Green and facile synthesis of highly biocompatible carbon nanospheres and their pH-responsive delivery of doxorubicin to cancer cells†
Xiaoli Liu,
Hui Jiang,
Wei Ge,
Changyu Wu,
Donghua Chen,
Qiwei Li,
Yun Chen and
Xuemei Wang*
State Key Laboratory of Bioelectronics (Chien-Shiung Wu Lab), School of Biological Science and Medical Engineering, Southeast University, Nanjing, 210096, PR China. E-mail: xuewang@seu.edu.cn; Tel: +86-25-83792177
First published on 3rd February 2015
Abstract
Developing an efficient nanoparticulate drug-delivery system with a sub-100 nm diameter plays a crucial role in delivering antitumor drugs into cancer cells and improving their therapeutic efficacy. Carbon spheres, due to their large surface areas, unique surface properties and ease of functionalization, can generally deliver a large quantity of therapeutic agents to the target disease sites. In this study, spherical carbon nanoparticles with uniform size (71 nm) and regular shape were synthesized by hydrothermal reaction of bacterial cellulose nanofibers (30–50 nm), which had been achieved by a microorganism synthesis. Then using a simple acidification treatment, we could obtain carbon nanospheres with high drug loading capacity (the drug encapsulation efficiency was found to be about 93.4% and the drug loading efficiency (DL) reached about 52.3%). Meanwhile, the carbon nanospheres also exhibited good pH sensitivity in drug delivery. The results of in vitro experiments demonstrate that the carbon nanospheres prepared played an important part in the increase of the intracellular drug concentration and delayed-efficacy of the drug effect, which make them a promising platform for the delivery of other therapeutic agents beyond DOX.
Introduction
Cancer is a leading cause of death in most developing countries. Current means for treatment of cancer include surgery, radiotherapy and chemotherapy. Surgery has a great advantage for precancerous lesions and early cancer treatment, while most cancer patients are diagnosed when they are in the progression and metastasis stage of cancer, with no effective treatment. Radiotherapy is another primary cancer treatment choice, however, it is also not a cure for cancer, besides, it has some side effects. With the in-depth studies of etiology, disease mechanisms, biochemistry and immunology of cancer, chemotherapy becomes the most prevalent cancer treatment choice. Doxorubicin and paclitaxel are among the most common chemotherapeutic drugs in clinical use currently.1,2 As is well known, doxorubicin is a wide spectrum anticancer drug, and its water-soluble hydrochloride form is usually used to treat a range of cancers. However, the systemic administration of DOX causes significant toxicity and undesirable side effects because it acts on normal cells as well as tumor cells.3 The effective method to reduce the side effects of DOX is to control the drug concentration in the blood, therefore, the development of tumor-specific drug delivery carriers with a controlled release rate is very important.4Nowadays, many approaches have been reported for fabricating nanosized vehicles such as liposomes or polymersomes,5,6 dendrimers,7 nanogels,8 polymeric micelles,9–11 porous materials,12 and carbon nanomaterials as drug delivery carriers. Carbon nanomaterials with unique structure such as large pore volume, high specific surface areas, ease of functionalization and generally none cytotoxicity within a certain concentration range could facilitate larger amount of drug loadings.13–18 Besides, it is observed that carbon nanomaterials are capable of transporting covalently bonded drugs or molecular probes across cell membranes and releasing anticancer drugs effectively in cancer cells. Nowadays, carbon nanosphere carriers for drug delivery are mainly produced by carbonization of different chemical hydrocarbons (i.e., mesitylene, pentane, cyclohexane, camphorquinone, xylene, camphene, diphenylmethane, decahydronaphtalene, and anthracene, etc.).19,20 Due to the complexity of the prepared methods, the carbon spheres prepared generally present sizes larger than the ideal diameters (below 100 nm, an ideal size for transmembrane delivery carrier),21 besides, the resulting products may have some impurities because of residual chemical precursors. Therefore, recently much attention has been focused on the use of diverse biomass (i.e., glucose, xylose, maltose, sucrose, amylopectin, starch, plant cellulose and others) and biomass derivatives (i.e., hydroxylmethyl-furfural (HMF) and furfural) to produce functional carbon spheres; however, most of the prepared carbon spheres are with the diameter larger than 100 nm.22 Among the biomass, bacterial cellulose (BC) is a kind of natural polymer material with good biocompatibility, and it has good prospects in tissue engineering scaffolds, drug controlled release and artificial organs, etc.23,24 Recently we have successfully prepared bacterial cellulose nanofibers with the ultra-fine structure (i.e., with the diameter from 30 nm to 50 nm) by using microorganisms as nanorobot.25 It is evident that the antibiotics loaded on the bacterial cellulose nanofibers have an excellent drug release capacity and good anti-bacterial property. Based on these observations, we have further synthesized carbon nanospheres from bacterial cellulose with a mean particle size below 71 nm by using an easy hydrothermal method. In order to improve the dispersibility of carbon nanospheres in aqueous solutions for long circulation in vitro and in vivo, the as-prepared carbon nanospheres are acidified in a mixture of concentrated nitric and sulphuric acids. The physicochemical properties of carbon nanospheres were systematically characterized and the drug loading and release efficiency of carbon naonspheres for DOX were evaluated. The cytotoxicity effect of the carbon nanospheres in biological systems was investigated in vitro on cervical carcinoma HeLa cells. Moreover, we also observed the internalization and intracellular location of carbon nanospheres by confocal laser scanning microscopy (CLSM) and investigated the antitumor mechanism of them on HeLa cells in vitro. All the results above indicated that the as-prepared carbon nanospheres can deliver DOX into cancer cells effectively and realize intracellular release of DOX with lasting medicinal effect.
