Bacteria microarrays as sensitive tools for exploring pathogen surface epitopes and recognition by host receptors
María Asunción Campanero-Rhodes*ab,
Enrique Llobetbcd,
José Antonio Bengoecheabe and
Dolores Solísab
aInstituto de Química Física Rocasolano, CSIC, Madrid, Spain. E-mail: ma.campanero@iqfr.csic.es; d.solis@iqfr.csic.es; Fax: +34 915642431; Tel: +34 915619400
bCentro de Investigación Biomédica en Red de Enfermedades Respiratorias (CIBERES), Madrid, Spain
cPrograma Infección e Inmunidad, Fundación de Investigación Sanitaria de las Illes Balears Ramón Llull (FISIB), Palma, Spain. E-mail: llobet@caubet-cimera.es
dInstituto de Investigación Sanitaria de Palma, Palma, Spain
eCentre for Infection and Immunity, Queen's University, Belfast, UK. E-mail: j.bengoechea@qub.ac.uk
First published on 16th December 2014
Abstract
We describe a protocol for the generation and validation of bacteria microarrays and their application to the study of specific features of the pathogen's surface and interactions with host receptors. Bacteria were directly printed on nitrocellulose-coated glass slides, using either manual or robotic arrayers, and printing quality, immobilization efficiency and stability of the arrays were rigorously controlled by incorporating a fluorescent dye into the bacteria. A panel of wild type and mutant strains of the human pathogen Klebsiella pneumoniae, responsible for nosocomial and community-acquired infections, was selected as model bacteria, and SYTO-13 was used as dye. Fluorescence signals of the printed bacteria were found to exhibit a linear concentration-dependence in the range of 1 × 108 to 1 × 109 bacteria per ml. Similar results were obtained with Pseudomonas aeruginosa and Acinetobacter baumannii, two other human pathogens. Successful validation of the quality and applicability of the established microarrays was accomplished by testing the capacity of the bacteria array to detect recognition by anti-Klebsiella antibodies and by the complement subcomponent C1q, which binds K. pneumoniae in an antibody-independent manner. The biotin/AlexaFluor-647-streptavidin system was used for monitoring binding, yielding strain- and dose-dependent signals, distinctive for each protein. Furthermore, the potential of the bacteria microarray for investigating specific features, e.g. glycosylation patterns, of the cell surface was confirmed by examining the binding behaviour of a panel of plant lectins with diverse carbohydrate-binding specificities. This and other possible applications of the newly developed arrays, as e.g. screening/evaluation of compounds to identify inhibitors of host–pathogen interactions, make bacteria microarrays a useful and sensitive tool for both basic and applied research in microbiology, biomedicine and biotechnology.
1. Introduction
Searching for “microarray” in the NCBI search engine (http://www.ncbi.nlm.nih.gov/pubmed/) returns over 62![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 references (September 2014), showing the relevance of this tool in biology and biomedicine. The concept of microarray technology was first put forward by Ekins1 over 20 years ago. It was proposed that a miniature spot of a purified antibody or protein can enhance detection sensitivity. DNA microarrays were the first application of this concept and have been tremendously successful in gene expression profiling and related applications.2–6 Next, numerous formats of protein microarrays were developed for high-throughput studies of protein expression and functionalities, the four major types being proteome microarrays,7,8 antibody microarrays,9,10 reverse-phase protein arrays,11,12 and lectin microarrays.13,14 In the past decade, and as natural extension of the successful development and application of DNA and protein arrays, glycan microarrays revolutionized the analysis of biological systems that operate through carbohydrate recognition, publications on their design, optimization and applicability having grown exponentially.15–19 Thus, carbohydrate microarrays have been successfully used for detection and quantitation of disease-associated anti-carbohydrate antibodies and analysis of their binding specificity,20–22 and for identification of oligosaccharide signals recognized by endogenous lectins, as C-type lectins,23–25 siglecs26–28 and galectins, or by pathogens' glycan-binding proteins that mediate docking into host cells.29–31
000 references (September 2014), showing the relevance of this tool in biology and biomedicine. The concept of microarray technology was first put forward by Ekins1 over 20 years ago. It was proposed that a miniature spot of a purified antibody or protein can enhance detection sensitivity. DNA microarrays were the first application of this concept and have been tremendously successful in gene expression profiling and related applications.2–6 Next, numerous formats of protein microarrays were developed for high-throughput studies of protein expression and functionalities, the four major types being proteome microarrays,7,8 antibody microarrays,9,10 reverse-phase protein arrays,11,12 and lectin microarrays.13,14 In the past decade, and as natural extension of the successful development and application of DNA and protein arrays, glycan microarrays revolutionized the analysis of biological systems that operate through carbohydrate recognition, publications on their design, optimization and applicability having grown exponentially.15–19 Thus, carbohydrate microarrays have been successfully used for detection and quantitation of disease-associated anti-carbohydrate antibodies and analysis of their binding specificity,20–22 and for identification of oligosaccharide signals recognized by endogenous lectins, as C-type lectins,23–25 siglecs26–28 and galectins, or by pathogens' glycan-binding proteins that mediate docking into host cells.29–31
Bacterial surfaces are coated with distinct signature molecules, most prominently capsular polysaccharides and lipopolysaccharides (LPS), that are targeted by host receptors, including endogenous lectins, for triggering defense responses or as mechanism exploited by the pathogen for attachment.32–34 Recently, bacterial polysaccharide microarrays have been employed to examine the ability of three members of the galectin family for targeting the presented determinants.35 Inevitably, the potential of these and analogous microarray set-ups for detecting pathogen recognition by host receptors is limited by the library of probes included in the array, often constrained by the laborious protocols required for their purification. In addition, the particular presentation of the probes in the array may substantially differ from their natural arrangement on the pathogen's surface, this factor potentially having a significant impact on recognition. Thus, the real accessibility on the bacterial surface of the recognized determinant is not taken into account and any operative synergetic contribution of other molecules to the recognition cannot be evaluated. To overcome these limitations, we have developed a novel application of the microarray technology based on the generation and validation of bacteria microarrays. Klebsiella pneumoniae, an important human pathogen,36 has been selected as model bacteria.
