IC50 was applied to express the sensitivity of cells to the drug treatment. All data were expressed as mean ± SD (standard deviation) unless otherwise stated. Statistical significance (P < 0.05) was performed by one-way ANOVA followed by an assessment of differences using SPSS 13.0 software. Three independent experiments were performed to represent the data.2.9 Hoechst 33342 staining
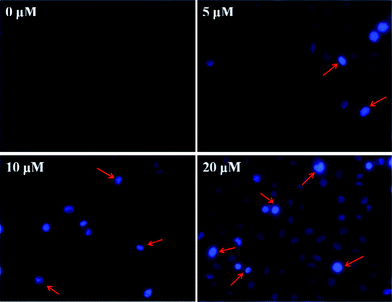
DNA-binding fluorochrome Hoechst 33342 staining was use to measure the changes in nucleic morphology of apoptotic cells. HepG-2 cells (8 × 103 per well) were incubated with complex 4 at 0, 5, 10 and 20 μM for 48 h. After the drug treatments, cells were stained with Hoechst 33342 (1 μg mL−1) for 15 min at 37 °C in the dark. Finally, the samples were visualized under a fluorescence microscopy (Nikon ECLIPSE Ti).3. Results and discussion
3.1 Description of the crystal structures
Mononuclear complexes 1–4 have been structurally characterized by X-ray crystallography (Fig. 1), the parameters of refinement process and the selected bond lengths and angles are given respectively in Tables 1 and 2. All Ni centers are hexacoordinated with N5Cl donor sets, and the geometry around metal centers can be best described as distorted octahedrons with the axial bonds longer than the equatorial ones.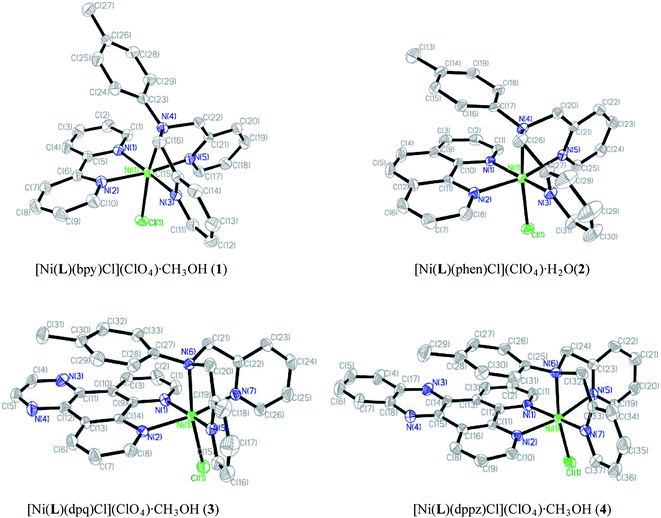 | ||
| Fig. 1 ORTEP view of the molecular structures and atom-labeling schemes of 1–4. Hydrogen atoms, negative ions and dissociative small molecules are omitted for clarity. | ||
The structural coordination models of the cationic mixed-ligand complexes 1–4 are essentially similar with the NiN5Cl coordination, but with not totally identical crystal systems and space groups. 1 crystallizes in a monoclinic cell with C2/c space group, and the diimine nitrogen atoms (N(1) and N(2)) of the bpy and two pyridine nitrogen atoms (N(3) and N(5)) of the tridentate ligand L occupy the corners of the basal plane. A chlorine atom (Cl(1)) and the tertiary amino nitrogen atom (N(4)) of the ligand L occupy the axial positions with distances longer (Ni(1)–Cl(1), 2.3759(10) Å; Ni(1)–N(4), 2.262(3) Å) than the equatorial ones (average Ni–N, 2.075(3) Å). The dihedral angle between benzene ring of the ligand L and the plane of the diimine (bpy) is 52.5°. Compounds 2 and 4 crystallize in monoclinic system with space group P2(1)/c, while complex 3 belongs to triclinic system with space group P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) . Similar to the bpy 1, four coordinated nitrogen atoms from diimine (phen for 2, dpq for 3, dppz for 4) and two pyridine nitrogen atoms located at the equator plane (Ni(1)–N: 2.064(3) to 2.106(4) Å), and chlorine atom (Cl(1)) and the tertiary amino nitrogen atom of the ligand occupy the axial positions at distances (Ni(1)–N: 2.255(4) to 2.284(3) Å, Ni(1)–Cl: 2.3607(12) to 2.3839(14) Å) longer than the equatorial ones. The dihedral angles between benzene ring of ligand L and the plane of the diimine (phen for 2, dpq for 3, dppz for 4) drop to 13.2°, 9.1°, 6.8°, respectively. The schematic diagram for the dihedral angle is shown in Scheme S1.† The encumbrance effects increase gradually for the phen, dpq and dppz species with the decrease of dihedral angles, however, in complex 4, the dppz ligand has an extended aromatic moiety stemmed partially outside ligand L. The result exhibits that the decrease of dihedral angles for 1–4 could be due to the regularly stronger π–π interaction between phenyl of the ligand L and the aromatic conjugated system of diimine (bpy, phen, dpq and dppz).
