Genome scanning inspired isolation of reedsmycins A–F, polyene-polyol macrolides from Streptomyces sp. CHQ-64†
Qian Che‡
a,
Tong Li‡a,
Xiaofang Liua,
Tingting Yaoa,
Jing Lib,
Qianqun Gua,
Dehai Li*a,
Wenli Li*a and
Tianjiao Zhu*a
aKey Laboratory of Marine Drugs, Chinese Ministry of Education, School of Medicine and Pharmacy, Ocean University of China, Qingdao 266003, People's Republic of China. E-mail: dehaili@ouc.edu.cn; liwenli@ouc.edu.cn; zhutj@ouc.edu.cn; Fax: +86-532-82033054; Tel: +86-532-82031632
bCollege of Marine Life Sciences, Ocean University of China, Qingdao 266003, People's Republic of China
First published on 23rd February 2015
Abstract
Genome scanning of the reed rhizosphere soil-derived Streptomyces sp. CHQ-64 revealed a partial gene cluster, putatively encoding a polyene-polyol compound. Inspired by this finding, six new polyene-polyol macrolides, reedsmycins A–F (1–6), were isolated guided by the characteristic NMR signals. Their structures were elucidated using mass spectrometry and extensive NMR spectroscopy. Among them, reedsmycin F (6) possessed a rare 31-membered macroring containing a tetrahydrofuran motif, and reedsmycin A (1) exhibited promising activity against Candida albicans with a MIC of 25–50 μM, in a comparable level to that of the positive control nystatin.
Introduction
The genome-guided strategy for isolation of new natural products was prompted by the development of the high throughput sequencing technique.1,2 Polyketide (PK) is one of the major family of natural products with structural and biological diversities. The backbones of PKs are assembled by polyketide synthetases (PKSs), and hence PKS genes are important molecular indicators for discovery of PK compounds. During our genome scanning program for the gene clusters encoding PKs, a 28 kb DNA fragment containing two partial type I PKSs genes was identified from the genome of the marine-derived Streptomyces sp. CHQ-64, and their functional domain composition indicated the potential producibility of polyene-polyol macrolides by this strain.Previously, two classes of cytotoxic hybrid isoprenoid alkaloids merging the amino acid and mavalonate pathways were obtained from this strain guided by the bioassay results.3,4 Inspired by genome scanning information, we checked the noncytotoxic fractions of the Streptomyces sp. CHQ-64 fermentation products by 1H NMR analysis. Among them, the characteristic signals related to polyene-polyol structures were observed in the fraction eluted by CH2Cl2/MeOH (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–1
1–1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) on the silica column. By further NMR-guided fractionation, six new polyene-polyol macrolides, named reedsmycins A–F (1–6), were isolated. Herein, we report the genome-guided discovery, isolation, structural elucidation, and antifungal activities of these new compounds.
1) on the silica column. By further NMR-guided fractionation, six new polyene-polyol macrolides, named reedsmycins A–F (1–6), were isolated. Herein, we report the genome-guided discovery, isolation, structural elucidation, and antifungal activities of these new compounds.
Results and discussion
A 28 kb DNA fragment, processing an overall G + C content of 68.5%, was obtained by genome scanning of Streptomyces sp. CHQ-64. Two partial open reading frames (orfs) were identified within the DNA fragment, which putatively encoded type I PKS. The module and domain organizations of ORF1-2 were deduced (Fig. 1), and their conserved motifs were analyzed. The KS domains contain the Cys–His–His catalytic triad required for the decarboxylative condensation.5 The ACP domains display the conserved L/IG(x)DS motif harboring the serine residue that was essential for 4′-phopspho-panthehenylation.6 The 13 active site residues in all the ATs are QQGHSIGRFHTHV, indicating that they are specific for malonyl-CoA.7 The KR domains contain the conserved consensus sequence GXGXXGXXA for NAD(P)H binding; KR1-3 harbor the conserved Trp141 and belong to A-type, catalyzing the formation of an S-configured alcohol; KR4-6 are B-type with the conserved LDD-motif being replaced with IDD in KR4 and LED in KR5-6, hence generating a R-configured alcohol (Table S1†).8 Functional DH domains featuring the conserved consensus sequence HXXXGXXXP are found for modules 4–6,9 which catalyze elimination of water from R-configured hydroxyl group to form E/Z-configured double bond.10 The above findings indicated that this DNA fragment was probably involved in the gene cluster biosynthesizing polyene-polyol compounds as predicted in Fig. 1.With the noncytotoxic subfractions of the fermentation product in hand, the 1H NMR was employed to detect whether the polyene-polyol-like compounds existed in the Streptomyces sp. CHQ-64 products. Delightedly, the fraction eluted by CH2Cl2/MeOH (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–1
1–1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) on silica column, indeed showed the characteristic signals related to polyene-polyol structures (Fig. 2). This fraction was further investigated by further fractionation, leading to the isolation of six new polyene-polyol macrolides, named reedsmycins A–F (1–6) (Fig. 3).