Results and discussion
Physicochemical characterization of carbon nanospheres (CNS)
The structural information for the CNS was obtained by the field emission scanning electron microscopy (FESEM) and transmission electron microscopy (TEM). It can be seen from Fig. 1a and b that the CNS were mainly in spherical shape and the mean diameter of particles was about to be 50 nm. The microstructure of the carbon nanospheres was observed by high resolution TEM (HRTEM) (Fig. 1c and d). The presence of disordered mesopores in the carbon nanospheres can be seen from the above two figures. To further characterize the size distribution and the stability of CNS in phosphate buffered saline (PBS), the dynamic light scattering (DLS) technique was performed to measure the hydrodynamic sizes of CNS. Fig. 1e shows the CNS exhibited an average diameter of about 71 nm with narrow size distributions (PDI < 0.149) in PBS, which was a little larger than the results estimated from the FESEM and TEM. This phenomenon is frequently observed since DLS measures the hydrodynamic diameter which is always larger than the geometric diameter of the particles.26 The DLS study revealed that the CNS were well dispersed in the aqueous solutions due to the introduction of hydroxyl/carboxylic/sulfonic groups onto the CNS surfaces. Besides, the zeta potential of CNS in the PBS at neutral pH of 7.0 was −37.2 mV, also indicating that CNS were surrounded by hydrophilic negatively-charged groups and well dispersed in PBS solution. The well dispersibility of CNS in PBS can also be directly recognized when we used a laser pointer to illuminate the CNS aqueous solution and a visible Tyndall phenomenon was clearly seen as depicted in Fig. 1f. From the above results we can see that CNS have a small and narrow size distribution and very good water dispersibility, which is vital for their application in anticancer drug delivery under the physiological environment.Fig. 2a shows the XRD patterns for BC and CNS. Broad peaks are observed at 2θ values of 14.5° and 22.5°, which are characteristic diffraction peaks for BC and can be assigned to the crystallographic plane of (1 0 0) and (2 0 0) reflection of BC, respectively.27 The XRD pattern associated to CNS exhibiting a broad but weak diffraction peaks C (002) graphite plane, at 2θ angles of 23°, which can be assigned to amorphous carbon-based materials consisting of disordered polycyclic aromatic carbon sheets.28 The carbon (101) diffraction peak (2θ = 35–50°) is not seen clearly in CNS, suggesting that CNS consist of small polycyclic aromatic carbon sheets containing several hexagonal carbon rings. Raman spectroscopy is an excellent tool to describe the degree of graphitization of carbon materials. Fig. 2b shows the Raman spectrum for CNS that carbonized by bacterial cellulose. From the Raman spectrum we can see that CNS present two main peaks at Raman shifts of ca. 1350 cm−1 and 1587 cm−1, historically named D- and G-bands, respectively. The D-band is due to the vibrations of carbon atoms with dangling bonds for the in-plane terminations of defect-containing graphite. The G peak, however, corresponds to the doubly degenerate zone center E2g symmetry of graphite and is mainly assigned to the bond stretching of all pairs of sp2-bonded carbon atoms in a two-dimensional hexagonal lattice,29–32 which also indicate the presence of polycyclic aromatic carbon sheets in the CNS. The intensity ratio of D to G band (I(D)/I(G)) is 0.75, suggesting that the CNS produced are of a certain degree of graphitization. While most anticancer drugs with aromatic structure such as DOX can interact strongly with the basal plane of graphite via π–π stacking,33 therefore, the materials with graphitic character are very suitable for used as anticancer drug delivery carriers. Both the Raman and XRD analyses reveal that the as-prepared CNS are of some graphitic characteristics, which can be used as anticancer drug carriers.
The chemical transformations that occur when the BC was converted into CNS were examined by several spectroscopic techniques. From the FT-IR spectroscopy of BC and CNS shown in Fig. 3a, the characteristic bands of BC include the hydrogen-bonded O–H stretching at 3342 cm−1, the C–H stretching at 2895 cm−1, the O–H bending of adsorbed water at 1648 cm−1, the H–C–H and O–C–H in-plane bending vibrations at 1429 cm−1, the C–O–C, C–C–O, and C–C–H deformation modes and stretching vibrations in which the motions of the C-5 and C-6 atoms are at 898 cm−1. After the hydrothermal carbonization of BC, the above mentioned bands become weak or completely disappeared, which indicates that the chemical bonds of the BC were broken during the reaction. In addition, several new peaks at 1708 cm−1, 1604 cm−1, 1377 cm−1 and 1045 cm−1 were observed in spectra of the CNS, which were due to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration, C
O stretching vibration, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching vibration, O
C stretching vibration, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S
S![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration and stretching mode of –SO3H groups, respectively.34,35 Thus, carbonized product with polycyclic aromatic rings and multifunctional groups was prepared by continuous treatment of BC. In accordance with the above results, X-ray photoelectron spectroscopy (XPS) measurements further confirmed the composition of the CNS. Fig. 3b presents the wide scan XPS spectrum of CNS showing the presence of C, O, S and N on the surface. A representative C1s spectrum obtained for CNS is shown in Fig. 3c. The C1s peak shows a broad feature signal due to the disorder in the CNS. The detailed fitting of the C1s spectrum contains four peaks, which are assigned to the C–C/C