. Similar to the bpy 1, four coordinated nitrogen atoms from diimine (phen for 2, dpq for 3, dppz for 4) and two pyridine nitrogen atoms located at the equator plane (Ni(1)–N: 2.064(3) to 2.106(4) Å), and chlorine atom (Cl(1)) and the tertiary amino nitrogen atom of the ligand occupy the axial positions at distances (Ni(1)–N: 2.255(4) to 2.284(3) Å, Ni(1)–Cl: 2.3607(12) to 2.3839(14) Å) longer than the equatorial ones. The dihedral angles between benzene ring of ligand L and the plane of the diimine (phen for 2, dpq for 3, dppz for 4) drop to 13.2°, 9.1°, 6.8°, respectively. The schematic diagram for the dihedral angle is shown in Scheme S1.† The encumbrance effects increase gradually for the phen, dpq and dppz species with the decrease of dihedral angles, however, in complex 4, the dppz ligand has an extended aromatic moiety stemmed partially outside ligand L. The result exhibits that the decrease of dihedral angles for 1–4 could be due to the regularly stronger π–π interaction between phenyl of the ligand L and the aromatic conjugated system of diimine (bpy, phen, dpq and dppz).
3.2 UV-visible spectrum
The electronic spectral features of complexes 1–4 (2.44 × 10−5 M, 2.4% CH3CN) exhibit distinct absorption spectra which are shown in Fig. 2, the aqueous acetonitrile exhibiting absorption at ∼190 nm will not interfere with the progress of the work. In the spectra of all complexes, several high energy bands observed from 208 to 380 nm (ε = 107![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 018–10
018–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) are assigned to intraligand and π–π* transitions. Furthermore, complexes 1 (bpy) and 4 (dppz) show strong broad bands around 300 (ε = 17
000) are assigned to intraligand and π–π* transitions. Furthermore, complexes 1 (bpy) and 4 (dppz) show strong broad bands around 300 (ε = 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 011) and 380 nm (ε = 10
011) and 380 nm (ε = 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), and complexes 2 (phen) and 3 (dpq) exhibit moderately strong bands at 294 nm (ε = 9554) and 338 nm (ε = 3422), which could be due to π–π* transition involving the aromatic conjugated system like bipyridy, quinoxaline or phenazine moieties of the diimine ligands.52
000), and complexes 2 (phen) and 3 (dpq) exhibit moderately strong bands at 294 nm (ε = 9554) and 338 nm (ε = 3422), which could be due to π–π* transition involving the aromatic conjugated system like bipyridy, quinoxaline or phenazine moieties of the diimine ligands.52
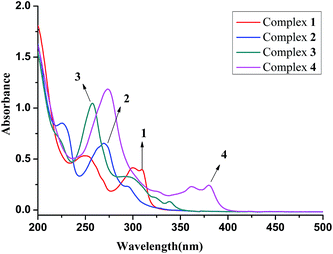 | ||
| Fig. 2 Overlay UV-vis spectra of complexes 1–4 (2.44 × 10−5 M) recorded in aqueous acetonitrile (2.4% CH3CN). | ||
3.3 DNA-binding and cleavage activities
| Complex | Complex/CT-DNA | Complex/BSA | ||||
|---|---|---|---|---|---|---|
| Kb (M−1) | Kapp (M−1) | KSV (M−1) | Kq (M−1 s−1) | K (M−1) | n | |
| 1 | 8.75 × 104 | 5.79 × 105 | 1.84 × 104 | 1.84 × 1012 | 6.09 × 102 | 0.71 |
| 2 | 9.99 × 104 | 6.26 × 105 | 2.66 × 104 | 2.66 × 1012 | 3.20 × 103 | 0.83 |
| 3 | 3.92 × 105 | 1.38 × 106 | 2.62 × 104 | 2.62 × 1012 | 4.27 × 103 | 0.84 |
| 4 | 5.91 × 105 | 1.66 × 106 | 7.04 × 104 | 7.04 × 1012 | 2.72 × 104 | 0.92 |
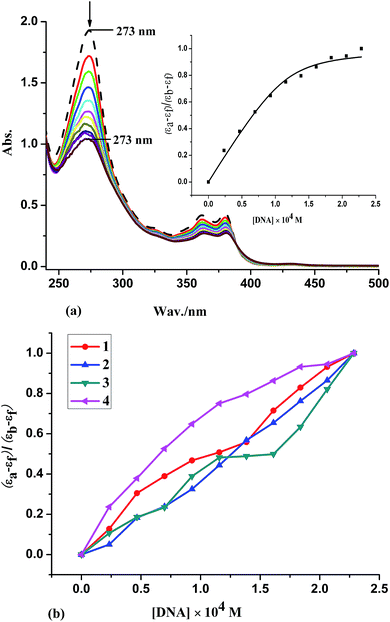
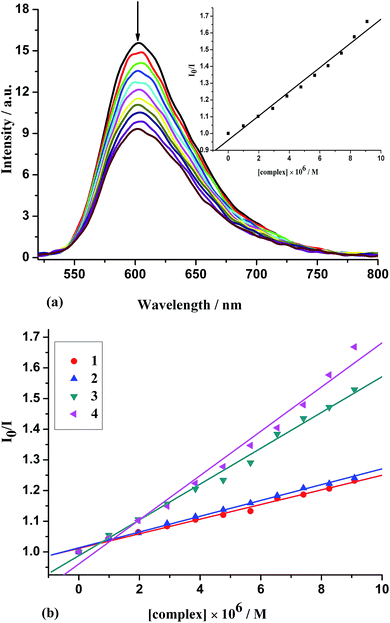
