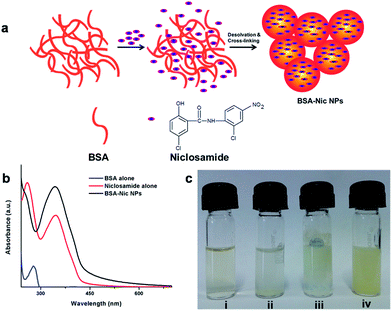
Bionanotherapeutics: niclosamide encapsulated albumin nanoparticles as a novel drug delivery system for cancer therapy†
Bharat Bhushana,
Poornima Dubeya,
S. Uday Kumara,
Abhay Sachdeva,
Ishita Mataia and
P. Gopinath*ab
aNanobiotechnology Laboratory, Centre for Nanotechnology, Indian Institute of Technology Roorkee, Roorkee, Uttarakhand-247667, India. E-mail: pgopifnt@iitr.ernet.in; genegopi@gmail.com; Fax: +91-1332-273560; Tel: +91-1332-285650
bDepartment of Biotechnology, Indian Institute of Technology Roorkee, Roorkee, Uttarakhand-247667, India
First published on 13th January 2015
Abstract
One of the major unresolved challenges among the scientific community is to develop anticancer drugs that are safe and effective. A large number of anticancer drugs have been screened so far in this campaign. Among them niclosamide has shown tremendous anti-cancer potential as demonstrated in a surfeit of human cancer cell lines and animal models. But the extreme hydrophobicity and consequently, minimal systemic bioavailability associated with this drug limited its widespread clinical applications. Nanoparticles based drug delivery systems have the potential for realizing water soluble formulation of highly hydrophobic anticancer drugs like niclosamide, thus evading the drawbacks of poor solubility. In this work niclosamide was encapsulated into albumin nanoparticles through a desolvation method to improve its scope of application in cancer therapy. Physico-chemical characterization confirms that the prepared nanoparticles are spherical, highly monodispersed, and stable in aqueous systems. These drug encapsulated albumin nanoparticles, unlike the free drug demonstrate better in vitro therapeutic efficacy against human lung and breast cancer cell lines, as assessed by cell viability assay and morphological analyses. Further, the proficient induction of apoptosis by these nanoparticles was confirmed by semi-quantitative RT-PCR. This work open up a new avenue to extend the clinical gamut of this effectual agent by enabling its aqueous dispersion.
Introduction
Biomacromolecule proteins have emerged as potential drug delivery systems, overcoming various limitations of conventional therapy such as poor solubility of the drug, poor bioavailability and therapeutic efficacy of the drug. These protein based systems have the inherent property of preferential uptake in tumor and inflamed tissue, lack of toxicity and immunogenicity and biodegradability. Moreover, the presence of charged groups in protein further assists in the physical entrapment of drug molecules. These properties make protein an ideal candidate for drug delivery.1,2During the last few decades albumin based nanoparticles have been explored for their clinical applications but a major breakthrough came with the development of food and drug administration (FDA) approved paclitaxel–albumin nanoparticles (Abraxane) by nanometer albumin-bound technology (nab™) for the treatment of advanced non-small-cell lung cancer and metastatic breast cancer.3–5 Since then there are around seven albumin based drugs or imaging agents in market and around ten such products are under clinical trials for various applications including oncology, diabetes, hepatitis C and rheumatoid arthritis.6 That augmented the interest in the use of bovine serum albumin (BSA) as nanocarrier for biomedical applications.7 Albumin is the most copious plasma protein constituting more than half of the human plasma protein having a molecular weight of 66.5 kDa, which remain stable over a wide range of pH from 4–9, and stay thermally stable when heated at 60 °C for up to 10 hours without deleterious effects.8,9 These plasma proteins constitute a crucial components in various biological processes such as maintaining colloidal osmotic pressure, delivery of nutrients to cells, balancing plasma pH and solubilizing long chain fatty acids. Moreover, the unique ligand-delivery property of serum albumin impart enhanced solubility for serum albumin conjugated hydrophobic drugs in plasma and help in improving pharmacokinetic property of drug molecules in biological environment.
Niclosamide ((5-chloro-N-2-chloro-4-nitrophenyl)-2-hydroxybenzamide), an anthelmintic drug recently reported as an effective human anticancer agent for brain cancer, colon cancer, breast cancer, lung cancer, prostate cancer and leukemia.10–15 However, despite its promising anti-cancer properties like many other drugs niclosamide suffer serious drawback due to its extremely low solubility in water (solubility as low as 230 ng mL−1 in water)16 and most organic solvent leading to their reduced bioavailability to cell that limits its clinical efficacy. To address these problems, efforts have been made in recent past for mounting the aqueous dispersion or solubility of niclosamide.17–20 But such systems were not designed keeping in mind their clinical aspects, therefore the biocompatibility and biotoxicity of such systems were remained questionable. So far, for in vitro or in vivo studies, niclosamide was dissolved in organic solvents such as dimethyl sulfoxide (DMSO) or cremophore and later mixed with PBS but implication of such organic solvent on biological systems lead to severe complications.12,21 Moreover, in vitro dissolution of organic solvent based niclosamide stock solution in water or PBS lead to the formation of insoluble curdy precipitate as shown in Fig. 1(c). The current work presents a feasible and highly reproducible method to form niclosamide encapsulated albumin nanoparticles (BSA–Nic NPs) with enhanced water solubility and improved therapeutic efficacy. Such nanoformulations were further characterized and investigated in vitro for their role in inducing programmed cell death in human breast and lung cancer cells.