O stretching vibration and stretching mode of –SO3H groups, respectively.34,35 Thus, carbonized product with polycyclic aromatic rings and multifunctional groups was prepared by continuous treatment of BC. In accordance with the above results, X-ray photoelectron spectroscopy (XPS) measurements further confirmed the composition of the CNS. Fig. 3b presents the wide scan XPS spectrum of CNS showing the presence of C, O, S and N on the surface. A representative C1s spectrum obtained for CNS is shown in Fig. 3c. The C1s peak shows a broad feature signal due to the disorder in the CNS. The detailed fitting of the C1s spectrum contains four peaks, which are assigned to the C–C/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C (284.5 eV), C–OH/C–O–C (285.5 eV), C
C (284.5 eV), C–OH/C–O–C (285.5 eV), C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (287.3 eV) and O
O (287.3 eV) and O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–O (289 eV), respectively.36–38 These results show that there is an abundance of oxygen groups on the surface of CNS. Similarly, the fitting of the O1s spectrum in Fig. 3d also includes four peaks, to the >S
C–O (289 eV), respectively.36–38 These results show that there is an abundance of oxygen groups on the surface of CNS. Similarly, the fitting of the O1s spectrum in Fig. 3d also includes four peaks, to the >S![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (531.1 eV), O–C (532.0 eV), >N–O (532.6 eV), C
O (531.1 eV), O–C (532.0 eV), >N–O (532.6 eV), C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O/O–C
O/O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O*/S–O (533.5 eV).36,39 The S2p peak attributable to SO3H groups at 168.8 eV can be seen in Fig. S1a.†40 The detailed N1s spectra of CNS in Fig. S1b† show that the binding energy of the N1s peak at 400.5 eV is assigned to N–C bonds.41 From the results of IR and XPS, we can see that the CNS prepared have a large amount of hydrophilic functional groups such as SO3H, phenolic OH, and COOH, which endow CNS with a good dispersibility in aqueous solution. All of these above consist with the results of Fig. 1e and f. Therefore the CNS prepared has wonderfully potential application in anticancer drug delivery.
O*/S–O (533.5 eV).36,39 The S2p peak attributable to SO3H groups at 168.8 eV can be seen in Fig. S1a.†40 The detailed N1s spectra of CNS in Fig. S1b† show that the binding energy of the N1s peak at 400.5 eV is assigned to N–C bonds.41 From the results of IR and XPS, we can see that the CNS prepared have a large amount of hydrophilic functional groups such as SO3H, phenolic OH, and COOH, which endow CNS with a good dispersibility in aqueous solution. All of these above consist with the results of Fig. 1e and f. Therefore the CNS prepared has wonderfully potential application in anticancer drug delivery.
Surface properties of carbon nanospheres are also one of the many factors that affect the drug carrier efficiency. Therefore, the surface structure parameters of CNS were determined by means of nitrogen adsorption–desorption tests. Fig. 4 is the nitrogen adsorption–desorption plot of CNS, which exhibits a typical type IV isotherm and a significant capillary condensation step occurring at a relative pressure (P/P0) of 0.4. The calculated Brunauer–Emmett–Teller (BET) surface area and the total pore volume of CNS are 208.17 m2 g−1 and 0.151 cm3 g−1, respectively, which are 152-fold and 25-fold higher than that of BC (Related data listed in Table 1). The average Barrett–Joyner–Halenda (BJH) pore diameters of the CNS calculated from the desorption branch of the isotherm is 3.5 nm. The large BET surface area and porous structure of the CNS are highly desirable for drug carrier applications.
| Sample | BET surface area [m2 g−1] | Total pore volume [cm3 g−1] | Mesoporous carbon [%] | Uniform pore diameter [nm] |
|---|---|---|---|---|
| BC | 1.37 | 0.006 | 63.1 | 35.4 |
| CNS | 208.17 | 0.151 | 96.5 | 3.5 |
The preparation mechanism of CNS proposed on the basis of all the above results is shown in Scheme 1. In the first step, when the bacterial cellulose aqueous dispersion is hydrothermally treated, the cellulose hydrolyzed and finally gave rise to fructose and glucose monomers. Then their monomers, which undergo dehydration and fragmentation reactions (i.e. ring opening and dissociation of –C–C–) leading to the formation of different soluble products, such as furans, 1,6-anhydroglucose, aldehydes, 1,2,4-benzenetriol, and erythrose.38 The subsequent reaction stages consist of intermolecular/intramolecular dehydration, aldol condensation/keto–enol tautomerism, polymerization and aromatization, which lead to the formation of small polycyclic aromatic carbon rings and the amorphous carbon structure.28,42–44 Finally, the as-prepared carbon material is modified and acidified in a mixture of concentrated nitric and sulfuric acids to improve its dispersion property in an aqueous solution. Therefore, the synthesized material is amorphous carbon consisting of –SO3H, –COOH, –NO2 and phenolic –OH bearing nanographene sheets in a considerably random fashion.