As a means for further clarifying the binding of the complexes, fluorescence spectral measurements were carried out on CT-DNA by varying the concentration of the complexes. No luminescence is observed for all complexes at room temperature, and therefore the binding of the complexes to CT-DNA is evaluated by the fluorescence emission intensity of EB bound to DNA as a probe. EB emits intense fluorescent light in the presence of DNA due to its strong intercalation between the adjacent DNA base pairs,59 which could be quenched by the addition of another molecule. The relative binding propensity of the complex 4 to CT-DNA studied with an EB bound CT-DNA solution in 5 mM Tris–HCl/50 mM NaCl buffer (pH = 7.2) is shown in Fig. 4(a) (similar spectra of 1–3 are presented as Fig. S2†), and a plot of I0/I versus [complex] for the quenched intensity of 1–4 to EB-DNA is shown in Fig. 4(b). Fluorescence intensities at 602 nm (510 nm excitation) were measured at different complex concentrations. The extent of reduction of the emission intensity gives a measure of the binding propensity of the complex to DNA. According to the classical Stern–Volmer equation,60 I0/I = 1 + K[Q], the quenching plot illustrates that the quenching of EB bound to CT-DNA by complex is in agreement with the linear Stern–Volmer equation, which also indicates the complexes bind to DNA. On the basis of the equation KEB[EB] = Kapp[complex], where the complex concentration was the value at a 50% reduction of the fluorescence intensity of EB and KEB = 1.0 × 107 M−1, ([EB] = 2.4 μM). The calculated apparent binding constant values (Kapp) (Table 3) follow the order: 4 (dppz) (1.66 × 106 M−1) > 3 (dpq) (1.38 × 106 M−1) > 2 (phen) (6.26 × 105 M−1) > 1 (bpy) (5.79 × 105 M−1), which is consistent with the results obtained Kb values by UV spectroscopy. On the whole, the binding constants are less than that of the classical intercalators and metallointercalators (107 M−1),61 indicating medium binding strength of the complexes with CT-DNA.
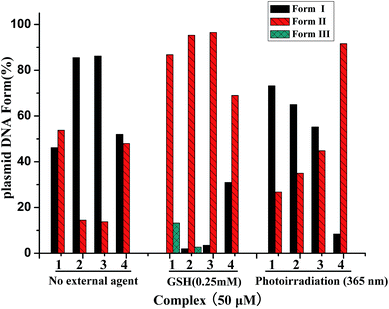 | ||
| Fig. 5 The histogram of relative amounts according to Fig. S3 (absence of external agent), S4† (presence of GSH) and S5† (photoirradiation at 365 nm) showing the cleavage of pBR322 DNA (0.1 μg μL−1) for complexes 1–4 (50 μM, 3 h) in Tris–HCl/NaCl buffer (pH = 7.2) and 37 °C. | ||
The concentration-dependent DNA cleavage activities by 1–4 were also performed in the presence of GSH (glutathione), the DNA cleavage efficiencies of all complexes exhibit remarkable increases at the same conditions (Fig. S4†), at a concentration of 5 μM Ni2+, all of them cause the extent of DNA cleavage, which implies that GSH as a revulsant or an activator plays a vital role. As shown in the Fig. 5, at 50 μM concentration, the DNA cleavage efficiencies (Form I into Form II and Form III) follow the order of 1 (86.8% Form II and 13.2% Form III) > 2 (95.3% Form II and 2.7% Form III) > 3 (96.5% Form II) > 4 (69% Form II).
In addition, the photo-induced concentration-dependent DNA cleavage activity of 1–4 has been also studied on irradiation with monochromatic UV light of 365 nm for 3 h. As shown in Fig. S5,† it has been observed that the dppz complex 4 is an efficient cleaver of SC DNA and produces ∼92% of NC DNA at 20 μM concentration. The percentages of SC DNA of phen 2 and dpq 3 both gradually reduced with the increase of concentration, whereas little NC DNA was found indicating undetectable minor fragments generation. The cleavage efficiency of bpy 1 shows more or less equal to the condition of without any external agents, which implies little impact on photoirradiation at 365 nm. As shown in the Fig. 5, at 50 μM concentration, the photo-induced DNA cleavage efficiencies (Form I into Form II) varied in the order dppz 4 (91.6%) > dpq 3 (44.8%)> phen 2 (35%)> bpy 1 (26.8% Form II). A. R. Chakravarty and co-workers36–38 have confirmed that the increase of steric hindrance has positive effects on the photoinduced DNA-cleavage activity of complexes and explained that the steric effect can increase its excited-state 3(n–π*) or 3(π–π*) lifetime in favor of causing the cleavage of DNA.
