Investigation of dendrimers functionalized with eosin as macrophotoinitiators for polymerization-based signal amplification reactions†
Abstract
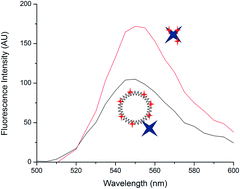
Polymerization-based signal amplification, a technique developed for use in rapid diagnostic tests, hinges on the ability to localize initiators as a function of interfacial binding events. A number of strategies are available for increasing this local concentration, including increasing the capture probe density, using higher affinity binding molecules, or, as presented here, directly conjugating additional initiators to the detection reagent through the use of functionalized polymers. We have previously considered poly(acrylic acid-co-acrylamide) backbones for this purpose; with eosin as the photoinitiator, these efforts were hindered by solubility limitations. Here, we use a poly(amidoamine) dendrimer as a scaffold to produce conjugates with enhanced solubility. Through an investigation into the surface binding and solution-phase properties of these conjugates, we show that quenching effects impact the efficacy of these conjugates as macrophotoinitiators.

 Please wait while we load your content...
Please wait while we load your content...