Results and discussion
Preparation of BSA–Nic NPs
BSA–Nic NPs used in the present study were prepared via desolvation method,22 using ethanol as desolvating agent and glutaraldehyde as the cross linking agent as shown in schematic Fig. 1(a). This method endorses to the creation of least aggregated particles with a homogeneous distribution, which remain stable in water and cell culture medium. Glutaraldehyde, a water soluble homo bifunctional reagent is capable of forming stable inter and intra-subunit covalent bonds. Its reaction with protein leads to the formation of Schiff bases between the positively charged amino groups of the protein and two carbonyl ends of glutaraldehyde. Thus, the amino moieties in lysine and arginine residues constituting around 10 percent of the total amino acids of BSA play crucial role in the formation of nanoparticles. As prepared NPs were analyzed by UV-visible spectroscopy and the absorption maximum assessed by a spectral scan from 190 to 800 nm as shown in Fig. 1(b). The UV-visible absorption spectrum of albumin shows a characteristic absorption peak of protein at 280 nm corresponding to the aromatic amino acids present inside the protein. The niclosamide drug alone (dissolved in ethanol) had two absorption maximum at 260 nm and 346 nm. While drug encapsulated albumin NPs in aqueous medium shows characteristic peak of drug at 346 nm. A slight change in spectra has been observed, which might be a result of chemical cross linking between the amino acids of protein and the formation of protein–drug nanoparticles complex.Characterization: surface morphology, particle size and zeta potential
The surface morphology of the BSA–Nic NPs was determined by the field emission-scanning electron microscopy (FE-SEM). The images of the drug loaded albumin NPs proclaimed a spherical morphology with uniform size distribution as shown in Fig. 2(b). While, the micrometer size rod shaped flakes were observed for pristine niclosamide powder as shown in Fig. 2(a). The BSA NPs and rhodamine conjugated niclosamide encapsulated BSA NPs showed similar spherical surface morphology corresponding to BSA–Nic NPs as shown in Fig. S1.†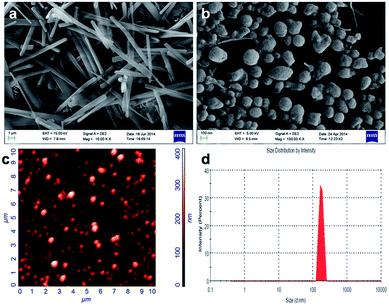 | ||
| Fig. 2 FE-SEM images of (a) raw niclosamide powder and (b) BSA–Nic NPs, showing their typical morphology (c) AFM and (d) DLS images of BSA–Nic NPs showing the distribution and size of nanoparticles. | ||
The stability of the NPs could be ascribed to the considerably higher zeta potential. The zeta potential value of the BSA–Nic NPs was found to be −34.2 mV as compared to −17.9 mV of pristine BSA. The value lies within the stable range, signifying that the nanoparticles formation lead to the formation of stable system. Moreover, the electrostatic repulsive force among the negatively charged surface of the nanoparticles affords high stability to the colloidal solution by preventing them from agglomerating in the colloid state.23
Fig. 2(d) illustrates the particle size distribution for the prepared BSA–Nic NPs. The size distribution for the prepared nanoparticles was acquired by using dynamic light scattering (DLS). The average hydrodynamic size for BSA–Nic NPs was found to be 199.9 nm. In contrast, the BSA NPs and rhodamine conjugated BSA–Nic NPs (Fig. S2†) synthesized for the carrier cytotoxicity and cellular uptake studies were found to be 264.4 nm and 273.2 nm respectively in size. The size of the prepared BSA–Nic NPs was also estimated by AFM as shown in the Fig. 2(c). The images were processed using NOVA software and the average grain size was found to be 188.27 nm. The size of the nanocarrier system plays a crucial role in drug delivery, as the nanoparticles up to 400 nm get preferentially accrue in the tumor microenvironment via “enhanced permeability and retention (EPR)” effect.24 The characteristics of BSA–Nic NPs were also compared to the previously reported drug encapsulated BSA based nanoparticles as summarized in the Table S1.†
X-ray diffraction (XRD) studies
XRD studies were conducted in order to check the physical nature of the prepared nanoparticles. On evaluating the drug encapsulated protein NPs with the control drug, BSA–Nic NPs is found to be much more amorphous in nature as compared to the niclosamide drug alone. The cross-linking mechanism occurring among the reactive functional groups of protein and drug molecules along with the prominent hydrogen bonding interaction between them may perhaps ascribed to the amorphous nature of the drug loaded albumin nanoparticles.25 Fig. 3(a) represents the XRD pattern of niclosamide and BSA–Nic NPs. Moreover, the sharp characteristic diffraction peaks of drug molecules at 25.6° and 26.7° of 2θ disappeared in the BSA–Nic NPs indicating that the drug were entrapped and no significant amount of free drug left in the system after interaction with protein.26,27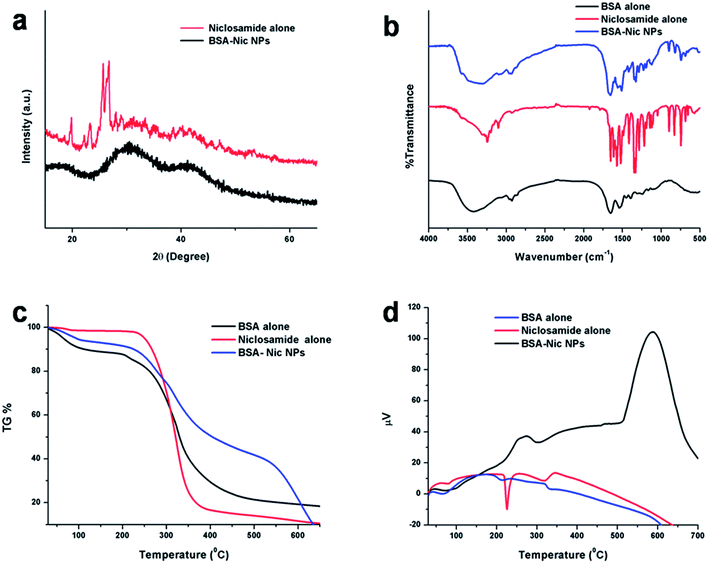 | ||