pH-dependent drug loading and in vitro drug release
In order to verify the π–π interaction between DOX and CNS, the fluorescence and UV-Vis absorption spectra of the DOX–CNS and free DOX were tested. The interaction between DOX and other molecules can be monitored because DOX bears a characteristic chromophore with an aromatic structure. As shown in Fig. 5a, free DOX exhibited two main absorptions at 233 nm and 490 nm, respectively. After DOX was loaded to CNS, the two peaks of DOX absorbance were shifted to 238 nm and 506 nm. The red shifts of the absorbance peaks of DOX in the CNS were due to the π–π stacking interaction between DOX and CNS. The π–π stacking interaction was further characterized by fluorescence spectroscopy study, which presented in Fig. 5b. The fluorescence intensity of DOX decreased significantly (by 80%) after it was loaded to CNS. The significant quenching was also the evidence of π-stacked DOX.45 DOX is one of the most valuable anticancer drugs widely used in the treatment of solid malignant tumors, such as cervical carcinoma, carcinomas of the breast, lung and ovarian carcinoma.46–48 It is known that DOX interferes with the topoisomerase II–DNA complex, leading to the formation of double–stranded breaks of DNA. So it is vital to deliver and release DOX in the cytoplasm and right into the cell nucleus. Therefore, in this study the drug-loading capacity of DOX and in vitro release behaviour of CNS were investigated. Under the optimized conditions, the drug encapsulation efficiency (EE) was found to be about 93.4%. Meanwhile, the drug loading efficiency (DL) reaches about 52.3% (Fig. 5c). From the results we can see that the CNS have a high loading capability of DOX, attributed to the supramolecular π–π stacking and hydrophobic interactions between DOX and CNS;49,50 besides, the large surface area and porous structure of CNS are also contributed to the high drug loading efficiency. The in vitro drug release study was carried out by re-dissolving DOX–CNS in PBS of different pH values at 37 °C for about two days (for accumulated release measurement). The effect of different medium pH on the DOX release rate is shown in Fig. 5d. Evidently, DOX–CNS exhibited a much faster release rate in the acidic medium (i.e., pH 5.5, pH 6.0) than that in pH 7.4 and 8.0. At pH 5.5, the cumulative release efficiency of DOX from CNS was 47.58% after 9 h, and 61.56% after two days, respectively. However, at pH 7.4, it only released 12.41% of the total DOX in CNS after 9 h and 18.96% after two days. This trend was attributed to the higher solubility of DOX at lower pH caused by the increased protonation, thereby reducing the hydrophobic interaction between DOX and CNS.51 The slower controlled release rate of DOX from CNS at pH 7.4 indicates that DOX–CNS is stable during the circulation in the physiological blood, which can greatly reduce the systemic toxicity in the neutral body fluids. The pH sensitivity of the DOX–CNS may benefit DOX release in cancer cells with an intracellular pH lower than that of the normal cells.52 Indeed, DOX–CNS were localized partly to lysosomal compartments (pH 4–5), where DOX could be released quickly. Compared with the original DOX in solution (i.e., DOX-I). Fig. S2† also shows the HPLC chromatograms of the DOX released from CNS (i.e., DOX-R). It was noted that for all the two samples there is a single sharp peak at 5.07 min. This suggests that the DOX released from CNS retains its original chemical structure.In vitro cytotoxicity
Then the cytotoxicity of free DOX, CNS, and DOX–CNS was investigated via MTT assay. HeLa cells were treated with free DOX and DOX–CNS with the same concentrations of DOX (see details in Table S2†) for 24 h (Fig. 6a) or 48 h (Fig. 6b). The results reveal that CNS show almost no obvious cytotoxic effect on HeLa cells at all five concentrations, indicating that CNS is an excellent biocompatible nanocarrier. HeLa cells treated with free DOX and DOX–CNS show a DOX dosage-dependent and time-dependent behavior. But it is worth noting that the antiproliferative action between the two groups (DOX–CNS and free DOX) shows some difference, as indicated in Fig. 6. Compared to the free DOX at the same concentrations, the DOX–CNS exhibits a lower cytotoxicity in most concentration range of DOX. This can be attributed to the sustained release property of the DOX–CNS and the results are in good agreement with the in vitro drug release profile. When used at a high concentration (1 μg mL−1) and for a long incubation time (48 h), DOX–CNS exhibited a similar cytotoxic effect to free DOX on HeLa cells, with about 82% decrease in cell viability (Fig. 6d). However, DOX–CNS exhibited an obviously enhancement in killing HeLa cells at a low DOX concentration (0.07 μg mL−1) after incubating for 48 h (Fig. 6c), indicating that DOX–CNS played an important part in the increase of the intracellular drug concentration and delayed-efficacy of the drug effect.Cellular uptake efficiency