As proof of principle, the binding of anti-Klebsiella antibodies to microarrays containing wild type K. pneumoniae and a panel of mutant strains lacking different surface epitopes has been tested. In addition, based on the reported binding of C1q to K. pneumoniae,37 the behaviour of this complement subcomponent in the Klebsiella microarray set-up has been examined. The results confirm the usefulness of bacteria microarrays for investigating pathogen–host counter-receptor interactions. Furthermore, the strain-specific selective binding of plant lectins to the arrays illustrates the potential of the newly developed microarrays for typifying bacterial surface features.
2. Methods
2.1. Bacterial strains
Bacterial strains used in this study are listed in Table 1. Strains were grown in lysogeny broth (LB) at 37 °C on an orbital shaker (180 rpm). When appropriate, antibiotics (Sigma) were added to the growth medium at the following concentrations: rifampicin (Rif) 25 μg ml−1, kanamycin (Km) 100 μg ml−1, and chloramphenicol (Cm) 12.5 μg ml−1. Bacteria were fixed as previously described,38 suspended in 500 μl of 10 mM Tris–HCl at pH 7.8, containing 0.15 M NaCl (TBS), and labelled by incubation with 5 μl of 5 mM SYTO-13 solution (Invitrogen), for 5 min in the dark. After thorough washing with TBS, the concentration of bacteria was adjusted to optical density 1 at 600 nm, equivalent to approximately 1 × 109 bacteria per ml. Labelling efficiency was assessed by measuring the fluorescence intensity of the different bacteria strains, using a Horiba Jobin Yvon Fluoromax-4 spectrofluorometer.| Bacterial strain | Genotype or comments | Identifier | Source or references |
|---|---|---|---|
| a Rif, rifampicin; Km, kanamycin; Cm, chloramphenicol. | |||
| K. pneumoniae | |||
| Kp52145 | Wild type; clinical isolate (serotype O1:K2), RifR | WT | 39 and 40 |
| 52O21 | Kp52145, wbbM gene inactivated; no OPS expression; RifR, KmR | Δops | 40 |
| 52145-ΔwcaK2 | Kp52145, ΔwcaK2; the wcaK2 gene inactivated, no CPS expression; RifR | Δcps | 41 |
| 52145-ΔwcaK2–ΔwaaL | 52145-ΔwcaK2, ΔwaaL; the waaL gene inactivated; nonpolar mutant; no CPS no OPS expressions; RifR | ΔcpsΔops | 42 |
| 52145-ΔwcaK2–ΔwabM | 52145-ΔwcaK2, ΔwabM; the wabM gene inactivated; nonpolar mutant; no CPS no OPS no first core sugar expressions; RifR | ΔcpsΔopsΔwabM | 42 |
| 52145-ΔwabM | Kp52145, ΔwabM; the wabM gene inactivated, nonpolar mutant, no OPS no first core sugar expression; RifR | ΔopsΔwabM | 43 |
| 52OmpA2 | Kp52145, ompA gene inactivated by insertion of pKNOCKIntKpnOmpA; no OmpA expression; RifR, CmR | ΔompA | 44 |
| 52OmpA2Com | Kp52145 ompA mutant harbouring mini-Tn7TKmKpnOmpA; OmpA levels restored, RifR, CmR, KmR | ΔompACom | 44 |
| 52145-ΔwcaK2ompA | 52145-ΔwcaK2; ompA gene inactivated by insertion of pKNOCKIntKpnOmpA; no CPS no OmpA expressions; RifR, CmR | ΔcpsΔompA | 44 |
| 52145-ΔwcaK2ompACom | 52145-ΔwcaK2, ompA mutant harbouring mini-Tn7TKmKpnOmpA; no CPS expression but OmpA levels restored RifR, CmR, KmR | ΔcpsΔompACom | 44 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Pseudomonas aeruginosa | |||
| PAO1 | Wild type; ATCC 15692 | ATCC | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Acinetobacter baumannii | |||
| ATCC 19606 | Wild type | ATCC | |
2.2. Protein targets
Two different rabbit polyclonal anti-K. pneumoniae antibodies, hereafter referred to as anti-Kp1 and anti-Kp2, were used without labelling as described in section 2.4. Anti-Kp1 (Abcam, ab20947) was obtained by immunization with an extracellular toxic complex produced by the bacteria and composed of 63% capsular polysaccharide, 30% lipopolysaccharide, and 7% protein. Anti-Kp2, kindly provided by Dr S. Albertí (Instituto Universitario de Investigaciones en Ciencias de la Salud, Universidad de las Islas Baleares, Palma, Mallorca, Spain), was raised against the whole bacterium using the wild type strain Kp52145.The complement component C1q from human serum (Sigma-Aldrich) was biotinylated by incubation at 4 °C overnight with biotinamidocaproate ester derivative (GE) in 10 mM HEPES pH 7.2, containing 0.3 M NaCl.
Commercial biotinylated lectins used were Aleuria aurantia lectin (AAL), Arachis hypogaea (peanut) agglutinin (PNA), Concanavalin A (ConA), Glycine max (soybean) agglutinin (SBA), Hippeastrum hybrid lectin (HHL), Pisum sativum agglutinin (PSA), Ricinus communis agglutinin (RCA), Sambucus nigra lectin (SNL) and Wisteria floribunda lectin (WFL) from VECTOR labs, and Lycopersicon esculentum agglutinin (LEA) from Sigma.
2.3. Preparation of bacteria microarrays
Manual arrays were prepared by printing bacteria on single-pad nitrocellulose-coated glass slides (FAST-slides, Whatman) using a manual glass-slide arraying system (V&P Scientific). Each probe was printed in a four-level dose-response format by applying approximately 8 nl per spot of SYTO-13-labelled bacteria suspensions typically ranging from 1 × 108 to 1 × 109 bacteria per ml in PBS (5 mM sodium phosphate, pH 7.2, 0.2 M NaCl). Spots were printed as quadruplicates. For preparation of robotic arrays, bacteria were printed on 16-pad nitrocellulose-coated glass slides (FAST-slides, Maine manufacturing) using a non-contact arrayer (Sprint, Arrayjet Ltd.). Each probe was also printed in a four-level dose–response format at the indicated concentrations by applying 100 pl per spot of bacteria suspensions in PBS diluted with two volumes of 70.5% glycerol, 0.09% Triton X100 (final concentration 47% and 0.06% respectively). Spots were printed as triplicates. Manual and robotic arrays were scanned for SYTO-13 signals with a GenePix 200-AL scanner (Axon, Molecular Devices), using an excitation wavelength of 488 nm (blue laser) and blue emission detection. Fluorescence signals were quantified with the GenePix Pro 6.0 software (Molecular Devices).Control (glyco)proteins (the highly glycosylated fetuin/asialofetuin as positive control and non-glycosylated ribonuclease A as negative control, all of them from Sigma) were similarly printed at concentrations ranging from 0.03 to 1 mg ml−1. 1 μl ml−1 of Cy3 fluorophore (GE Healthcare) was added to the protein solutions to enable post-array monitoring of the spots,27 by scanning fluorescence signals upon excitation at 532 nm (green laser).