1) on silica column, indeed showed the characteristic signals related to polyene-polyol structures (Fig. 2). This fraction was further investigated by further fractionation, leading to the isolation of six new polyene-polyol macrolides, named reedsmycins A–F (1–6) (Fig. 3).
 | ||
Fig. 2 The 1H NMR spectrum (600 MHz, in DMSO-d6) of the fraction eluted by CH2Cl2/MeOH (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–1 1–1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) on silica column. 1) on silica column. | ||
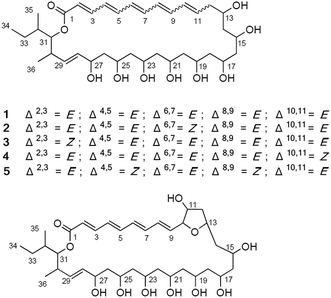
Reedsmycin A (1) was purified as a yellow powder and was determined to have the molecular formula C36H58O10 by HRESIMS. The IR absorption at 1696 cm−1, as well as the carbon signal observed at δC 166.1 in 13C NMR spectrum, revealed the presence of an ester group in 1 (Table 1). The 1H NMR spectrum of 1 clearly showed the feature of polyunsaturated and polyhydroxylated compounds, with 12 olefinic protons from 7.19 ppm and 5.41 ppm, and 9 protons of oxygenated methines between 4.77 and 3.55 ppm (Table 2). Further analysis of the 1H NMR spectrum of 1 identified 20 aliphatic protons in the region of δH 2.58–1.02, three methyl groups at δH 0.98, 0.89, and 0.80 and eight exchangeable protons in the region of δH 4.71–4.08, which illustrated that reedsmycin A (1) possessed at least 8 hydroxyl groups.
| Position | 1a | 2b | 3b | 4b | 5a | 6b |
|---|---|---|---|---|---|---|
| δC, type | δC, type | δC, type | δC, type | δC, type | δC, type | |
| a Spectra were recorded at 100 MHz for 13C NMR using TMS as internal standard.b Spectra were recorded at 150 MHz for 13C NMR using TMS as internal standard.c Interchangeable within column.d Signals could not be individually assigned. | ||||||
| 1 | 166.1, qC | 166.4, qC | 166.0, qC | 166.8, qC | 166.0, qC | 166.7, qC |
| 2 | 120.4, CH | 120.9, CH | 117.7, CH | 120.9, CH | 121.2, CH | 121.3, CH |
| 3 | 144.5, CH | 137.5, CH | 139.5, CH | 145.3, CH | 138.9, CH | 144.9, CH |
| 4 | 129.8, CH | 130.5, CH | 124.3, CH | 131.3, CH | 125.8, CH | 130.6, CH |
| 5 | 140.9, CH | 145.3, CH | 138.2, CH | 141.6, CH | 137.3, CH | 141.3, CH |
| 6 | 135.7, CH | 128.4, CH | 126.9, CH | 132.6, CH | 126.8, CH | 132.1, CH |
| 7 | 130.6, CH | 134.7, CH | 138.6, CH | 138.2, CH | 137.7, CH | 137.4, CH |
| 8 | 137.6, CH | 127.1, CH | 131.1, CH | 133.1, CH | 130.9, CH | 129.6, CH |
| 9 | 131.4, CH | 136.4, CH | 136.5, CH | 131.4, CH | 135.8, CH | 137.2, CH |
| 10 | 132.0, CH | 132.8, CH | 132.6, CH | 130.5, CH | 132.6, CH | 86.3, CH |
| 11 | 133.7, CH | 133.9, CH | 134.3, CH | 130.8, CH | 133.1, CH | 75.7, CH |
| 12 | 42.2, CH2 | 39.7, CH2 | 42.7, CH2 | 44.3, CH2 | 38.7, CH2 | 39.6, CH2 |
| 13 | 67.3, CH | 66.7, CH | 68.6, CH | 66.4, CH | (66.8–63.5)d, CH | 75.8, CH |