The cellular uptake of DOX–CNS was verified by CLSM photographs of HeLa cells that were incubated with the nanoparticles for 24 h. As shown in Fig. 7, when no nanoparticles were used to treat HeLa cells (control group), only the nuclei were observed in blue colour after staining with DAPI. DAPI was a blue-fluorescent dye known to complex with double-stranded DNA. The same phenomenon was also found in the group treated with CNS. However, there are some differences of the distribution of DOX in the nucleus and the cytoplasm between free DOX and DOX–CNS. when the cells incubated with free DOX, a strong red fluorescence in cytoplasm and a weak red fluorescence in nucleus were observed, which is understandable because free DOX enters into cells quickly via a membrane diffusion pathway and accumulated in the nuclei after incubation, while some of them were rapidly efflux from the nuclei into the cytoplasm within a few hours.53 In contrast, when the cells incubated with DOX–CNS, a weak red fluorescence is concentrated in the cytoplasm and much weaker red fluorescence in the nucleus, in accordance with the solution-phase results on the presence of intramolecular quenching in the DOX–CNS conjugate (Fig. 5b). However, the CNS cannot be internalized into the nucleus,47 the above slight red fluorescence in the nucleus might be due to the unbound DOX that required for DOX–CNS to be taken up by the cells via endocytosis, along with gradual intracellular release of DOX from DOX–CNS in the acidic micro-environment of HeLa cells. As illustrated in Scheme 2, all the results above suggest that the drug delivery mechanism includes the cellular internalization of DOX–CNS, followed by its release in endosomes/lysosomes, and the subsequent penetration of the DOX into the nucleus.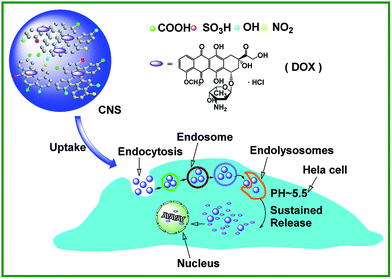 | ||
| Scheme 2 Schematic representation of the endocytosis of CNS and the pH-triggered intracellular controlled release of DOX molecules. | ||
Conclusions
In summary, carbon nanospheres with size below 71 nm was successfully prepared by using a new one-pot hydrothermal synthesis method. The porous structures, large surface area and hydrophilic surface functional groups endow the CNS with high drug loading efficiency for DOX and excellent dispersibility in aqueous solutions. The DOX–CNS show minimal drug release in basic solutions or the physiological pH range, while release drugs rapidly and quantitatively in acidic environment. The synthesized CNS could be easily internalized into HeLa cells and demonstrated good biocompatibility in vitro. The carried DOX can be released inside the cells and inhibited the growth of HeLa cells efficiently. Combining the general nontoxicity within a certain concentration range and low cost of the carbon materials, it is convincing that the as-prepared CNS holds great promise for advanced nanoplatforms for high efficacy therapy towards tumors.Experimental
Materials
All chemicals used in the experiments were of analytic-grade purity. Sulfuric acid, nitric acid, thiazolyl blue tetrazolium bromide (MTT), and paraformaldehyde (PFA) were purchased from Sigma-Aldrich (St. Louis, MO, USA). 2-(4-Amidinophenyl)-6-indole carbamidine dihydrochloride (DAPI) was purchased from Beyotime Institute of Biotechnology (Nantong, China). Doxorubicin hydrochloride (DOX) was purchased from Shanghai Hualan Chemical Corp. (Shanghai, China). The HeLa cell line was received from the Institute of Biochemistry and Cell Biology, Shanghai Institute of Biological Science, Chinese Academy of Science (Shanghai, China). All chemicals were used as received without any further purification unless otherwise stated.Synthesis of carbon nanospheres
The bacterial cellulose (BC) nanofibers used here were produced by Acetobacter xylinum according to an approach reported in our previous work.54,55 The carbon nanospheres were synthesized under hydrothermal conditions. In a typical synthesis, bacterial cellulose (1.5 g) was dissolved in deionized water (60 mL) with the assistance of ultrasonication for 30 min, and then sealed in a Teflon autoclave and hydrothermally treated at 200 °C for 8 h. The autoclave was gradually cooled down to the room temperature when the reaction was complete. The black products were collected by using five cycles of centrifugation/wash/re-dispersion repeated in water and in alcohol, and dried under vacuum at 80 °C for 5 h.Surface modification for carbon nanospheres
In order to improve the water dispersibility of carbon nanospheres in aqueous solutions, the as-prepared parent carbon nanospheres (2 g) were first dispersed in 90 mL mixed acid solution (formed as v(concentrated H2SO4)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v(concentrated HNO3) = 3
v(concentrated HNO3) = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) at 60 °C for 7 h with ultrasonic treatment to purify and acidize carbon nanospheres. The functionalized carbon nanospheres solution was cooled to room temperature. Then the suspension was diluted and filtrated several times to yield a black precipitate until the impurities such as sulfate ions and nitrate ions were no longer detected in the wash water and the pH value of the rinsing water reached a value of 7.4. The as-prepared carbon nanospheres are named as CNS in this paper. The resultant surface-functionalized CNS were collected by centrifugation, and dried at 383 K in a vacuum oven over night.
1) at 60 °C for 7 h with ultrasonic treatment to purify and acidize carbon nanospheres. The functionalized carbon nanospheres solution was cooled to room temperature. Then the suspension was diluted and filtrated several times to yield a black precipitate until the impurities such as sulfate ions and nitrate ions were no longer detected in the wash water and the pH value of the rinsing water reached a value of 7.4. The as-prepared carbon nanospheres are named as CNS in this paper. The resultant surface-functionalized CNS were collected by centrifugation, and dried at 383 K in a vacuum oven over night.