2.4. Microarray binding and inhibition assays
The arrayed slides were blocked for 1 h with 0.25% (v/v) Tween-20 in PBS, at 20 °C. Then, the microarrays were rinsed with PBS and overlaid with a solution containing the protein target of interest. Antibodies were tested at a working dilution of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000 in PBS containing 0.1% (v/v) Tween-20. After incubation for 1 h at 20 °C, microarrays were washed 4 times with PBS and overlaid for 1 h at 20 °C with the respective biotin-labelled secondary antibody (Sigma, working dilution 1
2000 in PBS containing 0.1% (v/v) Tween-20. After incubation for 1 h at 20 °C, microarrays were washed 4 times with PBS and overlaid for 1 h at 20 °C with the respective biotin-labelled secondary antibody (Sigma, working dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000). Slides were washed again and binding was monitored by incubating with AlexaFluor-647 (AF647)-labelled streptavidin (Invitrogen) at 1 μg ml−1 in overlay buffer, for 35 min at 20 °C. Finally, the slides were first washed thoroughly with PBS and then with water.
2000). Slides were washed again and binding was monitored by incubating with AlexaFluor-647 (AF647)-labelled streptavidin (Invitrogen) at 1 μg ml−1 in overlay buffer, for 35 min at 20 °C. Finally, the slides were first washed thoroughly with PBS and then with water.
Biotin-labelled lectins were tested at 20 μg ml−1 in either PBS containing 0.1% (v/v) Tween-20 (for RCA, AAL, SBA, LEA, SNL, HHL and WFL), 10 mM Tris–HCl at pH 7.8, 0.15 M NaCl (TBS) containing 1 mM CaCl2 and 0.5 mM MnCl2 (for ConA and PSA), or TBS containing 10 mM CaCl2 and 1 mM MgCl2 (for PNA). After incubation for 2 h at 20 °C, slides were washed 4 times with PBS and incubated with AF647-streptavidin, as described above.
C1q binding assays were carried out in a similar way by incubating the microarrays with 30 μg ml−1 biotin-labelled C1q in PBS containing 0.1% (v/v) Tween-20.
Following the binding assay, arrays were scanned for both AF647 (excitation at 635 nm, red laser) and SYTO-13 fluorescence signals. A significant reduction of the SYTO-13 signal of printed bacteria was observed when exposed to light during the overlay protocol. Therefore, all above-described incubation steps were strictly carried out in the dark.
3. Results and discussion
3.1. Preparation and validation of bacteria arrays
Bacteria microarrays were prepared by printing bacteria on nitrocellulose-coated glass slides. In order to spot any potential printing defects or reproducibility problems, a fluorescent dye was incorporated into the bacteria, also enabling evaluation of the immobilization efficiency and retention of the bacteria in the array. SYTO-13, which binds to DNA/RNA of Gram-negative and -positive bacteria and does not harm interactions taking place at the surface, was chosen to this aim. Moreover, its fluorophore properties, with λmax-excitation at 488/491 nm and λmax-emission at 509/514 nm, facilitate detection with a conventional microarray scanner, and the signal is clearly distinguishable from those of AlexaFluor-647 (AF647) and Cy5 (λEx-650 nm and λEm-668 nm), commonly used for read-out of protein binding.24 Labeling efficiency was similar for all K. pneumoniae strains (10% Standard deviation). A. baumannii yielded lower fluorescence intensity (80% compared to K. pneumoniae strains) and P. aeruginosa higher intensity (160% compared to K. pneumoniae).Serial dilutions (from 1 × 107 to 1 × 1010 bacteria per ml) of fluorescently labelled K. pneumoniae O1:K2 strain 52145, a clinically relevant serotype, and a panel of isogenic mutants lacking different surface epitopes of relevance to host–pathogen interactions, namely the capsule (CPS), LPS O-polysaccharide (OPS) and major outer membrane protein OmpA (see Fig. 1 and Table 1 for a description of the mutant strains), were initially printed on nitrocellulose-coated single-pad glass slides, using a manual glass-slide arraying system. Each spot was scanned for printing quality, accurate localization of bacteria spots and quantification of printed bacteria. Fluorescence signals were reliably detectable at concentrations of 1 × 108 bacteria per ml, and the concentration-dependent increment was linear up to 1 × 109. Therefore, this range was selected for subsequent analyses. Similar immobilization and retention studies were carried out in parallel with two other SYTO-13-labelled Gram-negative human pathogens, i.e. A. baumannii and P. aeruginosa (see Table 1), with comparable results. When stored at room temperature in a dry and dark place, SYTO-13 fluorescence signals of printed bacteria remained stable for at least 1 year. Furthermore, newly prepared and stored bacteria microarrays gave comparable results in binding assays (data not shown). Thus, bacteria microarrays generated using the described protocol can be stored and supplied as ready-to-use chips.