| Fig. 3 (a) XRD plot of raw niclosamide powder and BSA–Nic NPs (b) FTIR spectra (c) TG data (d) DTA curve of BSA (control), raw niclosamide powder and BSA–Nic NPs. | ||
FTIR studies
In FTIR spectrum of pristine BSA showed characteristic peaks at 1539.04, 1651.54, 3070, and 3418.56 cm−1 are allotted to the amide II (mostly from the in plane bending vibrate of N–H and stretching vibrate of C–N), amide A (mostly stretching vibrate of –NH), amide I (mostly due to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibrate of the peptides linkages) and stretching vibrate of –OH bands, respectively. While, the major characteristic peaks of drug alone were listed in Table S2.†27 On comparing the IR spectra of BSA alone, niclosamide alone and BSA–Nic NPs as shown in Fig. S3,† a major shifting of peaks were observed from 1192.09 to 1195.12, 1219.19 to 1227.90 and 1522.03 to 1511.15 indicating the involvement of the C–OH, C
O stretching vibrate of the peptides linkages) and stretching vibrate of –OH bands, respectively. While, the major characteristic peaks of drug alone were listed in Table S2.†27 On comparing the IR spectra of BSA alone, niclosamide alone and BSA–Nic NPs as shown in Fig. S3,† a major shifting of peaks were observed from 1192.09 to 1195.12, 1219.19 to 1227.90 and 1522.03 to 1511.15 indicating the involvement of the C–OH, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, NO2 groups of drug molecules in the formation of protein–drug complex. Moreover, as a result of possible interaction between the nanoparticles, the peaks were emerged as a wide spectrum. In Fig. 3(b), a typical reduction in the stretching frequency was observed in case of BSA–Nic NPs. The amide I peak of BSA shifted from 1651.54 to 1656.64 cm−1, amide II shifted from 1539.04 to 1561.02 cm−1 and amide III shifted from 1395.92 to 1417.80 cm−1, respectively which may be accredited to the cross-linking mechanism among the positively charged amino groups of protein and interaction of niclosamide with the residual amide species.25,28,29
O, NO2 groups of drug molecules in the formation of protein–drug complex. Moreover, as a result of possible interaction between the nanoparticles, the peaks were emerged as a wide spectrum. In Fig. 3(b), a typical reduction in the stretching frequency was observed in case of BSA–Nic NPs. The amide I peak of BSA shifted from 1651.54 to 1656.64 cm−1, amide II shifted from 1539.04 to 1561.02 cm−1 and amide III shifted from 1395.92 to 1417.80 cm−1, respectively which may be accredited to the cross-linking mechanism among the positively charged amino groups of protein and interaction of niclosamide with the residual amide species.25,28,29
Several interactions including electrostatic interactions, hydrogen bonds, hydrophobic force and van der Waals interactions play key role in the binding process of protein with small molecules like drug. Niclosamide move into the hydrophobic microenvironment of serum proteins including human serum albumin and interacted with the hydrophobic pockets of the protein. Interaction of niclosamide with serum proteins mainly involves hydrophobic interaction, hydrogen bonding and van der Waals interaction. Like many other drug molecules, niclosamide interact with the tryptophan residues (Trp 212) located inside the hydrophobic pocket of the BSA.30,31 To understand the effect of niclosamide on the tryptophan environment of BSA, the intrinsic fluorescence of BSA in the presence of increasing concentrations of niclosamide was studied as shown in Fig. S4.† Intrinsic fluorescence of BSA is due to two tryptophan residues: (1) Trp 134, situated on the exterior in the hydrophilic region of protein and typified by a longer wavelength emission maximum (2) Trp 212, situated within a hydrophobic binding pocket that is typified by a shorter wavelength emission maximum around 340 nm. Hence, the fluorescence emission of BSA with maximum around 340 nm corresponding to the Trp residue at 212 was monitored. The fluorescence intensity of BSA decreases regularly with increasing niclosamide concentrations. This decrease in the intensity might be a result of fluorescence quenching ascribed to the changes in the microenvironment around tryptophan residues suggesting interaction of niclosamide with BSA. Hence the results obtained from FTIR and fluorescence spectroscopy were in accordance with previous observation and clearly indicate the involvement of aryl groups of drug and aromatic residues of protein in the drug carrier interactions.
