High-efficient inhibition of recognition in allorejection via a pMyD88/liposomes complex†
Abstract
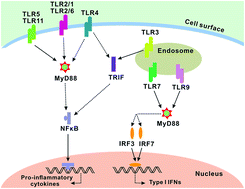
Data are emerging that the recognition of foreign antigens by Toll/like receptors (TLRs) was predominant in skin graft rejection. More interestingly, most of the TLRs recruit myeloid differentiation factor 88 (MyD88) as an adaptor during signaling transduction. Design of an efficient shRNA/vector complex to suppress the expression of MyD88 protein seems to be a potential approach in preventing allorejection of skin. In this study, we prepared a complex composed of a MyD88-shRNA plasmid (pMyD88) and a cationic polymeric vector. The results showed that the pMyD88 and vectors with an optimal mass ratio (1 : 2) was selected by agarose gel electrophoresis (AGE) experiments; moreover, the cationic liposome vectors increased the transfection efficiency of naked pMyD88, and the gene transcription of MyD88 mRNA and expression of MyD88 protein was significantly suppressed by the pMyD88/liposomes complex. Notably, the lipopolysaccharide (LPS) stimulation test showed that the recognition of foreign antigen in DCs, which are treated with the pMyD88/liposomes complex, was significantly inhibited. This attempt promises to solve skin allorejection problems in the future.

 Please wait while we load your content...
Please wait while we load your content...