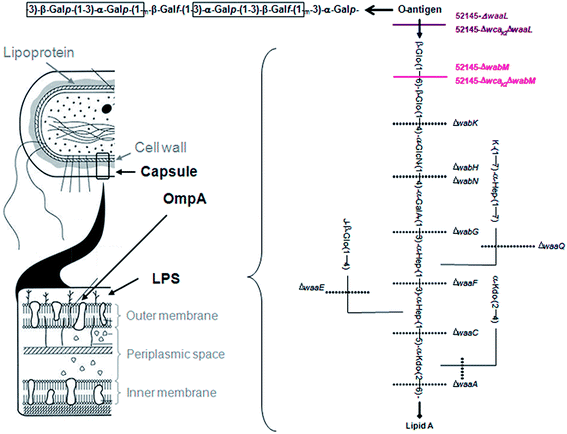 | ||
| Fig. 1 Description of K. pneumoniae surface structures. Kp52145 LPS structure is based on a published study.39,40 Lines denote the truncation level for the different core biosynthetic gene mutations. Residue K can be H or GalA; Hep, Heptose; Kdo, 3-deoxy-D-manno-oct-2-ulosonic acid. The repeating unit domains of the OPS are boxed. | ||
As first validation of the quality and applicability of the established microarrays, the extent of binding of two different anti-Klebsiella polyclonal antibodies, anti-Kp1 and anti-Kp2 (please, see Methods for description), was determined (Fig. 2). SYTO-13 fluorescence signals were intense and detectable after the binding assay (Fig. 2b), with a decrease in intensity of less than 10% with respect to fluorescence values measured before the assay, i.e. close to the error of the system. Furthermore, they did not interfere with monitoring of AF647, which yielded robust signals in strain- and dose-dependent manners (Fig. 2c–f). Of note, no binding to A. baumannii or P. aeruginosa was detected (Fig. 2c and d), supporting selectivity in the recognition of Klebsiella. Furthermore, the results confirmed the expected polyvalency of the antibodies against different Klebsiella antigenic determinants, as, with the single exception specified below, binding to the mutants bearing deletion of only one surface component was comparable or even stronger than to the wild type strain, the enhancement of binding plausibly resulting from increased exposure of recognized epitopes or from possible rearrangements in the bacterial surface upon mutation to compensate for the deletion. Excluding suppression of OmpA expression, all other combinations of double/triple mutations clearly resulted in decreased binding for both antibodies. Still, differences between anti-Kp1 and anti-Kp2 were observed. In particular, while decapsulation affected the binding of anti-Kp2 to various degrees, it had no significant impact on the binding of anti-Kp1, unless in combination with OPS deletion, likely reflecting predominance of LPS OPS-targeted antibodies in this polyclonal antibody. Indeed, as exception to the general rule mentioned above, anti-Kp1 showed almost negligible binding to the Δops single mutant. Overall, the developed K. pneumoniae microarrays proved to be effective for detecting binding of the two anti-Klebsiella antibodies tested and, what is more, served to unveil subtle specificity differences between them, most likely deriving from the different immunization protocols used in each case (see Methods).
 | ||
Fig. 2 Binding of anti-Klebsiella antibodies to K. pneumoniae manual arrays. K. pneumoniae wild type (strain 52145) and mutant strains (marked in panels a–d with an asterisk; please see Table 1 and Fig. 1 for description) labelled with SYTO-13 were printed onto nitrocellulose-coated glass slides using a hand arrayer and single-pad nitrocellulose-coated glass slides and overlaid with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000 working dilution of anti-Kp1 (c,e) or anti-Kp2 (d,f) SYTO-13-labelled A. baumannii and P. aeruginosa (see Table 1) were also included in the array as negative controls. Bacteria were printed as quadruplicates at four different concentrations (from 1 × 108 to 1 × 109 bacteria per ml), and SYTO-13 fluorescence was measured before (a) and after (b) the binding assay. Binding was detected with AF647-labelled streptavidin (c and d), as described in the Methods section. Panels e and f plot the intensity of the fluorescence signals shown in panels c and d, respectively, vs. the concentration of bacteria printed in the array: wild type K. pneumoniae ( 2000 working dilution of anti-Kp1 (c,e) or anti-Kp2 (d,f) SYTO-13-labelled A. baumannii and P. aeruginosa (see Table 1) were also included in the array as negative controls. Bacteria were printed as quadruplicates at four different concentrations (from 1 × 108 to 1 × 109 bacteria per ml), and SYTO-13 fluorescence was measured before (a) and after (b) the binding assay. Binding was detected with AF647-labelled streptavidin (c and d), as described in the Methods section. Panels e and f plot the intensity of the fluorescence signals shown in panels c and d, respectively, vs. the concentration of bacteria printed in the array: wild type K. pneumoniae ( ), and mutants Δops ( ), and mutants Δops ( ), Δcps ( ), Δcps ( ), ΔcpsΔops ( ), ΔcpsΔops ( ), ΔcpsΔopsΔwabM ( ), ΔcpsΔopsΔwabM ( ), ΔopsΔwabM ( ), ΔopsΔwabM ( ), ΔompA ( ), ΔompA ( ), ΔompACom ( ), ΔompACom ( ), ΔcpsΔompA ( ), ΔcpsΔompA ( ), ΔcpsΔompACom ( ), ΔcpsΔompACom ( ) and A. baumanii ( ) and A. baumanii ( ). ). | ||
3.2. Binding to K. pneumoniae arrays of the host counter-receptor C1q
C1q, the first subcomponent of the classical complement pathway, is capable of recognizing a number of structurally diverse ligands. It has been reported that Gram-negative bacteria are bound by C1q in an antibody-independent manner, with ensuing activation of the classical pathway. Lipopolysaccharides and outer-membrane proteins of the porin class are examples of molecules targeted by C1q.45–47 Furthermore, some bacteria, such as Escherichia coli or P. aeruginosa, express a specific receptor for C1q aimed to protect them from complement-mediated damage.48 Regarding K. pneumoniae, in vivo binding of C1q to porin OmpK36, present in most clinical isolates including the Kp52145 strain used in this work, has been reported, whereas binding to other components of the bacterial outer membrane, in particular the lipopolysaccharide lacking OPS could not be demonstrated.37,49,50 In order to check whether bacteria microarrays are also suitable to detect recognition of Klebsiella by C1q, as relevant example of pathogen–host counter–receptor interaction, we tested the binding behaviour of biotinylated C1q in the Klebsiella microarray set-up. As shown in Fig. 3, binding to the whole panel of wild-type and mutant K. pneumoniae strains was observed, the binding pattern also confirming the reported observation that strains possessing the LPS OPS bind less C1q37 (please compare wt vs. Δops in Fig. 3, and intermediate bacteria concentrations of Δcps vs. ΔcpsΔops), possibly due to a smaller accessibility to the porin. In addition, the results unveiled a previously unnoticed increase in C1q binding to strains lacking the capsular polysaccharide, also attributable to a higher exposure of the porin in these strains, with the single exception of the ΔompACom mutant. On the other hand, in striking contrast to the behaviour exhibited by anti-Klebsiella antibodies, binding of C1q to P. aeruginosa and A. baumannii was detected. As mentioned above, the occurrence in P. aeruginosa of a C1q-binding protein has been described. However, to the best of our knowledge, C1q binding to A. baumannii had not been reported so far. Thus, the results of the C1q binding assays demonstrate that bacteria microarrays are a powerful tool for detecting pathogen–host counter-receptor interactions. To examine the usefulness of the newly developed microarrays for exploring other bacterial surface features, e.g. presence of carbohydrate epitopes, we next performed binding assays with a panel of plant lectins with known sugar-binding specificities.3.3. Bacteria microarrays for exploring surface glycoepitopes