In order to obtain the information about the active oxygen species (ROS) which was responsible for the DNA damage, we investigated the potential mechanism of DNA cleavage mediated by the complexes in the presence of GSH. A series of DNA cleavage experiments (Fig. S6†) were carried out using reagents like NaN3 as singlet oxygen (1O2) quencher, KI as hydroxyl radical scavengers (OH˙), superoxide dismutase (SOD) as O2− radical scavenger, catalase as hydrogen peroxide scavenger and EDTA as the chelator of complexes. Fig. 6 showed the extent of DNA cleavage after adding standard radical scavengers, addition of NaN3 to SC DNA completely inhibited the DNA cleavage activity, and D2O enhanced the DNA cleavage,64 which suggested the possible involvement of singlet oxygen as the reactive species. Also, the complexes showed complete or partial inhibition in the DNA-cleavage activity in the presence of the hydroxyl radial scavenger KI, while no obvious inhibitions were observed for other radical scavengers. Therefore, the data suggest the involvement of both singlet oxygen (1O2) and hydroxyl radicals (OH˙) as ROS.
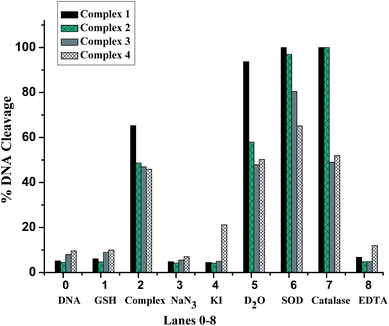 | ||
| Fig. 6 The histogram of relative amounts according to Fig. S5† shows the cleavage of plasmid pBR322 DNA (0.1 μg μL−1) in presence of 15 μM complexes 1–4 and different inhibitors after 3 h incubation at 37 °C. Lane 0: DNA control; lane 1: DNA + 0.25 mM GSH; lane 2: DNA + 0.25 mM GSH + complex; lane 3–8: DNA + 0.25 mM GSH + complex + inhibitors (0.1 M NaN3, 0.1 M KI, 25% (v/v) D2O, 2 U mL−1 SOD, 0.2 U mL−1 catalase, 0.5 mM EDTA. | ||
3.4 Protein binding studies
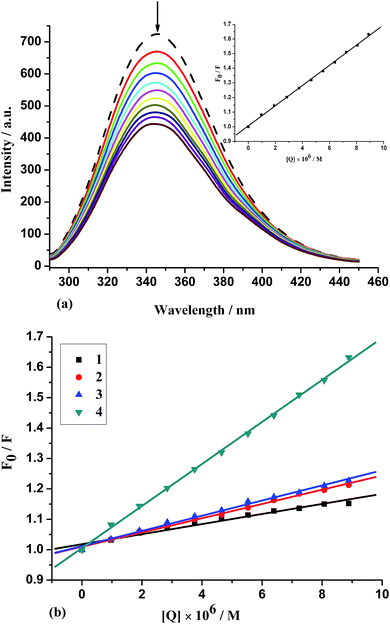
The interactions between serum albumin and chemicals have attracted increasing research interest in recent years, since serum albumin constitutes ∼55% of the total protein in blood plasma and it plays a pivotal role in drug transport and drug metabolism.65,66 Bovine serum albumin (BSA) is the most extensively studied serum albumin, due to its structural homology with human serum albumin (HSA). The intrinsic fluorescence of BSA is associated with the tryptophan, tyrosine and phenylalanine residues, although tryptophan is the most significant contributor.66 Fig. 7(a) shows the effect of increasing the concentration of added complex 4 on the fluorescence emission of BSA (similar spectra of 1–3 are presented as Fig. S7†). The intensity of the characteristic broad emission band at 342 nm decreases regularly with the increasing concentration of complexes, which confirms that the interaction between complexes and BSA have occurred. The fluorescence quenching is described by the Stern–Volmer equation, F0/F = 1 + Kqτ0[Q] = 1 + KSV[Q]. Where F0 and F represent the fluorescence intensities in the absence and in the presence of quencher, kq is the quenching rate constant, τ0 the average life-time of the biomolecule without quencher (about 10−8 s),60 KSV the Stern–Volmer quenching constant and [Q] the concentration of quencher. Fig. 7(b) displays the Stern–Volmer plots of the quenching of BSA fluorescence by different complexes, and KSV can be obtained by a slope from the plot of F0/F vs. [Q]. The calculated values of KSV and kq for the interaction of the complexes with BSA are given in Table 3 and the KSV values follow the order: 4 (dppz) (7.04 × 104 M−1) > 2 (phen) (2.66 × 104 M−1) ≈ 3 (dpq) (2.62 × 104 M−1) > 1 (bpy) (1.84 × 104 M−1).Quenching mechanisms are usually classified as dynamic quenching and static quenching; dynamic quenching refers to a process in which the fluorophore and the quencher come into contact during the transient existence of the exited state. Static quenching refers to fluorophore-quencher complex formation. The kq values (∼1012 M−1 s−1) of 1–4 are higher than the maximum scatter collision-quenching constant of diverse kinds of quenchers for biopolymers fluorescence (2 × 1010 M−1 s−1) indicating the existence of static quenching mechanism.67
