Highly active 4-aminoquinoline–pyrimidine based molecular hybrids as potential next generation antimalarial agents†
Abstract
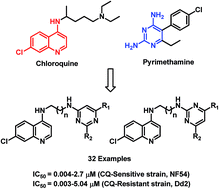
In order to overcome the problem of emerging drug resistance in malarial chemotherapy, a series of highly active 4-aminoquinoline–pyrimidine hybrids were synthesized and evaluated for their antimalarial activity against CQ-sensitive (NF54) and CQ-resistant (Dd2) strains of P. falciparum in an in vitro assay. The most active hybrid 19f exhibited 74-fold better potency than chloroquine and 4-fold better potency than artesunate against the drug-resistant strain of P. falciparum. Compound 19e, when evaluated for in vivo activity in the P. berghei-mouse malaria model showed 93.9% parasite suppression at 30 mg kg−1 dose on Day 4 with a mean survival time of 11 days. To gain insights towards the mechanism of action of these hybrids, heme binding and molecular modelling studies were performed on the most active hybrids. It was observed that inhibition of formation of β-hematin and dihydrofolate reductase-thymidylate synthase Pf-DHFR-TS enzyme could be associated with the observed antimalarial activity of these compounds.

 Please wait while we load your content...
Please wait while we load your content...