Pathogens' surfaces are coated with a variety of carbohydrate-rich structures, mainly CPS and LPS, which confer specific properties. For example, the virulence degree appears to correlate with the recognition, or lack of recognition, of specific glycoepitopes by endogenous lectins of the innate immune system.32 In particular, the K. pneumoniae strain Kp52145 (serotype O1:K2) used in this study presents a galactose-containing LPS OPS (Fig. 1) and a capsular polysaccharide built by a branched Glc/Man-based tetrasaccharide repeating unit.51 While other Klebsiella strains containing mannose-rich O-antigens and/or a repetitive D-mannose-α-2,3-D-mannose or L-rhamnose-α-2,3-L-rhamnose sequence in the capsular polysaccharide are less virulent due to pathogen clearance triggered by pattern recognition molecules of the immune system, strains not exhibiting these glycoepitopes, such as strain Kp52145, are the most commonly found in isolates from infected individuals. By testing the binding to the K. pneumoniae arrays of a panel of 10 plant lectins of known binding specificity (see Methods), we examined the applicability of the bacteria microarray set-up for exploring the presence of carbohydrate structures on the bacterial surface. To facilitate a high-throughput screening, bacteria were printed on 16-pad nitrocellulose-coated slides using a robotic arrayer. Of note, 47% glycerol was included in the printing buffer to increase the density of bacteria suspensions, what enables accurate printing of bacteria by reducing the sedimentation velocity. In addition, the presence of 0.06% Triton X100 (see Methods) diminishes bacteria aggregation, thereby avoiding that the microarraying needles get blocked. Printing quality, immobilization and retention were controlled as described above, and the extent of binding of the biotinylated lectins to the bacteria arrays was then examined.A strain-dependent and lectin-specific binding was observed (Table 2). First, no significant binding signals were detected for the GalNAc (WFL, SBA) and GlcNAc (LEA) specific lectins tested, although they clearly bound to control glycoproteins. Thus, no ligands with the appropriate structure or accessibility for detectable recognition are available to the tested lectins on the bacterial surface. In contrast, the Gal-specific lectins RCA and PNA showed binding in a strain-dependent manner (Table 2), as exemplarily illustrated for RCA in Fig. 4. In detail, a strong preference for non-capsulated vs. capsulated OPS-containing strains was evident, pointing to the Gal-containing O-antigen as the primary recognized epitope. Interestingly, deletion of OmpA also resulted in increased binding, what could tentatively be explained by a reorganization of the bacterial surface upon deletion of this major outer membrane protein, with increased LPS expression.
| Printed material | Overlay material and preferred sugar specificity | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| LEA (GlcNAc)2–4 | SBA α < βGalNAc | WFL GalNAc | RCA Gal | PNA Galβ3GalNAc | AAL Fucα6GlcNAc | SNL Neu5Acα6Gal | HHL αMan | PSA αMan, αGlc | ConA αMan, αGlc | ||
a Lectin binding was tested at 20 μg ml−1. Fluorescence intensity: ++++ > 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 > +++ > 20 000 > +++ > 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 > ++ > 10 000 > ++ > 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 > + > 5000 > (+) > 1000 > −. For definition of K. pneumoniae mutants, see Table 1. 000 > + > 5000 > (+) > 1000 > −. For definition of K. pneumoniae mutants, see Table 1. |
|||||||||||
| K. pneumoniae strain | WT | − | − | − | + | ++ | (+) | + | − | − | + |
| Mutants | |||||||||||
| Δops | − | + | − | (+) | + | + | ++ | (+) | (+) | ++ | |
| Δcps | − | (+) | − | +++ | +++ | + | + | − | − | + | |
| ΔcpsΔops | (+) | + | (+) | + | ++ | ++ | ++ | (+) | + | +++ | |
| ΔcpsΔopsΔwabM | − | + | (+) | (+) | ++ | ++ | ++ | (+) | (+) | +++ | |
| ΔopsΔwabM | − | (+) | − | − | + | + | + | − | − | + | |
| ΔompA | (+) | + | (+) | ++ | +++ | + | ++ | (+) | + | +++ | |
| ΔompACom | − | − | − | − | + | − | (+) | − | − | − | |
| ΔcpsΔompA | (+) | + | (+) | +++ | +++ | + | ++ | (+) | (+) | ++ | |
| ΔcpsΔompACom | − | + | (+) | +++ | +++ | + | ++ | (+) | (+) | ++ | |
| Protein | Asialofetuin | ++ | +++ | ++++ | ++++ | ++++ | ++++ | ++ | +++ | ++ | ++++ |
| Fetuin | ++ | +++ | + | ++ | ++ | ++++ | ++++ | ++ | +++ | +++ | |
| Ribonuclease A | (+) | − | − | − | (+) | − | (+) | − | (+) | (+) | |
 | ||
| Fig. 4 Binding of Ricinus communis agglutinin to K. pneumoniae robotic microarrays. K. pneumoniae wild type and mutant strains (see Table 1 and Fig. 1) labelled with SYTO-13 were printed onto 16-pad nitrocellulose-coated glass slides using a robotic arrayer. Bacteria were printed as triplicates at four different concentrations (from 1 × 108 to 1 × 109 bacteria per ml) and binding of biotin-labelled RCA (20 μg ml−1) was assayed in the absence (upper panel) and presence (lower panel) of 100 mM lactose. The extent of RCA binding was assessed by subsequent incubation with AF647-labelled streptavidin, as described in the Methods section. | ||
Second, intermediate signals were detected for fucose-specific AAL and sialic acid-binding SNL. The presence of fucose in the capsular polysaccharide of a clinical hepatic K. pneumoniae isolate had also been detected by AAL binding and confirmed by capillary high-performance liquid chromatography.52 However, our results showed a significant increase in AAL binding to the two ΔcpsΔops mutants, indicating that other glycoepitopes, different from the CPS and OPS and presumably more accessible in these strains, are recognized by this lectin. Bacterial glycoconjugates such as glycoproteins or glycolipids of the outer membrane are potential candidates. A similar conclusion can be drawn for SNL, for which binding was moderately increased in mutants lacking the OPS or the membrane protein OmpA. The presence of sialic acid on the surface of capsulated and non-capsulated K. pneumoniae strain 591, also of serotype O1:K2, had been proposed based on the results of specific topo-optical reactions.53 Still, SNL binding may not necessarily denote the presence of sialic acid, since cross-recognition of structurally similar pseudaminic acid moieties could be taking place. Such cross-reactivity has actually been reported for the sialic acid-specific lectin Limax flavus agglutinin when binding to Campylobacter jejuni flagellins.54