Characterizations
The TEM images were obtained with a JEM-2100 electron microscope (JEOL, Japan). The FESEM image was performed on JSM6700F electron microscope (JEOL, Japan). The dynamic light scattering (DLS) and zeta potential experiments were performed at 25 °C in PBS solution using a Malvern Zetasizer NanoZS (Malvern Instruments, England). The X-ray diffraction (XRD) patterns were recorded on an X-ray diffractometer (D8 Advance, Bruker, Germany), with an area detector using a Cu Kα radiation operating at 40 kV and 30 mA. The laser Raman spectroscopic images were carried out with a Renishaw (inVia) Raman spectrometer (Renishaw, England). The nitrogen adsorption–desorption isotherms of samples were obtained at a Micromeritics ASAP2020 volumetric adsorption analyzer (Micromeritics Instrument Corporation, USA). Prior to the measurement, the sample was degassed at 473 K for at least 8 h on a vacuum line. The pore size distributions were derived from the adsorption branch of the isotherm by using the Barrett–Joyner–Halenda (BJH) method. The Brunauer–Emmett–Teller (BET) method was used to calculate the specific surface areas. The total pore volume was calculated from the amount of nitrogen adsorbed at a maximum relative pressure (P/P0 = 0.99). The Fourier transform infrared (FT-IR) spectra were obtained using a Nicolet iS10 FT-IR Spectrometer (Thermo, USA). The X-ray photoelectron spectroscopy (XPS) measurements were performed on a PHI Quantera II X-ray photoelectron spectrometer (Ulvac-Phi, Japan) with an Mg Kα excitation source (1253.6 eV).Drug loading
The loading of DOX into CNS was carried out by mixing DOX with the CNS in a PBS buffered solution (pH 7.4). After shaking at 120 rpm at 37 °C for 2 hours, the mixtures were kept in a 4 °C fridge overnight under dark conditions. After adsorption, the DOX–CNS was collected by repeated centrifugation with PBS until no UV/Vis adsorption of DOX at 490 nm was found from the final PBS solution. The DOX–CNS obtained was used for subsequent tests of the in vitro DOX release and cytotoxicity assay against HeLa cells. To evaluate the DOX-loading efficiency, the supernatant and washed solutions were collected and the residual DOX content was measured by using a UV-2450 (UV-Vis spectrophotometer, Shimadzu). The loading amount of DOX can be calculated based on the original and residual DOX concentrations and volumes. Finally, the drug loading efficiency (DL) and drug encapsulation efficiency (EE) can be calculated according to eqn (1) and (2), in which DOX-loaded means DOX that were loaded on the CNS, DOX means the original DOX content. In order to further verify the interaction between DOX and CNS, the fluorescence and UV-Vis absorption spectra of the DOX–CNS and free DOX were tested. Fluorescence measurements of the DOX–CNS and free DOX were also performed on a fluorescence detector (F700, HITACHI, Japan).
 | (1) |
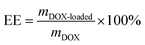 | (2) |
Drug release
For drug release testing, the DOX–CNS prepared above were dispersed in 10 mL of PBS buffer with different pH values (i.e., at pH 5.5 (approximate pH value in endosomes or lysosomes), pH 6.0 (pH value of the environment around the tumor), pH 7.4 (pH value of physiological blood), and pH 8.0), and then put in a dialysis bag (MWCO 1 kDa). Afterwards, the dialysis bag was immersed in the release medium (10 mL) and shaken at 100 rpm at 37 °C. At predetermined time intervals, aliquots of release medium (10 mL) were withdrawn for analysis of the released DOX by UV/Vis adsorption at 490 nm and the same volume of fresh medium was added. The amount of drug (%) released at specific time intervals was calculated according to eqn (3), in which DOX-release means the DOX that release from the DOX-loaded CNS. Evidence of the interaction of the DOX and CNS and the chemical stability of the DOX was followed using Agilent high-performance liquid chromatography (HPLC) equipped with an UV detector at 490 nm. The HPLC was run on a C18 column (150 × 4.6 mm I.D.). The mobile phase consisted of acetonitrile–water (35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 65, v/v), with a flow rate of 0.5 mL min−1.
65, v/v), with a flow rate of 0.5 mL min−1.
 | (3) |
Cell culture
HeLa cells (i.e., human cervical carcinoma cell line) were cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin at 37 °C in a humidified atmosphere with 5% CO2. The medium was routinely changed every two days and the cells were separated by trypsinisation before reaching confluence.Cytotoxicity assay
The cell growth inhibitory effect of DOX–CNS was determined by MTT assay. HeLa cells harvested in logarithmic growth phase were seeded in 96-well plates at a density of 1 × 104 cells per well, respectively. After incubating for 12 h, free DOX, DOX–CNS and CNS were added to the cells in DMEM medium at different DOX concentrations (0.0625, 0.125, 0.25, 0.5, 1.0 μg mL−1), and the CNS concentrations were 0.069, 0.14, 0.28, 0.56, 1.11 μg mL−1, respectively. After cultured in the media containing drug in 5% CO2 at 37 °C for 24 or 48 h, the cells were washed with PBS twice, shaken for 10 s and followed by incubation with MTT solution (20 μL, 5 mg mL−1) for 4 h at 37 °C. The resulting formazan dyes were dissolved in DMSO (150 μL), and the absorbance of solutions was monitored at 490 nm on a microplate reader (SpectraMax M5, Bio-Strategy, Ltd). The MTT assay was performed and the percentage of cell viability was determined by measuring optical density at 490 nm (OD490). Untreated HeLa cells were used as controls. Each sample was tested in five replicates.Confocal laser scanning microscopy (CLSM)
HeLa cells as described above were seeded on glass cover slips (14 mm) in six-well culture plates (1 × 105 cells per well) and allowed to attach overnight. After washing with PBS twice, the cells were incubated with a serial concentrations of free DOX, DOX–CNS and CNS in 3 mL of the serum-free DMEM medium in a CO2 incubator (5% CO2 at 37 °C) for 24 h. Subsequently, the cells were washed with PBS thrice to remove the remaining nanoparticles and dead cells. For fixed cell imaging, the cells were treated with 1 mL 4% PFA solution for 15 min at 4 °C, and their nuclei were stained with DAPI in 10% glycerol for 20 min in the dark. Finally, the cells were viewed under a confocal laser scanning microscope with a Leica confocal laser scanning microscope (SP5) and a 63× oil-immersion objective lens. DOX and DAPI were excited at 488 nm and 405 nm accordingly.Acknowledgements
This work was supported by the National High Technology Research and Development Program of China (2012AA022703) and National Natural Science Foundation of China (81325011, 21327902), National Basic Research Program (2010CB732404) as well as Suzhou Science & Technology Major Project (ZXY2012028).Notes and references
- F. Zheng, S. Wang, M. Shen, M. Zhu and X. Shi, Polym. Chem., 2013, 4, 933–941 RSC.