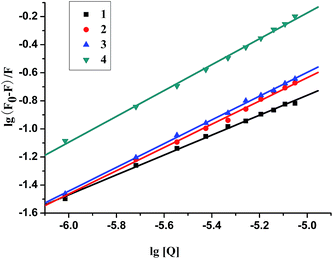
For the static quenching interaction, the binding constant (K) and the number of binding sites (n) can be determined according to the Scatchard equation:68 log(F0 − F)/F = log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K + n log[Q]. The n and K can be calculated by the slope and the intercept of the double logarithm regression curve of log(F0 − F)/F versus log[Q] (Fig. 8). Table 3 shows that 4 exhibits higher binding constants for BSA than 1–3, and the K and n values follow the order: 4 (dppz) (2.72 × 104 M−1, 0.92) > 3 (dpq) (4.27 × 103 M−1, 0.84) > 2 (phen) (3.20 × 103 M−1, 0.83) > 1 (bpy) (6.09 × 102 M−1, 0.71). As expected, the values of n are associated with binding constants K, which verify the conclusion35 that a direct relation between the binding constant and number of binding sites.
K + n log[Q]. The n and K can be calculated by the slope and the intercept of the double logarithm regression curve of log(F0 − F)/F versus log[Q] (Fig. 8). Table 3 shows that 4 exhibits higher binding constants for BSA than 1–3, and the K and n values follow the order: 4 (dppz) (2.72 × 104 M−1, 0.92) > 3 (dpq) (4.27 × 103 M−1, 0.84) > 2 (phen) (3.20 × 103 M−1, 0.83) > 1 (bpy) (6.09 × 102 M−1, 0.71). As expected, the values of n are associated with binding constants K, which verify the conclusion35 that a direct relation between the binding constant and number of binding sites.
3.5 MTT assay
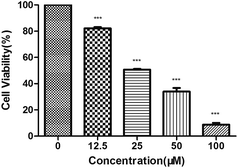
MTT assay is a colorimetric assay based on the conversion of the yellow tetrazolium salt to purple formazan crystals by metabolically active cells. The cytotoxic effects of complex 1–4 on the viability of several human cancer cell lines (MCF-7, HepG-2 and SGC-7901) were examined by MTT assay. A variety of tumor cells were treated with 1–4 and incubated for 48 h at increasing concentration, respectively. As shown in Table 4, we observe that 1–4 are all cytotoxic to tested tumor cells and inhibit the growth of cells in a dose-dependent manner, while 3 and 4 show better antitumor effect than 1, 2. Based on the comparison of IC50 values, complex 4 is most likely to become potential anticancer drugs among these complexes. In particular, 4 exhibited a strong anti-proliferative effect on HepG-2 cells (Fig. 9) with the IC50 value of 28.9 ± 2.1 μM.| Complexes (μM)/cell lines | MCF-7 | HepG-2 | SGC-7901 |
|---|---|---|---|
| 1 | >100 | >100 | >100 |
| 2 | >100 | >100 | >100 |
| 3 | 89.2 ± 5.7 | 50.0 ± 3.2 | 97.8 ± 6.5 |
| 4 | 34.4 ± 3.3 | 28.9 ± 2.1 | 69.9 ± 4.9 |
| Cisplatin | — | 25 ± 3.1 | <6.5 |
In order to give a comparison, NiCl2 standards were used in the biological experiment of MTT assay. We found that NiCl2 itself showed a very weak inhibitory effect on HepG-2 cells (IC50 = 326.9 ± 45.1 μM), as 11 fold as complex 4 (IC50 = 28.9 ± 2.1 μM), 6 fold for complex 3 (IC50 = 50.0 ± 3.2 μM), suggesting that the observed differences were not decided simply by the releasing the Ni2+ in the solution solely.