| 14 | 45.7, CH2 | 46.7, CH2 | 46.8, CH2 | (47.8–44.5)d, CH2 | (45.8–40.3)d, CH2 | 41.9, CH2 |
| 15 | 65.6, CH | 67.9, CH | 66.9, CH | 65.5, CH | (66.8–63.5)d, CH | 65.7, CH |
| 16 | 47.5, CH2 | 46.2c, CH2 | 46.2, CH2 | (47.8–44.5)d, CH2 | (45.8–40.3)d, CH2 | (47.2–46.0)d, CH2 |
| 17 | 64.3, CH | 63.6, CH | 65.5, CH | 67.0, CH | (66.8–63.5)d, CH | 65.9, CH |
| 18 | 45.9, CH2 | 46.0c, CH2 | 45.6, CH2 | (47.8–44.5)d, CH2 | (45.8–40.3)d, CH2 | (47.2–46.0)d, CH2 |
| 19 | 66.1, CH | 67.5, CH | 67.0, CH | 63.2, CH | (66.8–63.5)d, CH | 64.3, CH |
| 20 | 46.5, CH2 | 46.2c, CH2 | 45.1, CH2 | (47.8–44.5)d, CH2 | (45.8–40.3)d, CH2 | (47.2–46.0)d, CH2 |
| 21 | 66.6, CH | 66.5, CH | 63.6, CH | 66.4, CH | (66.8–63.5)d, CH | 66.5, CH |
| 22 | 42.3, CH2 | 44.2, CH2 | 46.5, CH2 | (47.8–44.5)d, CH2 | (45.8–40.3)d, CH2 | (47.2–46.0)d, CH2 |
| 23 | 65.4, CH | 68.1, CH | 66.6, CH | 65.2, CH | (66.8–63.5)d, CH | 66.8, CH |
| 24 | 46.1, CH2 | 45.2, CH2 | 47.8, CH2 | (47.8–44.5)d, CH2 | (45.8–40.3)d, CH2 | (47.2–46.0)d, CH2 |
| 25 | 62.9, CH | 65.7, CH | 64.9, CH | 66.1, CH | (66.8–63.5)d, CH | 65.2, CH |
| 26 | 45.6, CH2 | 45.6, CH2 | 45.9, CH2 | (47.8–44.5)d, CH2 | (45.8–40.3)d, CH2 | (47.2–46.0)d, CH2 |
| 27 | 69.2, CH | 69.7, CH | 69.6, CH | 69.4, CH | 69.3, CH | 69.1, CH |
| 28 | 133.3, CH | 134.7, CH | 134.0, CH | 134.1, CH | 134.1, CH | 134.2, CH |
| 29 | 129.6, CH | 132.2, CH | 130.7, CH | 130.8, CH | 131.5, CH | 130.5, CH |
| 30 | 35.6, CH | 38.7, CH | 36.1, CH | 36.2, CH | 37.6, CH | 36.5, CH |
| 31 | 77.8, CH | 78.7, CH | 78.5, CH | 79.0, CH | 78.6, CH | 78.3, CH |
| 32 | 34.7, CH | 36.3, CH | 35.4, CH | 35.5, CH | 35.8, CH | 35.3, CH |
| 33 | 24.9, CH2 | 26.4, CH2 | 25.3, CH2 | 25.1, CH2 | 25.5, CH2 | 25.7, CH2 |
| 34 | 10.7, CH3 | 11.8, CH3 | 11.1, CH3 | 11.1, CH3 | 11.1, CH3 | 11.3, CH3 |
| 35 | 15.5, CH3 | 16.4, CH3 | 16.0, CH3 | 15.9, CH3 | 15.1, CH3 | 16.0, CH3 |
| 36 | 11.6, CH3 | 14.3, CH3 | 12.2, CH3 | 12.3, CH3 | 14.0, CH3 | 12.7, CH3 |
| Position | 1b | 2a | 3a | 4a | 5b | 6a |
|---|---|---|---|---|---|---|
| δH (J in Hz) | δH (J in Hz) | δH (J in Hz) | δH (J in Hz) | δH (J in Hz) | δH (J in Hz) | |
| a Spectra were recorded at 600 MHz for 1H NMR using TMS as internal standard.b Spectra were recorded at 400 MHz for 1H NMR using TMS as internal standard.c Signals could not be individually assigned. | ||||||
| 2 | 5.87, d (14.8) | 5.93, d (15.4) | 5.65, d (11.5) | 5.94, d (15.4) | 5.98, d (15.0) | 5.95, d (15.4) |
| 3 | 7.19, dd (11.5, 15.4) | 7.36, dd (11.0, 14.9) | 7.24, t (11.5, 12.1) | 7.24, dd (11.6, 15.4) | 7.71, dd (12.3, 15.3) | 7.21, dd (11.5, 15.4) |
| 4 | 6.44, dd (11.6, 14.8) | 6.46, dd (11.0, 14.3) | 7.16, t (10.4, 12.7) | 6.46, dd (11.0, 15.4) | 6.20, t (11.4, 11.5) | 6.47, m |
| 5 | 6.73, dd (11.0, 14.3) | 7.35, dd (11.5, 15.4) | 6.44, t (11.0, 13.2) | 6.77, dd (11.6, 14.6) | 6.47, dd (11.5, 11.9) | 6.72, dd (11.0, 14.9) |