Finally, disparate results were obtained for the Man/Glc-specific lectins ConA, PSA and HHL. While the latter two lectins gave negligible binding signals, ConA exhibited a strain-dependent behaviour, giving stronger binding to strains lacking the O chain or the OmpA protein. These divergences probably result from differences in the fine carbohydrate-binding specificity of these lectins, which in the case of ConA and PSA have been found to be translated into a different binding behaviour towards glycoconjugates, cells and tissues.55–57 ConA has been reported to bind and agglutinate cells of the clinical hepatic K. pneumoniae NK-5 strain (K2 serotype) without affecting their growth.58 Although the tetrasaccharide repeating unit of the capsular K2 polysaccharide is composed of Glc and Man residues, the results here reported clearly indicate that the CPS is not the epitope recognized by ConA, as there is no preference for capsulated over non-capsulated strains. Apparently, the particular configuration of glycosidic linkages between Glc and Man residues in the tetrasaccharide does not fit the topological requirements of ConA's binding site. As discussed above for AAL and SNL, other glycoepitopes, different from the CPS and OPS, are recognized by ConA. In addition to possible bacterial glycoconjugates, the L-glycero-α-D-manno-heptoses present in the LPS inner core (see Fig. 1) appear as potential candidates. Indeed, binding of ConA to the seven-carbon monosaccharides L-glycero-α-D-manno- and D-glycero-α-D-manno-heptose has been demonstrated.59 Interestingly, the inner core manno-heptoses are also the primary site of interaction of surfactant protein D (SP-D), an endogenous lectin of the innate immune system, in binding to rough LPSs of Gram-negative bacteria.60 Furthermore, the presence of the O1-antigen in K. pneumoniae LPS has been shown to significantly reduce the binding efficiency of SP-D compared to rough LPS,61 fully in line with the behaviour exhibited by ConA.
Overall, the screening of lectin binding to the K. pneumoniae microarrays reveals the presence of different lectin ligands on the bacterial surface. In order to verify that the detected binding is carbohydrate-mediated, inhibition assays were carried out in parallel in the presence of specific haptens for the tested lectins (GlcNAc, GalNAc, Gal, Lac, Fuc, Man). As illustrated for RCA in Fig. 4, the binding signals were in all cases reduced down to background levels, thereby proving that lectin binding takes place via carbohydrate recognition and confirming the value of the microarray approach for exploring the presence of glycoepitopes on the bacterial surface. Additionally, these inhibition assays illustrate the usefulness of bacteria microarrays for the screening/evaluation of inhibitors of pathogen–host counter-receptor interactions.
4. Conclusions
We have developed a readily adaptable microarray technology for high-throughput screening of pathogen binding biomolecules. The results reveal applicability and binding selectivity, illustrating the potential of this approach for investigating the diversity of interactions occurring between pathogens and host cells during infection. Of general importance, it is expected that the described technology can be applied to the screening of any type of pathogen-binding biomolecule, beyond those tested here as model targets, i.e. antibodies, endogenous counter–receptors and lectins. The versatility of the microarray set-up facilitates the adjustment of assay conditions (e.g. amount of printed bacteria, thus surface density, and concentration of overlaying target) for detection of previously unnoticed binding activities. In addition, the natural presentation of surface molecules can be exploited for high-throughput screening/evaluation of compounds to identify inhibitors of pathogen–host counter-receptor interactions, thereby helping in the development of new classes of anti-infective drugs. Other applications of the newly developed bacteria arrays include delineation of antigenic determinants recognized by anti-bacteria antibodies, and exploration of carbohydrate structures present at the bacterial surface, the latter accomplishable by testing the binding of a panel of lectins with well-defined carbohydrate-binding specificities. Characterization of the glycosylation patterns may be particularly relevant when comparing different strains and mutant libraries or characterizing clinical isolates, aiding to the establishment of functional correlations. Furthermore, the above listed microarray-derived information may facilitate the development and optimization of on-chip devices for specific detection of pathogenic bacteria in the clinical field, as well as in the environmental or agri-food sectors.62,63Acknowledgements
This work was supported by the Marie Curie Initial Training Networks DYNANO (PITN-GA-2011-289033) and GLYCOPHARM (PITN-GA-2012-317297), the MICINN (Grant BFU2012-36825), the Biomedicine Program (SAF2009-07885) from Ministerio de Economía y Competitividad (Spain), Marie Curie Career Integration Grant U-KARE (PCIG13-GA-2013-618162), and the CIBER of Respiratory Diseases (CIBERES), an initiative from the Spanish Institute of Health Carlos III (ISCIII). The research leading to these results has also received funding from the European Union Seventh Framework Programme (FP7/2007–2013) under grant agreement N. 260600 (“GlycoHIT”). M.A.C-R is fellow of the CSIC, programme “Junta para la Ampliación de Estudios” (JaeDoc/098/2011), co-financed by the FSE.References
- R. P. Ekins, J. Pharm. Biomed. Anal., 1989, 7, 155 CrossRef CAS.
- A. C. Pease, D. Solas, E. J. Sullivan, M. T. Cronin, C. P. Holmes and S. P. Fodor, Proc. Natl. Acad. Sci. U. S. A., 1994, 91, 5022 CrossRef CAS.
- M. Schena, D. Shalon, R. W. Davis and P. O. Brown, Science, 1995, 270, 467 CAS.
- M. Morley, C. M. Molony, T. M. Weber, J. L. Devlin, K. G. Ewens, R. S. Spielman and V. G. Cheung, Nature, 2004, 430, 743 CrossRef CAS PubMed.
- E. Servoli, H. Feitsma, B. Kaptheijns, P. J. van der Zaag and R. Wimberger-Friedl, Lab Chip, 2012, 12, 4992 RSC.
- B. Uszczynska, T. Ratajczak, E. Frydrych, H. Maciejewski, M. Figlerowicz, W. T. Markiewicz and M. K. Chmielewski, Lab Chip, 2012, 12, 1151 RSC.