- M. E. Godsey, S. Suryaprakash and K. W. Leong, RSC Adv., 2013, 3, 24794–24811 RSC.
- F. Fu, Y. Wu, J. Zhu, S. Wen, M. Shen and X. Shi, ACS Appl. Mater. Interfaces, 2014, 6, 16416–16425 CAS.
- N. Doshi and S. Mitragotri, Adv. Funct. Mater., 2009, 19, 3843–3854 CrossRef CAS.
- F. Wang and J. Liu, Nanoscale, 2013, 5, 12375–12382 RSC.
- S. D. Koker, R. Hoogenboom and B. G. D. Geest, Chem. Soc. Rev., 2012, 41, 2867–2884 RSC.
- Y. Wang, X. Cao, R. Guo, M. Shen, M. Zhang, M. Zhu and X. Shi, Polym. Chem., 2011, 2, 1754–1760 RSC.
- J. K. Oh, D. J. Siegwart, H. Lee, G. Sherwood, L. Peteanu, J. O. Hollinger, K. Kataoka and K. Matyjaszewski, J. Am. Chem. Soc., 2007, 129, 5939–5945 CrossRef CAS PubMed.
- K. Xiao, J. Luo, Y. Li, J. S. Lee, G. Fung and K. S. Lam, J. Controlled Release, 2011, 155, 272–281 CrossRef CAS PubMed.
- W. Cao, Y. Li, Y. Yi, S. Ji, L. Zeng, Z. Sun and H. Xu, Chem. Sci., 2012, 3, 3403–3408 RSC.
- N. V. Cuong, Y. L. Li and M. F. Hsieh, J. Mater. Chem., 2012, 22, 1006–1020 RSC.
- S. Jambhrunkar, S. Karmakar, A. Popat, M. Yu and C. Yu, RSC Adv., 2014, 4, 709–712 RSC.
- S. Wen, H. Liu, H. Cai, M. Shen and X. Shi, Adv. Healthcare Mater., 2013, 2, 1267–1276 CrossRef CAS PubMed.
- L. Zhang, J. Xia, Q. Zhao, L. Liu and Z. Zhang, Small, 2010, 6, 537–544 CrossRef CAS PubMed.
- X. Ma, H. Tao, K. Yang, L. Feng, L. Cheng, X. Shi, Y. Li, L. Guo and Z. Liu, Nano Res., 2012, 5, 199–212 CrossRef CAS.
- Z. Liu, X. Sun, N. Nakayama-Ratchford and H. Dai, ACS Nano, 2007, 1, 50–56 CrossRef CAS PubMed.
- N. G. Sahoo, H. Bao, Y. Pan, M. Pal, M. Kakran, H. K. Cheng, L. Li and L. P. Tan, Chem. Sci., 2011, 47, 5235–5237 CAS.
- T. W. Kim, P. W. Chung, I. I. Slowing, M. Tsunoda, E. S. Yeung and V. S. Y. Lin, Nano Lett., 2008, 8, 3724–3727 CrossRef CAS PubMed.
- V. G. Pol, M. Motiei, A. Gedanken, J. Calderon-Moreno and M. Yoshimura, Carbon, 2004, 42, 111–116 CrossRef CAS PubMed.
- X. B. Zhang, H. W. Tong, S. M. Liu, G. P. Yong and Y. F. Guan, J. Mater. Chem. A., 2013, 1, 7488–7493 CAS.
- S. Nie, Y. Xing, G. J. Kim and J. W. Simons, Annu. Rev. Biomed. Eng., 2007, 9, 257–288 CrossRef CAS PubMed.
- B. Hu, K. Wang, L. Wu, S. H. Yu, M. Antonietti and M. M. Titirici, Adv. Mater., 2010, 22, 813–828 CrossRef CAS PubMed.
- D. Klemm, F. Kramer, S. Moritz, T. Lindstrom, M. Ankerfors, D. Gray and A. Dorris, Angew. Chem., Int. Ed., 2011, 50, 5438–5466 CrossRef CAS PubMed.
- N. Petersen and P. Gatenholm, Appl. Microbiol. Biotechnol., 2011, 91, 1277–1286 CrossRef CAS PubMed.
- J. Yang, W. Tang, X. Liu, C. Chao, J. Liu and D. Sun, Int. J. Hydrogen Energy, 2013, 38, 10813–10818 CrossRef CAS PubMed.
- Z. Y. Qiao, S. L. Qiao, G. Fan, Y. S. Fan, Y. Chen and H. Wang, Polym. Chem., 2014, 5, 844–853 RSC.
- C. Tokoh, K. Takabe, M. Fujita and H. Saiki, Cellulose, 1998, 5, 249–261 CrossRef CAS.
- M. Hara, T. Yoshida, A. Takagaki, T. Takata, J. N. Kondo, S. Hayashi and K. Domen, Angew. Chem., Int. Ed., 2004, 43, 2955–2958 CrossRef CAS PubMed.
- I. Y. Kim, J. M. Lee, T. W. Kim, H. N. Kim, H. I. Kim, W. Choi and S. J. Hwang, Small, 2012, 8, 1038–1048 CrossRef CAS PubMed.
- K. Kato, K. Ishioka, M. Kitajima, J. Tang, R. Saito and H. Petek, Nano Lett., 2008, 8, 3102–3108 CrossRef CAS PubMed.
