Metabolic transformation evidence of caffeic acid derivatives in male rats after the oral administration of functional food by UPLC coupled with a hybrid quadrupole-orbitrap mass spectrometer
Abstract
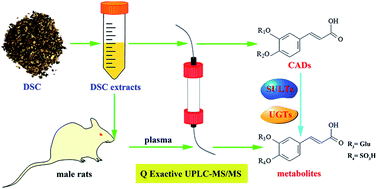
Caffeic acid (CaA) and caffeic acid derivatives (CADs) in plant-derived foods or common diet have a variety of biological functions, which rely on their absorption and metabolism. However, their metabolic pathways and transformation mechanisms are unclear due to the co-existence of multi- and micro-CADs in functional foods. We report an efficient and comprehensive analytical methodology based on ultra-performance liquid chromatography coupled with a Q Exactive hybrid quadrupole-orbitrap mass spectrometer (Q Exactive UPLC-MS/MS) to simultaneously determine the CADs and their metabolites in male rat plasma after the oral administration of a type of typical Chinese functional food, danshencha (DSC), as well as the transformation of the phase II enzymes. A total of 20 phenolic acid components were obtained and identified from the DSC extracts. This method was successfully applied to the simultaneous identification of 15 prototype and 13 metabolites of CADs in rat plasma. The biotransformation of DSC was a phase II metabolic process, especially sulfation and glucuronidation. The sulfation and glucuronidation were further confirmed by evaluating the mRNA expression of sulfotransferases (SULTs) and UDP-glucoronosyltransferases (UGTs) in rat liver. SULT1A1 and SULT1B1 were induced significantly (P < 0.001). The present study provides a suitable and convenient method to analyze and identify the CADs and metabolites of DSC in complex biological matrices. In addition, this study also reveals the possible phase II metabolic process of DSC mediated by SULTs and UGTs in male rats.

 Please wait while we load your content...
Please wait while we load your content...