In addition, cytotoxicity was measured on human normal liver cell line LO2 for complex 4, and cisplatin was tested as control. The result indicates that complex 4 and cisplatin are both cytotoxic to LO2 cells. In HepG-2 cells and LO2 cells, the IC50 values at 48 h were 28.9 μM and 29.0 μM for complex 4 while 25.0 μM and 21.2 μM for cisplatin, respectively. We used “cytotoxic selection index (SI)” to evaluate the selectivity between cancer cells and normal cells for our synthetic complex. The SI value of complex 4 is 1.00 and cisplatin is 0.85. It suggests that the selectivity of 4 on HepG-2 cells is stronger than cisplatin. The results show that study on the anti-tumor effect of complex 4 is valuable.69,70
3.6 Hoechst 33342 staining
Hoechst 33342 staining was applied to investigate the influence of complex 4 on apoptosis in HepG-2 cells. After 48 h of exposure to 4, cells were checked for cell morphology changes under fluorescence microscopy. In Fig. 10, a stronger blue fluorescence can be observed in apoptotic cells compared with non-apoptotic cells. Moreover, with the increasing dosage of 4, significant morphological changes in nucleus were evidently observed, such as reduction in nuclear size, chromatin condensation, and DNA fragmentation, which are typical characteristics of apoptosis. The results show that complex 4 induced apoptosis in HepG2 cells.4. Conclusion
A series of mononuclear mixed-ligand nickel(II) complexes have been synthesized and characterized. Crystal structures of 1–4 show that the dihedral angles between benzene ring of ligand L and the plane of the diimine (bpy, phen, dpq and dppz) drop considerably (52.5–6.8°) and cause the increase in steric hindrance. The steric hindrance has a profound effect on the DNA interaction with the complexes. Partial intercalation between the complexes and CT-DNA has been confirmed by using absorption and emission spectral methods and medium binding strength follow the order: 4 (dppz) > 3 (dpq) > 2 (phen) > 1 (bpy). With the single condition change such as absence of an external agent, presence of GSH and photoirradiation at 365 nm, the DNA cleavage abilities of complexes exhibit remarkable changes. The oxidative mechanism was demonstrated via a pathway involving formation of both singlet oxygen (1O2) and hydroxyl radicals (OH˙) as active oxygen species. Further, the ability to bind to proteins (BSA) also has been explored, and the quenching mechanisms of BSA by the complexes are static procedures. The vitro cytotoxicity of the complexes on tumor cells lines (MCF-7, HepG-2 and SGC-7901) have been assessed by MTT, the results indicate that 4 shows better antitumor effect than others and induces apoptosis in HepG-2 cells.Acknowledgements
This work was supported by the National Natural Science Foundation of China (no. 21171101, 21471085 and 21371135), MOE Innovation Team (IRT13022) of China, Scientific Research Foundation of Shanxi Agricultural University (no. 2013YJ40 and 2014013) and NFFTBS (no. J1103306).References
- K. E. Erkkila, D. T. Odom and J. K. Barton, Chem. Rev., 1999, 99, 2777–2796 CrossRef CAS PubMed.
- B. Armitage, Chem. Rev., 1998, 98, 1171–1200 CrossRef CAS PubMed.
- W. K. Pogozelski and T. D. Tullius, Chem. Rev., 1998, 98, 1089–1107 CrossRef CAS PubMed.
- C. J. Burrows and J. G. Muller, Chem. Rev., 1998, 98, 1109–1152 CrossRef CAS PubMed.
- D. R. McMillin and K. M. McNett, Chem. Rev., 1998, 98, 1201–1220 CrossRef PubMed.
- D. S. Sigman, T. W. Bruice, A. Mazumder and C. L. Sutton, Acc. Chem. Res., 1993, 26, 98–104 CrossRef CAS.
- G. Pratviel, J. Bernadou and B. Meunier, Adv. Inorg. Chem., 1998, 45, 251–312 CrossRef CAS.
- A. M. Pyle and J. K. Barton, Prog. Inorg. Chem., 1990, 38, 413–475 CrossRef CAS.
- J. K. Barton, Science, 1986, 233, 727–734 CrossRef CAS.
- J. K. Barton and A. L. Raphael, Proc. Natl. Acad. Sci. U. S. A., 1985, 82, 6460–6464 CrossRef CAS.
- S. Delaney, M. Pascaly, P. K. Bhattacharya, K. Han and J. K. Barton, Inorg. Chem., 2002, 41, 1966–1974 CrossRef CAS PubMed.
- P. A. Sontz, N. B. Muren and J. K. Barton, Acc. Chem. Res., 2012, 45, 1792–1800 CrossRef CAS PubMed.
- D. S. Sigman, A. Mazumder and D. M. Perrin, Chem. Rev., 1993, 93, 2295–2316 CrossRef CAS.
- D. S. Sigman, Acc. Chem. Res., 1986, 19, 180–186 CrossRef CAS.
- D. S. Sigman, D. R. Graham, V. D'Aurora and A. M. Stern, J. Biol. Chem., 1979, 254, 12269–12272 CAS.
- O. Zelenko, J. Gallagher and D. S. Sigman, Angew. Chem., Int. Ed. Engl., 1997, 36, 2776–2778 CrossRef CAS.
- O. Zelenko, J. Gallagher, Y. Xu and D. S. Sigman, Inorg. Chem., 1998, 37, 2198–2204 CrossRef CAS PubMed.