Thermal stability of the nanoparticles
The thermogravimetric (TG) analysis of niclosamide, BSA–Nic NPs and BSA alone (control) were conducted, which depicted slower degradation rate of protein–drug nanoformulation indicating their improved stability as compare to pristine BSA and niclosamide particles. From Fig. 3(c), it is clearly visible that the particles start degrading from 200 °C and beyond 250 °C, an abrupt decrease in weight was found which could be as a result of loss of small molecule such as carbon dioxide, ammonia etc. At 400–500 °C there is considerable difference in weight loss was observed, as 21% was remaining for control BSA, 41% was remaining for BSA–Nic NPs, whereas only 13% was remaining for niclosamide drug alone which confirm the slower degradation rate for BSA–Nic NPs as compared to niclosamide drug alone. Beyond 500 °C, a faster rate of degradation of BSA–Nic NPs was observed as compared to BSA (control), which may be due to the crystalline nature of entrapped niclosamide drug molecules in BSA–Nic NPs. Whereas, no significant change was observed in BSA (control) due to the char formation in nitrogen atmosphere. Recently, Gebregeorgis et al. demonstrated that no char formation take place in air atmosphere as compared to nitrogen atmosphere, an additional step is required related to the combustion of the char product in air atmosphere.32The differential thermal analysis (DTA) of the bare niclosamide drug showed an endothermic peak at 225 °C which was not there in case of BSA–Nic NPs depicting the high amorphous nature of the as prepared nanoparticles and thus has better stability. Moreover, the exothermic peak also moved toward higher temperature range which once more substantiates the high amorphous nature of protein–drug nanoformulation as evident from Fig. 3(d). The higher amorphous nature of the therapeutic system, the more likely is its delivery efficiency.33
Encapsulation efficiency (EE) of nanoparticles
Various concentrations of the drug with the albumin nanoparticles were taken and synthesis was done by desolvation method. The encapsulation efficiency of the BSA NPs was analyzed by using different concentrations of niclosamide, while the carrier concentration was kept constant. The protein–drug combination with the preeminent encapsulation efficiency was taken for rest of the studies. As shown in Fig. 4(a), 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 ratio of niclosamide
25 ratio of niclosamide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BSA provide the paramount encapsulation efficiency of 92.36%.
BSA provide the paramount encapsulation efficiency of 92.36%.
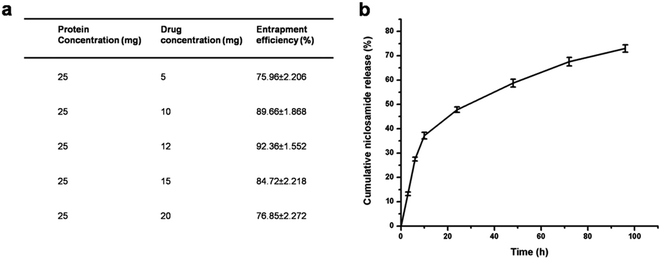 | ||
| Fig. 4 (a) Entrapment efficiency of BSA NPs with varying niclosamide concentrations (b) niclosamide release profile from BSA NPs in PBS. | ||
In vitro drug release study
The in vitro drug release profiles of the BSA–Nic NPs over 96 hours was studied at pre defined time intervals in PBS at pH 7.4 as shown in Fig. 4(b). The in vitro drug release is a coalesced effect of diffusion of drug molecule out of the protein nanoparticles into the external environment and concurrent degradation of carrier itself. A biphasic drug release pattern was observed with an initial burst release of 37.15% in the first 10 hours pursued by a sustained release of the drug. The initial burst release is because of the surface bound drug molecules, while the controlled release is attributed to the encapsulated drug molecules. In the following 24 hours, cumulative release reached 47.81%, in a controlled manner, making the protein based nanoparticles an effective drug carrier to fight repeatedly against cancer cells for an effective cancer therapy. After 4 days cumulative release reached approximately 73%.Generally, the sustained release profile of niclosamide makes the potential application of these nanoparticles in the field of nanomedicine. The improved stability of the BSA–Nic NPs in the physiological buffer is accredited to the cross linking mechanism of the protein nanoparticles via glutaraldehyde. The results demonstrated that slow controlled release of the drug in the media proved it as better delivery system for cancer treatment. Moreover, as compared to the extremely slow release profile of raw niclosamide powder as reported earlier,34 the continuous release of niclosamide from the BSA–Nic NPs solution can be accredited to the solubilizing capability of the BSA carrier. The erosion and degradation of BSA NPs and the insolubility of niclosamide in aqueous medium were attributed to the drug release mechanism.