| 6 | 6.37, dd (10.4, 14.8) | 6.12, t (11.0, 11.0) | 6.89, t (13.7, 13.1) | 6.38, t (12.1, 14.3) | 6.84, t (12.8, 13.3) | 6.38, dd (11.0, 14.9) |
| 7 | 6.25, dd (11.5, 14.8) | 6.28, t (11.0, 11.6) | 6.50, t (12.1, 13.2) | 6.56, dd (11.0, 14.8) | 6.54, dd (9.9, 14.7) | 6.47, m |
| 8 | 6.49, dd (11.5, 14.8) | 6.89, dd (13.7, 12.7) | 6.30, t (12.1, 13.7) | 6.36, t (11.6, 14.3) | 6.40, t (10.0, 10.7) | 6.32, dd (11.5, 14.9) |
| 9 | 6.34, dd (11.0, 14.3) | 6.40, dd (10.4, 14.9) | 6.39, t (9.9, 16.5) | 6.64, dd (12.1, 14.3) | 6.40, t (10.0, 10.7) | 5.94, m |
| 10 | 6.16, dd (10.4, 14.8) | 6.20, dd (11.0, 15.4) | 6.17, t (11.6, 14.3) | 6.20, t (11.0, 11.5) | 6.16, m | 4.17, m |
| 11 | 6.47, m | 5.92, m | 5.82, m | 5.68, m | 5.90, m | 3.95, m |
| 12 | 2.47, m; 2.13, m | 2.31, m; 2.23, m | 2.44, m; 2.12, m | 2.66, m; 2.20, m | 2.36, m | 1.78, dd (6.6, 11.5); 1.62, m |
| 13 | 3.81, m | 3.86, m | 3.78, m | 3.79, m | (3.45–3.95)c, m | 4.34, d (5.0) |
| 14 | 1.54, m; 1.38, m | (1.51–1.20)c, m | 1.58, m; 1.33, m | (1.60–1.10)c, m | (1.60–1.10)c, m | 1.66, m; 1.54, m |
| 15 | 3.55, m | 3.70, m | (3.82–3.90)c, m | 3.67, m | (3.45–3.95)c, m | 3.70, m |
| 16 | 1.54, m; 1.23, m | (1.51–1.20)c, m | 1.10, m | (1.60–1.10)c, m | (1.60–1.10)c, m | (1.70–1.10)c, m |
| 17 | 3.71, m | 3.90, m | (3.82–3.90)c, m | 3.93, m | (3.45–3.95)c, m | 3.78, m |
| 18 | 1.14, m; 1.06, m | (1.51–1.20)c, m | 1.45, m; 1.16, m | (1.60–1.10)c, m | (1.60–1.10)c, m | (1.70–1.10)c, m |
| 19 | 3.88, m | 3.88, m | (3.82–3.90)c, m | 3.91, m | (3.45–3.95)c, m | 3.91, m |
| 20 | 1.14, m | (1.51–1.20)c, m | 1.19, m | (1.60–1.10)c, m | (1.60–1.10)c, m | (1.70–1.10)c, m |
| 21 | 3.74, m | 3.87, m | (3.82–3.90)c, m | 3.87, m | (3.45–3.95)c, m | 3.83, m |
| 22 | 1.31, m; 1.10, m | 1.43, m; 1.40, m | 1.29, m | (1.60–1.10)c, m | (1.60–1.10)c, m | (1.70–1.10)c, m |
| 23 | 3.75, m | 3.82, m | 3.62, m | 3.84, m | (3.45–3.95)c, m | 3.89, m |
| 24 | 1.28, m; 1.12, m | 1.49, m; 1.42, m | 1.52, m; 1.23, m | (1.60–1.10)c, m | (1.60–1.10)c, m | (1.70–1.10)c, m |
| 25 | 3.89, m | 3.57, m | 3.68, m | 3.41, m | (3.45–3.95)c, m | 3.72, m |
| 26 | 1.17, m | 1.51, m; 1.35, m | 1.34, m; 1.15, m | (1.60–1.10)c, m | (1.60–1.10)c, m | (1.70–1.10)c, m |
| 27 | 4.08, m | 4.05, m | 4.02, m | 4.01, m | 4.07, m | 4.07, m |
| 28 | 5.41, dd (4.4, 16.0) | 5.39, m | 5.45, m | 5.43, m | 5.42, m | 5.41, dd (4.9, 16.0) |
| 29 | 5.51, dd (5.0, 16.0) | 5.40, m | 5.45, m | 5.43, m | 5.42, m | 5.49, dd (6.0, 15.4) |
| 30 | 2.58, m | 2.49, m | 2.57, m | 2.56, m | 2.55, m | 2.57, m |
| 31 | 4.77, dd (2.8, 9.4) | 4.76, t (5.5, 6.6) | 4.76, d (9.3) | 4.76, dd (2.0, 12.1) | 4.77, t (5.8, 6.3) | 4.77, dd (2.8, 8.8) |
| 32 | 1.68, m | 1.65, m | 1.68, m | 1.67, m | 1.66, m | 1.63, m |
| 33 | 1.44, m; 1.15, m | 1.26, m; 1.10, m | 1.47, m; 1.15, m | 1.49, m; 1.15, m | 1.32, m; 1.10, m | 1.44, m; 1.52, m |