- J. Reedijk, in Comprehensive Coordination Chemistry, ed. G. Wilkinson, R. D. Gillard and J. A. McCleverty, Pergamon Press, Oxford, 1987, vol. 2, p. 73 Search PubMed.
- G. R. Newkome, D. K. Kohli and F. Fronczek, Chem. Commun., 1980, 9–11 RSC.
- T. Garber, S. V. Wallendael, D. P. Rillema, M. Kirk, W. E. Hatfield, J. H. Welch and P. Singh, Inorg. Chem., 1990, 29, 2863–2868 CrossRef CAS.
- M. Athar Masood and P. S. Zacharias, Dalton Trans., 1991, 111–114 RSC.
- R. J. Crutchley, R. Hynes and E. J. Gabe, Inorg. Chem., 1990, 29, 4921–4928 CrossRef CAS.
- D. H. Lee, N. N. Murthy and K. D. Karlin, Inorg. Chem., 1997, 36, 5785–5792 CrossRef CAS PubMed.
- G. A. V. Albada, A. Mohamadou, I. Mutikainen, U. Turpeinen and J. Reedijk, Eur. J. Inorg. Chem., 2004, 3733–3742 Search PubMed.
- T. Pintauer, J. Qiu, G. Kickelbick and K. Matyjaszewski, Inorg. Chem., 2001, 40, 2818–2824 CrossRef CAS PubMed.
- S. Itoh, N. Kishikawa, T. Suzuki and H. D. Takagi, J. Chem. Soc., Dalton Trans., 2005, 1066–1078 RSC.
- T. Joshi, G. J. Barbante, P. S. Francis, C. F. Hogan, A. M. Bond and L. Spiccia, Inorg. Chem., 2011, 50, 12172–12183 CrossRef CAS PubMed.
- S. Dhar, D. Senapati, P. K. Das, P. Chattopadhyay, M. Nethaji and A. R. Chakravarty, J. Am. Chem. Soc., 2003, 125, 12118–12124 CrossRef CAS PubMed.
- T. Kojima, T. Morimoto, T. Sakamoto, S. Miyazaki and S. Fukuzumi, Chem.–Eur. J., 2008, 14, 8904–8915 CrossRef CAS PubMed.
- R. Loganathan, S. Ramakrishnan, E. Suresh, A. Riyasdeen, M. A. Akbarsha and M. Palaniandavar, Inorg. Chem., 2012, 51, 5512–5532 CrossRef CAS PubMed.
- Y. Jin and J. A. Cowan, J. Am. Chem. Soc., 2005, 127, 8408–8415 CrossRef CAS PubMed.
- N. Valls, I. Usón, C. Gouyette and J. A. Subirana, J. Am. Chem. Soc., 2004, 126, 7812–7816 CrossRef CAS PubMed.
- S. Arounaguiri and B. G. Maiya, Inorg. Chem., 1996, 35, 4267–4270 CrossRef CAS.
- Q. L. Zhang, J. G. Liu, J. Liu, G. Q. Xue, H. Li and J. Z. Liu, J. Inorg. Biochem., 2001, 85, 291–296 CrossRef CAS.
- P. Sathyadevi, P. Krishnamoorthy, R. R. Butorac, A. H. Cowley, N. S. P. Bhuvanesh and N. Dharmaraj, Dalton Trans., 2011, 9690–9702 RSC.
- S. Dhar, P. A. N. Reddy, M. Nethaji, S. Mahadevan, M. K. Saha and A. R. Chakravarty, Inorg. Chem., 2002, 41, 3469–3476 CrossRef CAS PubMed.
- S. Dhar, M. Nethaji and A. R. Chakravarty, Inorg. Chem., 2005, 44, 8876–8883 CrossRef CAS PubMed.
- S. Roy, A. K. Patra, S. Dhar and A. R. Chakravarty, Inorg. Chem., 2008, 47, 5625–5633 CrossRef CAS PubMed.
- M. C. Rodŕıguez-Arguelles, M. B. Ferrari, F. Biscegli, C. Pellizi, G. Pelosi, S. Pinelli and M. Sassi, J. Inorg. Biochem., 2004, 98, 313–321 CrossRef PubMed.
- F. Liang, P. Wang, X. Zhou, T. Li, Z. Li, H. Lin, D. Gao, C. Zheng and C. Wu, Bioorg. Med. Chem. Lett., 2004, 14, 1901–1904 CrossRef CAS PubMed.
- Z. Afrasiabi, E. Sinn, W. Lin, Y. Ma, C. Campana and S. Padhye, J. Inorg. Biochem., 2005, 99, 1526–1531 CrossRef CAS PubMed.
- S. Ramakrishnan, E. Suresh, A. Riyasdeen, M. A. Akbarsha and M. Palaniandavar, Dalton Trans., 2011, 3245–3256 RSC.
- C. Y. Gao, X. Qiao, Z. Y. Ma, Z. G. Wang, J. Lu, J. L. Tian, J. Y. Xu and S. P. Yan, Dalton Trans., 2012, 12220–12232 RSC.