In vitro stability of BSA–Nic NPs
In order to evaluate the physiological stability of BSA–Nic NPs in aqueous and 0.9% saline solution, the change of particle size were monitored by DLS in vitro for more than 96 h. As shown in Fig. S5,† when BSA–Nic NPs were placed in aqueous and 0.9% saline solution, there was no obvious size change even after 96 h at 25 °C. This phenomenon demonstrated the high stability of the BSA–Nic NPs. Since, stability always remains a decisive feature for the clinical use of drug formulation as the drug could be slowly escape from the nanoparticles or may degrade with time. Therefore, the stability of encapsulated niclosamide in BSA–Nic NPs was evaluated for 96 h at 25 °C by analytical measurement of drugs contents. The spectrophotometric spectra remained unchanged after 96 h, showing that no change in drug content took place.Cell viability assay
The effect of BSA–Nic NPs on the viability of A549 and MCF-7 cells was assessed quantitatively by MTT assay as shown in the Fig. 5. The results illustrate the concentration-dependent inhibition of cell viability by BSA–Nic NPs. This might be due to the sustained release of drug molecule from the nanocarrier. The IC50 concentration of BSA–Nic NPs was found to be 5 μM and 2.6 μM for A549 and MCF-7 cells, respectively. On the other hand, bare niclosamide (in water) showed a nontoxic effect on both the cell lines, probably it could be due to the practical insoluble nature of the bare niclosamide drug in aqueous medium, due to which most of the drug molecules were not taken up by the cells. However, as the nanoformulation increase the niclosamide solubility in aqueous medium, it can be easily taken up by the cells and resulted in concentration dependent cell inhibition. Thus BSA–Nic NPs emerged as an effective drug carrier with enhanced anticancer activity in contrast to the free drug. However, it should be taken into account as previously reported that free niclosamide (dissolved in DMSO) also exhibited cytotoxicity against A549 and MCF-7 cell lines as shown in Fig. S6(b).† However, implication of organic solvent DMSO as a solubilizing agent lead to severe complication as it itself produce cytotoxic effect, thereby not suitable for future clinical applications. We confirm that at each concentration the BSA–Nic NPs were taken such that the concentration of niclosamide in the free drug is comparable to the entrapped drug, thus making a through evaluation possible.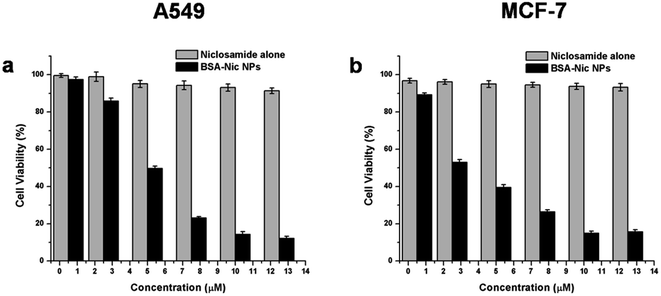 | ||
| Fig. 5 Cell viability assay (MTT assay) of raw niclosamide (in water) and BSA–Nic NPs on (a) A549 cells (b) MCF-7 cells. | ||
The bare BSA NPs were also analyzed for their cell toxicity and found to be non-toxic against both the cells line as shown in the Fig. S6(a),† more than 90% cell viability was found after 24 hours confirming the biocompatibility of bare nanoparticles. These results clearly signifying that the therapeutic efficacy is not due to the carrier BSA NPs but because of the drug encapsulated BSA NPs.
Acridine orange/ethidium bromide (AO/EB) staining
In order to examine the mode of cell death (via apoptosis or necrosis), EB and AO were used in combination to stain the BSA–Nic NPs treated A549 and MCF-7 cells and observed under the fluorescence microscope. EB stain was excluded by the healthy live cell's nucleus and it appears green due to the presence of AO alone (Fig. 6(a)). The cells treated with half the IC50 concentration of BSA–Nic NPs showed no significant changes in the morphology of the cell and it appears similar to untreated cells with uniformly green and normal morphology (Fig. 6(b)). However, typical morphological changes correlated with apoptosis (programmed cell death) including nuclear fragmentation, membrane blebbing, and cytoplasmic constrictions35,36 were observed in treated cells at IC50 and 2× IC50 concentrations. Fig. 6(c) and (d) clearly depict the occurrence of early apoptosis in the cells characterized by their condensed chromatin in addition to that late apoptosis were also observed in the cells typified by their fragmented chromatin and apoptotic bodies. The results of AO/EB staining clearly demonstrated the induction of apoptosis in A549 and MCF-7 cells by BSA–Nic NPs.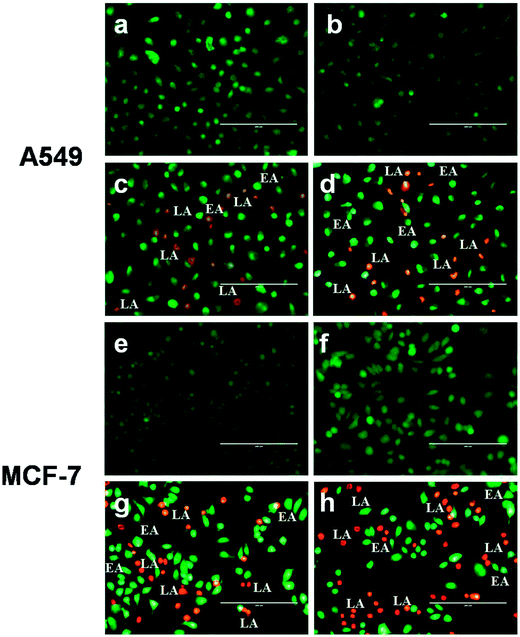 | ||