| 34 | 0.89, t (7.1, 7.7) | 0.86, t (7.1, 7.7) | 0.89, t (6.5, 7.7) | 0.89, t (7.1, 7.7) | 0.87, t (7.4, 8.8) | 0.88, t (7.2, 7.7) |
| 35 | 0.80, d (6.6) | 0.86, d (4.4) | 0.80, d (6.0) | 0.77, d (6.6) | 0.85, d (5.8) | 0.79, d (6.6) |
| 36 | 0.98, d (6.6) | 0.92, d (7.1) | 0.97, d (6.1) | 0.97, d (6.6) | 0.93, d (6.7) | 0.99, d (6.6) |
The combination of COSY (Fig. 4) and HMQC experimental data indicated a conjugated pentaene moiety from H-2 (δH 5.87) to H-11 (δH 6.47), which was connected to a repeating 1,3-hydroxy group moiety from H-12 (δH 2.47, 2.13) to H-27 (δH 4.08), and further extended the spin system to H-34 (δH 0.89). The two methyls CH3-35 (δH 0.80) and CH3-36 (δH 0.98) were located on C-32 (δC 34.7) and C-30 (δC 35.6) based on the COSY correlations (H-35/H-32, H-36/H-30) and HMBC correlations from H-35 to C-31 and from H-36 to C-29, C-30 and C-31. Finally, the planar structure of 1 was determined as a macrolide by the HMBC correlations from H-2, H-3 (δH 7.19) and H-31 (δH 4.77) to the carbonyl carbon C-1 (δC 166.1) and named as reedsmycin A (1).
The geometries of the six double bonds (Δ2 to Δ10 and Δ28) were determined as E on the basis of their characteristically large coupling constants (J ≥ 14.0 Hz) (Table 1) together with the NOESY correlations for H-3/H-5, H-5/H-7, H-7/H-9, H-2/H-4, H-4/H-6, H-6/H-8 and H-27/H-29.
The complex polyol segments and overlap of the 1H NMR signals make the elucidation for the stereochemistry of the polyene-polyol macrolides a serious challenge. A literature survey showed that the attempts to assign the relative configuration of the polyol system were carried out including X-ray crystallography,11,12 application of Kishi's Universal NMR Database13,14 or 13C NMR acetonide analysis.15,16 Unfortunately, none of the above methods worked properly for 1, resulting in its configurations unclear.
Reedsmycin B (2) was obtained as a yellow powder. Its molecular formula was determined as C36H58O10 on the basis of HRESIMS, requiring 8 degrees of unsaturation. Compound 2 showed the same features of the NMR, UV, and mass spectra with those of 1, indicating a structurally related isomer of 1. Detailed analysis of 1D and 2D-NMR spectroscopy revealed that 2 had a smaller coupling constant (3JH-6,H-7 = 11.0 Hz) for Δ6,7, indicated the Z configuration of this double bond in 2 (Table 2). Similarly, reedsmycins C–E (3–5) were also structurally related isomers of 1 on the basis of IR, UV, HRESIMS, UV, 1D and 2D NMR spectroscopy. Comprehensive NMR analysis (Table 2) showed that the differences between these isomers (3–5) were the positions of Z geometric double bonds (Δ2,3 = Z (3), Δ10,11 = Z (4), and Δ4,5 = Z and Δ8,9 = Z (5), respectively). Compounds 3–5 were named as reedsmycins C–E, respectively.