- H. Zhu, M. Bilgin, R. Bangham, D. Hall, A. Casamayor, P. Bertone, N. Lan, R. Jansen, S. Bidlingmaier, T. Houfek, T. Mitchell, P. Miller, R. A. Dean, M. Gerstein and M. Snyder, Science, 2001, 293, 2101 CrossRef CAS PubMed.
- C. S. Chen, E. Korobkova, H. Chen, J. Zhu, X. Jian, S. C. Tao, C. He and H. Zhu, Nat. Methods, 2008, 5, 69 CrossRef CAS PubMed.
- B. B. Haab, M. J. Dunham and P. O. Brown, Genome Biol., 2001, 2 CrossRef CAS , research0004.1–research0004.13.
- J. H. Rho and P. D. Lampe, J. Proteome Res., 2013, 12, 2311 CrossRef CAS PubMed.
- C. P. Paweletz, L. Charboneau, V. E. Bichsel, N. L. Simone, T. Chen, J. W. Gillespie, M. R. Emmert-Buck, M. J. Roth, E. F. Petricoin III and L. A. Liotta, Oncogene, 2001, 20, 1981 CrossRef CAS PubMed.
- K. M. Sheehan, V. S. Calvert, E. W. Kay, Y. Lu, D. Fishman, V. Espina, J. Aquino, R. Speer, R. Araujo, G. B. Mills, L. A. Liotta, E. F. Petricoin III and J. D. Wulfkuhle, Mol. Cell. Proteomics, 2005, 4, 346 CAS.
- S. C. Tao, Y. Li, J. Zhou, J. Qian, R. L. Schnaar, Y. Zhang, I. J. Goldstein, H. Zhu and J. P. Schneck, Glycobiology, 2008, 18, 761 CrossRef CAS PubMed.
- J. Hirabayashi, M. Yamada, A. Kuno and H. Tateno, Chem. Soc. Rev., 2013, 42, 4443 RSC.
- Y. Liu, T. Feizi, M. A. Campanero-Rhodes, R. A. Childs, Y. Zhang, B. Mulloy, P. G. Evans, H. M. Osborn, D. Otto, P. R. Crocker and W. Chai, Chem. Biol., 2007, 14, 847 CrossRef CAS PubMed.
- S. Park and I. Shin, Org. Lett., 2007, 9, 1675 CrossRef CAS PubMed.
- O. Blixt, K. Allin, O. Bohorov, X. Liu, H. Andersson-Sand, J. Hoffmann and N. Razi, Glycoconjugate J., 2008, 25, 59 CrossRef CAS PubMed.
- R. Karamanska, J. Clarke, O. Blixt, J. I. Macrae, J. Q. Zhang, P. R. Crocker, N. Laurent, A. Wright, S. L. Flitsch, D. A. Russell and R. A. Field, Glycoconjugate J., 2008, 25, 69 CrossRef CAS PubMed.
- S. N. Narla and X. L. Sun, Lab Chip, 2012, 12, 1656 RSC.
- O. E. Galanina, M. Mecklenburg, N. E. Nifantiev, G. V. Pazynina and N. V. Bovin, Lab Chip, 2003, 3, 260 RSC.
- C. C. Wang, Y. L. Huang, C. T. Ren, C. W. Lin, J. T. Hung, J. C. Yu, A. L. Yu, C. Y. Wu and C. H. Wong, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 11661 CrossRef CAS PubMed.
- H. H. Wandall, O. Blixt, M. A. Tarp, J. W. Pedersen, E. P. Bennett, U. Mandel, G. Ragupathi, P. O. Livingston, M. A. Hollingsworth, J. Taylor-Papadimitriou, J. Burchell and H. Clausen, Cancer Res., 2010, 70, 1306 CrossRef CAS PubMed.
- O. Blixt, S. Head, T. Mondala, C. Scanlan, M. E. Huflejt, R. Alvarez, M. C. Bryan, F. Fazio, D. Calarese, J. Stevens, N. Razi, D. J. Stevens, J. J. Skehel, I. D. van, D. R. Burton, I. A. Wilson, R. Cummings, N. Bovin, C. H. Wong and J. C. Paulson, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 17033 CrossRef CAS PubMed.
- A. S. Palma, T. Feizi, Y. Zhang, M. S. Stoll, A. M. Lawson, E. Diaz-Rodriguez, M. A. Campanero-Rhodes, J. Costa, S. Gordon, G. D. Brown and W. Chai, J. Biol. Chem., 2006, 281, 5771 CrossRef CAS PubMed.
- L. M. Graham, V. Gupta, G. Schafer, D. M. Reid, M. Kimberg, K. M. Dennehy, W. G. Hornsell, R. Guler, M. A. Campanero-Rhodes, A. S. Palma, T. Feizi, S. K. Kim, P. Sobieszczuk, J. A. Willment and G. D. Brown, J. Biol. Chem., 2012, 287, 25964 CrossRef CAS PubMed.
- B. S. Bochner, R. A. Alvarez, P. Mehta, N. V. Bovin, O. Blixt, J. R. White and R. L. Schnaar, J. Biol. Chem., 2005, 280, 4307 CrossRef CAS PubMed.
- M. A. Campanero-Rhodes, R. A. Childs, M. Kiso, S. Komba, C. Le Narvor, J. Warren, D. Otto, P. R. Crocker and T. Feizi, Biochem. Biophys. Res. Commun., 2006, 344, 1141 CrossRef CAS PubMed.
- P. Redelinghuys, A. Antonopoulos, Y. Liu, M. A. Campanero-Rhodes, E. McKenzie, S. M. Haslam, A. Dell, T. Feizi and P. R. Crocker, J. Biol. Chem., 2011, 286, 34522 CrossRef CAS PubMed.
- T. M. Blumenschein, N. Friedrich, R. A. Childs, S. Saouros, E. P. Carpenter, M. A. Campanero-Rhodes, P. Simpson, W. Chai, T. Koutroukides, M. J. Blackman, T. Feizi, D. Soldati-Favre and S. Matthews, EMBO J., 2007, 26, 2808 CrossRef CAS PubMed.
- M. A. Campanero-Rhodes, A. Smith, W. Chai, S. Sonnino, L. Mauri, R. A. Childs, Y. Zhang, H. Ewers, A. Helenius, A. Imberty and T. Feizi, J. Virol., 2007, 81, 12846 CrossRef CAS PubMed.