| Fig. 6 Representative images of AO/EB dual staining of (a and e) untreated (b and f) half IC50 (c and g) IC50 and (d and h) 2× IC50 BSA–Nic NPs treated A549 and MCF-7 cells after 24 h of treatment. | ||
FE-SEM analysis
In addition, to the AO/EB staining, BSA–Nic NPs treated A549 and MCF-7 cells were too monitored under the FE-SEM in the pursuit of the typical morphological changes occurred during apoptosis. Fig. 7(a and c) shows the typical morphology of untreated A549 and MCF-7 cells that are well-adhere to the surface. However, the cells treated with BSA–Nic NPs are loosely attached cells with rounded morphology (Fig. 7(b and d)) as compared to untreated cells. In both the cells, the event of cytoplasmic constriction, membrane blebbing and formation of apoptotic bodies clearly indicate the apoptotic cell death.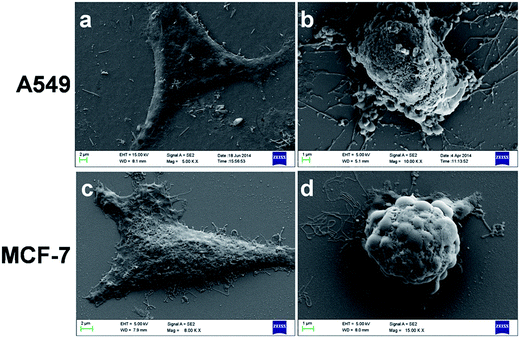 | ||
| Fig. 7 Representative FE-SEM images of (a and c) untreated and (b and d) treated A549 and MCF-7 cells. | ||
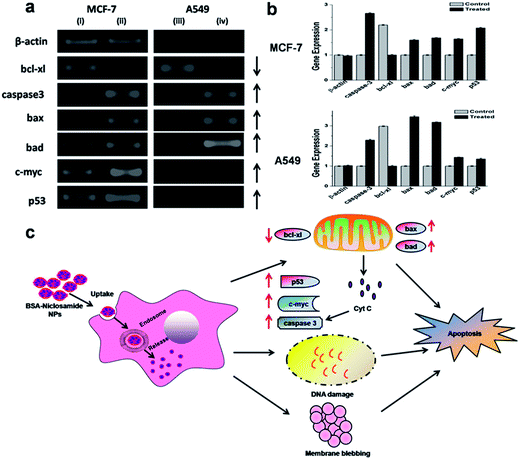
Semi-quantitative RT-PCR analysis
The potency of niclosamide encapsulated BSA NPs to induce apoptosis in human breast and lung cancer was assessed in vitro via semi-quantitative RT-PCR analysis, results suggested that various apoptotic signaling genes were involved in BSA–Nic NPs mediated cell death as shown in Fig. 8(c). The NPs treated cells were analyzed for their apoptotic signaling genes expression profile such as c-myc, bax, p53, bad and caspase-3 that indicated their up-regulation, while the expression of anti-apoptotic gene such as bcl-xl found to be down-regulated. Moreover, the expression of housekeeping gene β-actin, which was taken as an internal control stayed unchanged during the process (Fig. 8(a and b)).It is known that anticancer drug niclosamide induces mitochondrial fragmentation and promote apoptotic cell death.37 The destabilization of mitochondrial integrity precedes the release of cytochrome-c, a pro-apoptotic molecule into the cytoplasm that activates the caspases leading to apoptosis. Such phenomenon is intimately governed by a group of proteins belonging to the Bcl-2 (B-cell lymphoma 2) family, namely the pro-apoptotic proteins (e.g. bcl-2-associated X protein (Bax) and bcl-2-associated death promoter (Bad)) and the anti-apoptotic protein (e.g. basal cell lymphoma-extra large (Bcl-xl)).38 The apoptosis is regulated by both the anti-apoptotic proteins which block the mitochondrial release of cytochrome-c and the pro-apoptotic proteins which promote such release and thereby act as a crucial factor in the inhibition of anti-apoptotic function of bcl-xl.39,40 The members of the Bcl-2 family are situated on the outer mitochondrial membrane and regulate the release of cytochrome c into the cytosol by controlling the membrane permeability either by creating an ion channel or via formation of pores.41 In the present study, a substantial increase in expression of bax and bad was observed, suggesting their role in the downregulation of anti-apoptotic gene bcl-xl.
p53 gene, a key factor which play an important role in apoptosis was up regulated in the BSA–Nic NPs treated cells. It has been accounted that p53 protein up-regulates the pro-apoptotic gene bax.42 As, we observed an increment in the level of bax expression suggesting the role of p53 in their upregulation. Similarly, c-myc is also viewed as a promising target for anti-cancer drugs. The expression of c-myc, an identified inducer of apoptosis is found to be up-regulated, which further contributes in the down-regulation of anti-apoptotic genes bcl-xl and in the strengthening of the apoptotic signals.
In many cells, during apoptosis caspases (cysteine–aspartic acid proteases) get activated and are found to act as a crucial factor both in the initiation and execution of apoptosis. It was accounted that the actual cleavage of cellular components during apoptosis was accountable to caspase-3.43 We found that in BSA–Nic NPs treated cells gene expression level of caspase-3 was up-regulated, which suggested its role in BSA–Nic NPs mediated apoptosis. The above stated gene expression profiles clearly depicted that BSA–Nic NPs treatment of both the A549 and MCF-7 cells leads to apoptosis and consequent cell blebbing.