Reedsmycin F (6) was obtained as a pale yellow powder. Its molecular formula was determined as C36H58O11 according to the HRESIMS at m/z 689.3860 [M + Na]+, indicating eight degrees of unsaturations and possessing one more oxygen than that of 1. The general features of the NMR spectra of 6 were very similar to those of 1, indicating that they are structurally related compounds. The main differences were observed at the positions of C-10 and C-11, where the two olefinic methines of 1 were replaced by two oxygenated methines in 6. The UV absorption maximum at 327 nm also revealed the smaller conjugated system in 6. Since seven unsaturations were accounted for the five double bonds and the macrolide feature, 6 was inferred to contain an additional ring. The tetrahydrofuran (THF) ring was indicated by the 13C chemical shifts at C-10 (δC 86.3) and C-13 (δC 75.8), the COSY correlations (H-10/H-11(OH-11)/H-12/H-13) and the HMBC correlations from H-10 (δH 4.17) to C-13. The NOESY correlations of H-10/OH-11 (δH 5.09) and H-13 (δH 4.34)/H-15 (δH 4.34) while no correlation for H-11 (δH 3.95)/H-13 implied the H-10/OH-11-cis, H-13/H-15-cis and the C-11/C-13-trans configurations in the THF ring.
Reedsmycin A (1) possessed a similar polyene-polyol feature to those antibiotics such as bahamaolides,17 marinisporolides,14 mycoticins,18 dermostatins,19 roflamycoin,20 and roxaticin,11 which were all isolated from Streptomyces species. Previous biosynthetic studies conducted with these oxo polyenes showed that they share similar biosynthetic pathway.11 Based on the structure of reedsmycin A, it is proposed to be biosynthesized by 16 rounds of condensation probably using propionate as the start unit, 2 methylmalonyl CoA and 14 malonyl CoA as the extender units successively. As shown in Fig. 1, the 28 kb DNA fragment is possibly responsible for the biosynthesis of the C5–C18 fragment of reedsmycin A, generating 3 S-configured hydroxyl groups (at C13, C15 and C17) and 3 E/Z-configured double bonds (Δ6,7, Δ8,9, and Δ10,11).
Polyene macrolides are potent antifungal agents that also exhibit a range of promising biological activities against parasites, enveloped viruses, tumor cells, prion diseases.20,21 In our current research, all the compounds showed no cytotoxicities on K562, A549 and HL-60 tumor cell lines (IC50 > 50 μM). The antifungal activities of reedsmycins A–F (1–6) were also evaluated against Candida albicans using nystatin as the positive control22 (Table 3). Compound 1 showed pronounced activity with MIC of 25–50 μM, which was stronger than those of the compounds containing Z double bond. Additionally, reedsmycin F (6), lack of one double bond but containing THF ring, exhibited no inhibitory activity. The results indicated that the number and geometry of the double bonds are essential to the antifungal activity.
| Compounds | 1 | 2 | 3 | 4 | 5 | 6 | Nystatin |
|---|---|---|---|---|---|---|---|
| C. albicans/MIC (μM) | 25–50 | 100–200 | 50–100 | 50–100 | 50–100 | >200 | 25–50 |
Experimental section
General
Specific rotations were obtained on a JASCO P-1020 digital polarimeter. UV spectra were recorded on Beckman DU 640 spectrophotometer. CD spectra were measured on JASCO J-715 spectropolarimeter. IR spectra were taken on a Nicolet Nexus 470 spectrophotometer in KBr discs. NMR spectra were recorded on a JEOL JNM-ECP 600 and Bruker-400 spectrometers using TMS as internal standard, and chemical shifts were recorded as δ values. ESIMS utilized on a Thermo Scientific LTQ Orbitrap XL mass spectrometer. Semipreparative HPLC was performed using an ODS column [HPLC (YMC-Pack ODS-A, 10 × 250 mm, 5 μm, 4 mL min−1)]. TLC and column chromatography (CC) were performed on plates precoated with silica gel GF254 (10–40 μm) and over silica gel (200–300 mesh, Qingdao Marine Chemical Factory), and Sephadex LH-20 (Amersham Biosciences), respectively. Vacuum-liquid chromatography (VLC) was carried out over silica gel H (Qingdao Marine Chemical Factory). Marinum salt used is made from the evaporation of sea water collected in Laizhou Bay (Weifang Haisheng Chemical Factory).Strains and culture conditions
Streptomyces sp. CHQ-64 (Former name: AH1-5) was isolated from reed rhizosphere soil collected from the mangrove conservation area of Guangdong province, China. The voucher specimen is deposited in our laboratory at −80 °C. The producing strain was prepared on ISP-2 agar slants at 3.3% salt concentration and stored at 4 °C. For biological assays, Candida albicans was cultured overnight at 28 °C in MH broth.Bioinformatic analysis
The sequence was analyzed for putative orfs with the FramePlot 2.3.2 program.23 The proposed function of the orfs was accomplished by using the Blast programs.24 The module and domain organizations of ORF1–2 were deduced by SBSPKS analysis.25Nucleotide sequence accession number
The nucleotide sequence reported in this paper has been deposited in the GenBank database under accession number KM411608.Purification
The crude extract (35.5 g) was subjected to vacuum liquid chromatography over a silica gel (200–300 mesh) column using stepwise gradient elution with the mixtures of petroleum ether–CHCl3–MeOH to give eight fractions. Fraction 7 (0.5 g) was purified by repeated ODS CC to afford six subfractions (fractions 7.1–7.6). Fraction 7.3 was further purified on Sephadex LH-20 and semipreparative HPLC (75% MeOH) to give compound 6 (2 mg, tR 12 min), compound 5 (10 mg, tR 21 min), compound 2 (3 mg, tR 22 min), compound 3 (2 mg, tR 24 min), compound 4 (2 mg, tR 25 min) and compound 1 (20 mg, tR 29 min), respectively.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 369 (1.09); 1H and 13C NMR data, see Tables 1 and 2; HRESIMS m/z 673.3917 [M + Na]+ (calcd for C36H58O10Na, 673.3922).