- C. Casiraghi, A. Hartschuh, H. Qian, S. Piscanec, C. Georgi, A. Fasoli, K. S. Novoselov, D. M. Basko and A. C. Ferrari, Nano Lett., 2009, 9, 1433–1441 CrossRef CAS PubMed.
- B. Lu, T. Li, H. Zhao, X. Li, C. Gao, S. Zhang and E. Xie, Nanoscale, 2012, 4, 2978–2982 RSC.
- J. Huang, C. Zong, H. Shen, Y. Cao, B. Ren and Z. Zhang, Nanoscale, 2013, 5, 10591–10598 RSC.
- S. Suganuma, K. Nakajima, M. Kitano, D. Yamaguchi, H. Kato, S. Hayashi and M. Hara, J. Am. Chem. Soc., 2008, 130, 12787–12793 CrossRef CAS PubMed.
- K. Nakajima and M. Hara, ACS Catal., 2012, 2, 1296–1304 CrossRef CAS.
- T. I. T. Okpalugo, P. Papakonstantinou, H. Murphy, J. McLaughlin and N. M. D. Brown, Carbon, 2005, 43, 153–161 CrossRef CAS PubMed.
- M. M. Titirici, A. Thomas, S. H. Yu, J. O. Muller and M. Antonietti, Chem. Mater., 2007, 19, 4205–4212 CrossRef CAS.
- M. Sevilla and A. B. Fuertes, Carbon, 2009, 47, 2281–2289 CrossRef CAS PubMed.
- J. G. C. Shen, T. H. Kalantar, R. G. Herman, J. E. Roberts and K. Klier, Chem. Mater., 2001, 13, 4479–4485 CrossRef CAS.
- F. Liu, J. Sun, L. Zhu, X. Meng, C. Qi and F.-S. Xiao, J. Mater. Chem., 2012, 22, 5495 RSC.
- C. Karavasili, E. P. Amanatiadou, L. Sygellou, D. K. Giasafaki, T. A. Steriotis, G. C. Charalambopoulou, I. S. Vizirianakis and D. G. Fatouros, J. Mater. Chem. B, 2013, 1, 3167 RSC.
- Y. Wu, Z. Fu, D. Yin, Q. Xu, F. Liu, C. Lu and L. Mao, Green Chem., 2010, 12, 696 RSC.
- M. Okamura, A. Takagaki, M. Toda, J. N. Kondo, K. Domen, T. Tatsumi, M. Hara and S. Hayashi, Chem. Mater., 2006, 18, 3039–3045 CrossRef CAS.
- M. Toda, A. Takagaki, M. Okamura, J. N. Kondo, S. Hayashi, K. Domen and M. Hara, Green Chem., 2005, 438, 178–179 CAS.
- J. Zhu, L. Liao, X. Bian, J. Kong, P. Yang and B. Liu, Small, 2012, 8, 2715–2720 CrossRef CAS PubMed.
- F. Gao, L. Li, T. Liu, N. Hao, H. Liu, L. Tan, H. Li, X. Huang, B. Peng, C. Yan, L. Yang, X. Wu, D. Chen and F. Tang, Nanoscale, 2012, 4, 3365–3372 RSC.
- H. J. Cho, I. S. Yoon, H. Y. Yoon, H. Koo, Y. J. Jin, S. H. Ko, J. S. Shim, K. Kim, I. C. Kwon and D. D. Kim, Biomaterials, 2012, 33, 1190–1200 CrossRef CAS PubMed.
- H. Wang, Y. Zhao, Y. Wu, Y. L. Hu, K. Nan, G. Nie and H. Chen, Biomaterials, 2011, 32, 8281–8290 CrossRef CAS PubMed.
- K. Liu, J.-J. Zhang, F.-F. Cheng, T.-T. Zheng, C. Wang and J.-J. Zhu, J. Mater. Chem., 2011, 21, 12034–12040 RSC.
- Z. Liu, A. C. Fan, K. Rakhra, S. Sherlock, A. Goodwin, X. Chen, Q. Yang, D. W. Felsher and H. Dai, Angew. Chem., Int. Ed., 2009, 48, 7668–7672 CrossRef CAS PubMed.
- S. Wang, Y. Wu, R. Guo, Y. Huang, S. Wen, M. Shen, J. Wang and X. Shi, Langmuir, 2013, 29, 5030–5036 CrossRef CAS PubMed.
- S. J. Lee, K. H. Min, H. J. Lee, A. N. Koo, H. P. Rim, B. J. Jeon, S. Y. Jeong, J. S. Heo and S. C. Lee, Biomacromolecules, 2011, 12, 1224–1233 CrossRef CAS PubMed.
- S. Han, Y. Liu, X. Nie, Q. Xu, F. Jiao, W. Li, Y. Zhao, Y. Wu and C. Chen, Small, 2012, 8, 1596–1606 CrossRef CAS PubMed.
- J. Yang, J. Yu, J. Fan, D. Sun, W. Tang and X. Yang, J. Hazard. Mater., 2011, 189, 377–383 CrossRef CAS PubMed.
- J. Yang, D. Sun, J. Li, X. Yang, J. Yu, Q. Hao, W. Liu, J. Liu, Z. Zou and J. Gu, Electrochim. Acta, 2009, 54, 6300–6305 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: High-resolution S2p spectrum and high-resolution N1s spectrum, C18-HPLC chromatogram of DOX standard and DOX released from CNS, details of the concentrations of CNS, free DOX, and DOX–CNS in Fig. 6. See DOI: 10.1039/c4ra16359a |
| This journal is © The Royal Society of Chemistry 2015 |