Cellular uptake studies
Systematic cellular uptake studies of BSA–Nic NPs by A549 and MCF-7 cells were carried out by monitoring the fluorescence of rhodamine via fluorescence microscopy. As reported earlier, rhodamine conjugated protein nanoparticles were used in the cell uptake studies of drug loaded protein nanoparticles.25 Rhodamine B is a red fluorescent dye, thus the cells which uptake rhodamine conjugated nanoparticles would characteristically show bright red color fluorescence. After 8–12 hours of incubation, fluorescence microscopic images were taken and it was observed that there was a considerable internalization of nanoparticles inside the cells (Fig. S7†). In addition, no fluorescence was observed in case of control cells without any particles, which further confirms the study.Conclusion
A novel anticancer drug loaded delivery system based on biodegradable BSA NPs was designed in the present study. This study is the first report of the enhanced solubilization and stability of niclosamide in aqueous environment by preparing a protein–drug nanoformulation via desolvation technique. It also allows us to eliminate the use of toxic organic compounds such as cremphor and DMSO. The prepared BSA–Nic NPs were typified by AFM, FE-SEM and DLS. This corroborated that even the drug incorporation did not alter the size of the prepared nanoparticles and it stays within an optimal range applicable for drug delivery applications. BSA NPs showed improved sustained release of niclosamide in aqueous environment. Moreover, the MTT result revealed the non toxic nature of the nanocarrier and enhanced anti-proliferative ability of drug against breast and lung cancer cells in vitro than that of the free drug in aqueous medium. Morphological analysis of cells (via FE-SEM and AO/EB staining) and expression of apoptotic signaling genes (via semi-quantitative RT-PCR), further confirm the successful induction of apoptosis in BSA–Nic NPs treated cancer cells. The cellular uptake studies of BSA–Nic NPs conducted in A549 and MCF-7 cells revealed considerable internalization of these nanoparticles inside the cells, suggesting the utility of these nanoparticles system for drug delivery applications. However, it should be noted that this study is only a proof-of-concept investigation. In future, human serum albumin (HSA) would be taken in place of BSA so as to avoid any probable immunologic consequences in vivo. Thus our studies propose that the chemotherapeutic potential of niclosamide could be better utilized by loading it in a protein based carrier for future clinical applications.Acknowledgements
This work was supported by the Science and Engineering Research Board (no. SR/FT/LS-57/2012) and Department of Biotechnology (no. BT/PR6804/GBD/27/486/2012), Government of India. Sincere thanks to Department of Chemistry and Institute Instrumentation Centre, IIT Roorkee for the various analytical facilities provided.References
- B. Bhushan, U. K. Sukumar, I. Matai, A. Sachdev, P. Dubey and P. Gopinath, J. Biomed. Nanotechnol., 2014, 10, 2950–2976 CrossRef CAS PubMed.
- U. K. Sukumar, B. Bhushan, P. Dubey, I. Matai, A. Sachdev and P. Gopinath, Int. Nano Lett., 2013, 3, 45–53 CrossRef.
- N. K. Ibrahim, N. Desai, S. Legha, P. Soon-Shiong, R. L. Theriault, E. Rivera, B. Esmaeli, S. E. Ring, A. Bedikian, G. N. Hortobagyi and J. A. Ellerhorst, Clin. Cancer Res., 2002, 8, 1038–1044 CAS.
- E. Miele, G. P. Spinelli, E. Miele, F. Tomao and S. Tomao, Int. J. Nanomed., 2009, 4, 99–105 CrossRef CAS.
- M. R. Green, G. M. Manikhas, S. Orlov, B. Afanasyev and A. M. Makhson, Ann. Oncol., 2006, 17, 1263–1268 CrossRef CAS PubMed.
- K. Ren, A. Dusad, R. Dong and L. Quan, J. Nanomed. Nanotechnol., 2013, 4, 176 Search PubMed.
- M. Rahimnejad, M. Jahanshahi and G. D. Najafpour, Afr. J. Biotechnol., 2006, 5, 1918–1923 CAS.
- E. Neumann, E. Frei, D. Funk, M. D. Becker, H. H. Schrenk, U. Müller-Ladner and C. Fiehn, Expert Opin. Drug Delivery, 2010, 7, 915–925 CrossRef CAS PubMed.
- F. Kratz, J. Controlled Release, 2008, 132, 171–183 CrossRef CAS PubMed.
- A. Wieland, D. Trageser, S. Gogolok, R. Reinartz, H. Hofer, M. Keller, A. Leinhaas, R. Schelle, S. Normann, L. Klaas, A. Waha, P. Koch, R. Fimmers, T. Pietsch, A. T. Yachnis, D. W. Pincus, D. A. Steindler, O. Brustle, M. Simon, M. Glas and B. Scheffler, Clin. Cancer Res., 2013, 19, 4124–4136 CrossRef CAS PubMed.
- Y. Jin, Z. Lu, K. Ding, J. Li, X. Du, C. Chen, X. Sun, Y. Wu, J. Zhou and J. Pan, Cancer Res., 2010, 70, 2516–2527 CrossRef CAS PubMed.
- U. Sack, W. Walther, D. Scudiero, M. Selby, D. Kobelt, M. Lemm, I. Fichtner, P. M. Schlag, R. H. Shoemaker and U. Stein, J. Natl. Cancer Inst., 2011, 103, 1018–1036 CrossRef CAS PubMed.
- Y. C. Wang, T. K. Chao, C. C. Chang, Y. T. Yo, M. H. Yu and H. C. Lai, PLoS One, 2013, 8, e74538 CAS.