ε) 369 (1.09); 1H and 13C NMR data, see Tables 1 and 2; HRESIMS m/z 673.3917 [M + Na]+ (calcd for C36H58O10Na, 673.3922).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 364 (1.11); 1H and 13C NMR data, see Table 1; HRESIMS m/z 673.3911 [M + Na]+ (calcd for C36H58O10Na, 673.3922).
ε) 364 (1.11); 1H and 13C NMR data, see Table 1; HRESIMS m/z 673.3911 [M + Na]+ (calcd for C36H58O10Na, 673.3922).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 363 (1.04); 1H and 13C NMR data, see Table 1; HRESIMS m/z 673.3920 [M + Na]+ (calcd for C36H58O10Na, 673.3922).
ε) 363 (1.04); 1H and 13C NMR data, see Table 1; HRESIMS m/z 673.3920 [M + Na]+ (calcd for C36H58O10Na, 673.3922).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 366 (1.04); 1H and 13C NMR data, see Table 1; HRESIMS m/z 673.3912 [M + Na]+ (calcd for C36H58O10Na, 673.3922).
ε) 366 (1.04); 1H and 13C NMR data, see Table 1; HRESIMS m/z 673.3912 [M + Na]+ (calcd for C36H58O10Na, 673.3922).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 366 (1.09); 1H and 13C NMR data, see Table 1; HRESIMS m/z 673.3911 [M + Na]+ (calcd for C36H58O10Na, 673.3922).
ε) 366 (1.09); 1H and 13C NMR data, see Table 1; HRESIMS m/z 673.3911 [M + Na]+ (calcd for C36H58O10Na, 673.3922).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 327 (1.05); 1H and 13C NMR data, see Table 1; HRESIMS m/z 689.3860 [M + Na]+ (calcd for C36H58O11Na, 689.3817).
ε) 327 (1.05); 1H and 13C NMR data, see Table 1; HRESIMS m/z 689.3860 [M + Na]+ (calcd for C36H58O11Na, 689.3817).Bioassays
The antifungal activity of the compounds were tested against Candida albicans. Solutions of the compounds were made up in methanol and dispensed into 96 well plates using the 2× microdilution method to give seven concentrations in the range of 200–3.125 μmol for each compound. MH broth was used as a blank control, and methanol was used as a negative control, while nystatin was used as a positive control. The bioassay was replicated three times for each compound. The plates were incubated for 72 h at 28 °C in the dark. Fungal growth in the wells was visually examined and the lowest concentration that inhibited hyphal growth in all the replicates was recorded as the MIC.22Conclusions
In conclusion, inspired by the genome scanning result, six new skipped-polyol polyene macrolides were isolated from the noncytotoxic fractions of the fermentation products of Streptomyces sp. CHQ-64. Reedsmycin F (6) possessed a rare tetrahydrofuran ring in the structure compared with other known polyene-polyol macrolides. Reedsmycin A (1) showed promising antifungal activity. This genome-guided strategy was more efficient than the traditional methods, and would be applicable for other compounds with characteristic structures.Acknowledgements
This work was financially supported by National Natural Science Foundation of China (nos 21102137, 31171201, and 21372208), National High Technology Research and Development Program of China (no. 2013AA092901), Program for New Century Excellent Talents in University (nos NCET-12-0499), Public Projects of State Oceanic Administration (no. 2010418022-3) and the Basic Scientific Research Fund for Young Teachers of University (no. 201413013), the Special Financial Fund of Innovative Development of Marine Economic Demonstration Project (GD2012-D01-001).Notes and references
- C. Corre and G. L. Challis, Nat. Prod. Rep., 2009, 26, 977–986 RSC.