- A. Delgadillo, P. Romo, A. M. Leiva and B. Loeb, Helv. Chim. Acta, 2003, 86, 2110–2120 CrossRef CAS.
- M. Navarro, E. J. Cisneros-Fajardo, A. Sierralta, M. Fernandez-Mestre, P. Silva, D. Arrieche and E. Marchan, J. Biol. Inorg. Chem., 2003, 8, 401–408 CAS.
- G. M. Sheldrick, SHELXS-97, Program for the Solution of Crystal Structure, University of Göttingen, Germany, 1997 Search PubMed.
- G. M. Sheldrick, SHELXL-97, Program for the Refinement of Crystal Structure, University of Göttingen, Germany, 1997 Search PubMed.
- J. Marmur, J. Mol. Biol., 1961, 3, 208–218 CrossRef CAS.
- Y. Gultneh, A. R. Khan, D. Blaise, S. Chaudhry, B. Ahvazi, B. B. Marvey and R. J. Butcher, J. Inorg. Biochem., 1999, 75, 7–18 CrossRef CAS.
- J. Bermadou, G. Pratviel, F. Bennis, M. Girardet and B. Meunier, Biochemistry, 1989, 28, 7268–7275 CrossRef.
- A. Ghadersohi, D. Pan, Z. Fayazi, D. G. Hicks, J. S. Winston and F. Z. Li, Breast Cancer Res. Treat., 2007, 102, 19–30 CrossRef CAS PubMed.
- R. Gaur and L. Mishra, Inorg. Chem., 2012, 51, 3059–3070 CrossRef CAS PubMed.
- M. Baldini, M. Belicchi-Ferrari, F. Bisceglie, P. P. Dall'Aglio, G. Pelosi, S. Pinelli and P. Tarasconi, Inorg. Chem., 2004, 43, 7170–7179 CrossRef CAS PubMed.
- A. Wolf, G. H. Shimer Jr and T. Meehan, Biochemistry, 1987, 26, 6392–6396 CrossRef.
- J. D. McGhee and P. H. von Hippel, J. Mol. Biol., 1974, 86, 469–489 CrossRef CAS.
- M. T. Carter, M. Rodriguez and A. J. Bard, J. Am. Chem. Soc., 1989, 111, 8901–8911 CrossRef CAS.
- J. Talib, D. G. Harman, C. T. Dillon, J. Aldrich-Wright, J. L. Beck and S. F. Ralph, Dalton Trans., 2009, 504–513 RSC.
- T. Phillips, I. Haq, A. J. H. M. Meijer, H. Adams, I. Soutar, L. Swanson, M. J. Sykes and J. A. Thomas, Biochemistry, 2004, 43, 13657–13665 CrossRef CAS PubMed.
- F. J. Meyer-Almes and D. Porschke, Biochemistry, 1993, 32, 4246–4253 CrossRef CAS.
- J. R. Lakowicz and G. Webber, Biochemistry, 1973, 12, 4161–4170 CrossRef CAS.
- M. Cory, D. D. McKee, J. Kagan, D. W. Henry and J. Allen Miller, J. Am. Chem. Soc., 1985, 107, 2528–2536 CrossRef CAS.
- S. Ramakrishnan, D. Shakthipriya, E. Suresh, V. S. Periasamy, M. A. Akbarsha and M. Palaniandavar, Inorg. Chem., 2011, 50, 6458–6471 CrossRef CAS PubMed.
- T. K. Goswami, B. V. S. K. Chakravarthi, M. Roy, A. A. Karande and A. R. Chakravarty, Inorg. Chem., 2011, 50, 8452–8464 CrossRef CAS PubMed.
- P. B. Merkel and D. R. Kearns, J. Am. Chem. Soc., 1972, 94, 1029–1030 CrossRef CAS.
- D. Gibellini, F. Vitone, P. Schiavone, C. Ponti, M. L. Placa and M. C. Re, J. Clin. Virol., 2004, 29, 282–289 CrossRef CAS.
- J. R. Lakowicz, Principles of Fluorescence Spectroscopy, Springer, New York, 3rd edn, 2006 Search PubMed.
- W. R. Ware, J. Phys. Chem. B, 1962, 66, 455–458 CrossRef CAS.
- G. Scatchard, Ann. N. Y. Acad. Sci., 1949, 51, 660–672 CrossRef CAS PubMed.
- D. Inci, R. Aydın, D. Yılmaz, H. M. Genckal, O. Vatan, N. Cinkılıc and Y. Zorlu, Spectrochim. Acta, Part A, 2015, 136, 761–770 CrossRef CAS PubMed.
- M. Kvasnica, L. Rarova, J. Oklestkova, M. Budesinsky and L. Kohout, Bioorg. Med. Chem., 2012, 20, 6969–6978 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. CCDC 910520–910523. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c4ra16755d |
| ‡ Both authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2015 |