- X. M. Ren, L. Duan, Q. He, Z. Zhang, Y. Zhou, D. Wu, J. Pan, D. Pei and K. Ding, ACS Med. Chem. Lett., 2010, 1, 454–459 CrossRef CAS PubMed.
- W. Lu, C. Lin, M. J. Roberts, W. R. Waud, G. A. Piazza and Y. Li, PLoS One, 2011, 6, e29290 CAS.
- M. J. O'Neil, A. Smith, P. E. Heckelman and S. Budavari, in The Merck Index, Merck, New York, 13th edn, 2001 Search PubMed.
- J. Dai, W. Wang, Y. Liang, H. Li, X. Guan and Y. Zhu, Parasitol. Res., 2008, 103, 405–412 CrossRef PubMed.
- W. Yang and M. M. D. Villiers, AAPS J., 2005, 7, E241–E248 CrossRef CAS PubMed.
- E. Kenawy and E. Rizk, Macromol. Biosci., 2004, 4, 119–128 CrossRef PubMed.
- B. Devarakonda, R. A. Hill, W. Liebenberg, M. Brits and M. M. D. Villiers, Int. J. Pharm., 2005, 304, 193–209 CrossRef CAS PubMed.
- J. L. Hanslick, K. Lau, K. K. Noguchi, J. W. Olney, C. F. Zorumski, S. Mennerick and N. B. Farber, Neurobiol. Dis., 2009, 34, 1–10 CrossRef CAS PubMed.
- C. Weber, C. Coester, J. Keruter and K. Langer, Int. J. Pharm., 2000, 194, 91–102 CrossRef CAS.
- S. Sripriyalakshmi, C. H. Anjali, C. G. P. Doss, B. Rajith and A. Ravindran, PLoS One, 2014, 9, e86317 CAS.
- D. Peer, J. M. Karp, S. Hong, O. C. Farokhzad, R. Margalit and R. Langer, Nat. Nanotechnol., 2007, 2, 751–760 CrossRef CAS PubMed.
- N. S. Rejinold, M. Muthunarayanan, K. P. Chennazhi, S. V. Nair and R. Jayakumar, J. Biomed. Nanotechnol., 2011, 7, 521–534 CrossRef CAS PubMed.
- C. Li, L. Xing and S. Che, Dalton Trans., 2012, 41, 3714–3719 RSC.
- E. C. V. Tonder, T. S. P. Maleka, W. Liebenberg, M. Song, D. E. Wurster and M. M. Villiers, Int. J. Pharm., 2004, 269, 417–432 CrossRef PubMed.
- J. Kong and Y. U. Shaoning, Acta Biochim. Biophys. Sin., 2007, 39, 549–559 CrossRef CAS PubMed.
- P. Huang, Z. Li, H. Hu and D. Cui, J. Nanomater., 2010, 2010, 641545 Search PubMed.
- M. Hossain, A. Y. Khan and G. S. Kumar, PLoS One, 2011, 6, e18333 CAS.
- E. Maltas, J. Food Drug Anal., 2014, 22, 549–555 CrossRef CAS PubMed.
- A. Gebregeorgis, C. Bhan, O. Wilson and D. Raghavan, J. Colloid Interface Sci., 2013, 389, 31–41 CrossRef CAS PubMed.
- W. Abdelwahed, G. Degobert, S. Stainmesse and H. Fessi, Adv. Drug Delivery Rev., 2006, 58, 1688–1713 CrossRef CAS PubMed.
- M. Y. Bai and H. C. Yang, Colloids Surf., A, 2013, 419, 248–256 CrossRef CAS PubMed.
- S. Rello, J. C. Stockert, V. Moreno, A. G'amez, M. Pacheco, A. Juarranz, M. Canete and A. Villanueva, Apoptosis, 2005, 10, 201–208 CrossRef CAS PubMed.
- S. U. Kumar, I. Matai, P. Dubey, B. Bhushan, A. Sachdev and P. Gopinath, RSC Adv., 2014, 4, 38263–38272 RSC.
- S. J. Park, J. H. Shin, H. Kang, J. J. Hwang and D. H. Cho, BMB Rep., 2011, 44, 517–522 CrossRef CAS.
- R. S. Y. Wong, J. Exp. Clin. Cancer Res., 2011, 30, 87 CrossRef CAS PubMed.
- E. H. Yang, J. Zha, J. Jockel, L. B. Boise, C. B. Thompson and S. J. Korsmeyer, Cell, 1995, 80, 285–291 CrossRef CAS.
- L. H. Boise, M. Gonzalez-Garcia, C. E. Postema, L. Ding, T. Lindsten, L. A. Turka, X. Mao, G. Nunez and C. B. Thompson, Cell, 1993, 74, 597–608 CrossRef CAS.
- A. J. Minn, P. Vélez, S. L. Schendel, H. Liang, S. W. Muchmore, S. W. Fesik, M. C. Fill and B. Thompson, Nature, 1997, 385, 353–357 CrossRef CAS PubMed.
- K. G. Wolter, Y. Hsu, C. L. Smith, A. Nechushtan, X. Xi and R. J. J. Youle, Cell Biol., 1997, 139, 1281–1292 CrossRef CAS.
- S. L. Fink and B. T. Cookson, Infect. Immun., 2005, 73, 1907–1916 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4ra15233f |
| This journal is © The Royal Society of Chemistry 2015 |