- P. Monciardini, M. Iorio, S. Maffioli, M. Sosio and S. Donadio, Microb. Biotechnol., 2014, 7, 209–220 CrossRef CAS PubMed.
- Q. Che, T. J. Zhu, X. Qi, A. Mándi, T. Kurtán, X. M. Mo, J. Li, Q. Q. Gu and D. H. Li, Org. Lett., 2012, 14, 3438–3441 CrossRef CAS PubMed.
- Q. Che, T. J. Zhu, R. A. Keyzers, X. F. Liu, J. Li, Q. Q. Gu and D. H. Li, J. Nat. Prod., 2013, 76, 759–763 CrossRef CAS PubMed.
- W. Li, J. Ju, S. R. Rajski, H. Osada and B. Shen, J. Biol. Chem., 2008, 283, 28607–28617 CrossRef CAS PubMed.
- J. Staunton and K. J. Weissman, Nat. Prod. Rep., 2001, 18, 380–416 RSC.
- G. Yadav, R. S. Gokhale and D. Mohanty, J. Mol. Biol., 2003, 328, 335–363 CrossRef CAS.
- P. Caffrey, ChemBioChem, 2003, 4, 654–657 CrossRef CAS PubMed.
- D. J. Bevitt, J. Cortes, S. F. Haydock and P. F. Leadlay, Eur. J. Biochem., 1992, 204, 39–49 CrossRef CAS PubMed.
- C. Hertweck, Angew. Chem., Int. Ed., 2009, 48, 4688–4716 CrossRef CAS PubMed.
- H. Maehr, R. Yang, L. N. Hong, C. M. Liu, M. H. Hatada and L. J. Todaro, J. Org. Chem., 1989, 54, 3816–3819 CrossRef CAS.
- J. S. GUO, G. Schlingmann, G. T. Carter and N. Berova, Chirality, 2000, 12, 43–51 CrossRef CAS.
- Y. Kobayashi, C. H. Tan and Y. Kishi, Helv. Chim. Acta, 2000, 83, 2562–2570 CrossRef CAS.
- H. C. Kwon, C. A. Kauffman, P. R. Jensen and W. Fenical, J. Org. Chem., 2009, 74, 675–684 CrossRef CAS PubMed.
- S. D. Rychnovsky, G. Griesgraber and R. Schlegel, J. Am. Chem. Soc., 1995, 117, 197–210 CrossRef CAS.
- S. D. Rychnovsky, B. N. Rogers and T. I. Richardson, Acc. Chem. Res., 1998, 31, 9–17 CrossRef CAS.
- D. G. Kim, K. Moon, S. H. Kim, S. H. Park, S. Park, S. K. Lee, K. B. Oh, J. Shin and D. C. Oh, J. Nat. Prod., 2012, 75, 959–967 CrossRef CAS PubMed.
- S. L. Schreiber and M. T. Goulet, Tetrahedron Lett., 1987, 28, 6001–6004 CrossRef CAS.
- Y. C. Zhang, C. C. Arpin, A. J. Cullen, M. J. Mitton-Fry and T. Sammakia, J. Org. Chem., 2011, 76, 7641–7653 CrossRef CAS PubMed.
- P. Caffrey, J. F. Aparicio, F. Malpartida and S. B. Zotchev, Curr. Top. Med. Chem., 2008, 8, 639–653 CrossRef CAS.
- S. B. Zotchev, Curr. Med. Chem., 2003, 10, 211–223 CrossRef CAS.
- S. N. Aslam, P. C. Stevenson, T. Kokubun and D. R. Hall, Microbiol. Res., 2009, 164, 191–195 CrossRef CAS PubMed.
- J. Ishikawa and K. Hotta, FEMS Microbiol. Lett., 1999, 174, 251–253 CrossRef CAS PubMed.
- S. F. Altschul, W. Gish, W. Miller, E. W. Myers and D. J. Lipman, J. Mol. Biol., 1990, 215, 403–410 CrossRef CAS.
- S. Anand, M. V. Prasad, G. Yadav, N. Kumar, J. Shehara, M. Z. Ansari and D. Mohanty, Nucleic Acids Res., 2010, 38, 487–496 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: HRESIMS and NMR spectra of compounds 1–6. See DOI: 10.1039/c4ra15415k |
| ‡ These authors have contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2015 |