- R. A. Childs, A. S. Palma, S. Wharton, T. Matrosovich, Y. Liu, W. Chai, M. A. Campanero-Rhodes, Y. Zhang, M. Eickmann, M. Kiso, A. Hay, M. Matrosovich and T. Feizi, Nat. Biotechnol., 2009, 27, 797 CrossRef CAS PubMed.
- H. Sahly, Y. Keisari, E. Crouch, N. Sharon and I. Ofek, Infect. Immun., 2008, 76, 1322 CrossRef CAS PubMed.
- H. Kumar, T. Kawai and S. Akira, Int. Rev. Immunol., 2011, 30, 16 CrossRef CAS PubMed.
- R. C. Davicino, R. J. Elicabe, M. S. Di Genaro and G. A. Rabinovich, Int. Immunopharmacol., 2011, 11, 1457 CrossRef CAS PubMed.
- Y. A. Knirel, H.-J. Gabius, O. Blixt, E. M. Rapoport, N. R. Khasbiullina, N. V. Shilova and N. V. Bovin, Glycoconjugate J., 2014, 31, 7–12 CrossRef CAS PubMed.
- Y. Keynan and E. Rubinstein, Int. J. Antimicrob. Agents, 2007, 30, 385 CrossRef CAS PubMed.
- S. Alberti, G. Marques, S. Camprubi, S. Merino, J. M. Tomas, F. Vivanco and V. J. Benedi, Infect. Immun., 1993, 61, 852 CAS.
- R. I. Amann, B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux and D. A. Stahl, Appl. Environ. Microbiol., 1990, 56, 1919 CAS.
- X. Nassif, J. M. Fournier, J. Arondel and P. J. Sansonetti, Infect. Immun., 1989, 57, 546 CAS.
- G. Cortes, N. Borrell, B. De Astorza, C. Gomez, J. Sauleda and S. Alberti, Infect. Immun., 2002, 70, 2583 CrossRef CAS.
- E. Llobet, J. M. Tomas and J. A. Bengoechea, Microbiology, 2008, 154, 3877 CrossRef CAS PubMed.
- C. March, V. Cano, D. Moranta, E. Llobet, C. Perez-Gutierrez, J. M. Tomas, T. Suarez, J. Garmendia and J. A. Bengoechea, PLoS One, 2013, 8, e56847 CAS.
- M. Regue, L. Izquierdo, S. Fresno, N. Pique, M. M. Corsaro, T. Naldi, C. De Castro, D. Waidelich, S. Merino and J. M. Tomas, J. Bacteriol., 2005, 187, 4198 CrossRef CAS PubMed.
- E. Llobet, C. March, P. Gimenez and J. A. Bengoechea, Antimicrob. Agents Chemother., 2009, 53, 298 CrossRef CAS PubMed.
- F. Galdiero, M. A. Tufano, L. Sommese, A. Folgore and F. Tedesco, Infect. Immun., 1984, 46, 559 CAS.
- B. Aubert, S. Chesne, G. J. Arlaud and M. G. Colomb, Biochem. J., 1985, 232, 513 CAS.
- F. Clas, B. Euteneuer, F. Stemmer and M. Loos, Behring Inst. Mitt., 1989, 236 CAS.
- R. H. van den Berg, M. C. Faber-Krol, J. A. van de Klundert, L. A. van Es and M. R. Daha, J. Immunol., 1996, 156, 4466 CAS.
- S. Hernandez-Alles, S. Alberti, D. Alvarez, A. Domenech-Sanchez, L. Martinez-Martinez, J. Gil, J. M. Tomas and V. J. Benedi, Microbiology, 1999, 145(3), 673 CrossRef CAS PubMed.
- S. Alberti, G. Marques, S. Hernandez-Alles, X. Rubires, J. M. Tomas, F. Vivanco and V. J. Benedi, Infect. Immun., 1996, 64, 4719 CAS.
- M. M. Corsaro, C. De Castro, T. Naldi, M. Parrilli, J. M. Tomas and M. Regue, Carbohydr. Res., 2005, 340, 2212 CrossRef CAS PubMed.
- J. H. Wu, A. M. Wu, C. G. Tsai, X. Y. Chang, S. F. Tsai and T. S. Wu, Exp. Biol. Med., 2008, 233, 64 CrossRef CAS PubMed.
- Z. Tigyi, W. Gahrs, L. Emody and J. Makovitzky, Acta Histochem., 2009, 111, 300 CrossRef CAS PubMed.
- P. Thibault, S. M. Logan, J. F. Kelly, J. R. Brisson, C. P. Ewing, T. J. Trust and P. Guerry, J. Biol. Chem., 2001, 276, 34862 CrossRef CAS PubMed.
- K. Kornfeld, M. L. Reitman and R. Kornfeld, J. Biol. Chem., 1981, 256, 6633 CAS.
- I. De Dios, M. Manso, V. Leon and A. Lopez-Borrasca, Biochem. Med. Metab. Biol., 1986, 35, 12 CrossRef CAS.
- C. M. Griffith and E. J. Sanders, Anat. Rec., 1991, 231, 238 CrossRef CAS PubMed.
- C. F. Kuo, Y. H. Wang, H. Y. Lei, C. H. Wang and N. Tsao, Antimicrob. Agents Chemother., 2007, 51, 3122 CrossRef CAS PubMed.
- F. A. Jaipuri, B. Y. Collet and N. L. Pohl, Angew. Chem., Int. Ed. Engl., 2008, 47, 1707 CrossRef CAS PubMed.
- H. Wang, J. Head, P. Kosma, H. Brade, S. Muller-Loennies, S. Sheikh, B. McDonald, K. Smith, T. Cafarella, B. Seaton and E. Crouch, Biochemistry, 2008, 47, 710 CrossRef CAS PubMed.
- H. Sahly, I. Ofek, R. Podschun, H. Brade, Y. He, U. Ullmann and E. Crouch, J. Immunol., 2002, 169, 3267 CrossRef CAS.
- S. Bouguelia, Y. Roupioz, S. Slimani, L. Mondani, M. G. Casabona, C. Durmort, T. Vernet, R. Calemczuk and T. Livache, Lab Chip, 2013, 13, 4024 RSC.
- S. David, C. Polonschii, M. Gheorghiu, D. Bratu, A. Dobre and E. Gheorghiu, Lab Chip, 2013, 13, 3192 RSC.
| This journal is © The Royal Society of Chemistry 2015 |

