A non-immunosuppressive FK506 analogue with neuroregenerative activity produced from a genetically engineered Streptomyces strain†
Pramod B. Shinde‡§
a,
Yeon Hee Ban‡a,
Jae-yeon Hwanga,
Yumi Chob,
Yi-Ahn Chenb,
Eunji Cheongb,
Sang-Jip Nama,
Ho Jeong Kwonb and
Yeo Joon Yoon*a
aDepartment of Chemistry and Nano Science, Ewha Global Top 5 Program, Ewha Womans University, Seoul 120-750, Republic of Korea. E-mail: joonyoon@ewha.ac.kr; Fax: +82-2-3277-3419; Tel: +82-2-3277-4446
bDepartment of Biotechnology, The Translational Research Center for Protein Function Control, Yonsei University, Seoul 120-746, Republic of Korea
First published on 18th December 2014
Abstract
FK506 exhibits neuroprotective and neuroregenerative activities in addition to its clinically important immunosuppressant properties. The macrolide ring of FK506 is biosynthesized by a hybrid polyketide synthase/nonribosomal peptide synthetase system and is further modified via C-9 oxidation by FkbD and 31-O-methylation by FkbM. A new FK506 analogue, 9-deoxo-prolyl-FK506 (1), was isolated from the fkbD deletion mutant of Streptomyces sp. KCTC11604BP, and its biological activities were evaluated. The in vitro immunosuppressive activity was significantly reduced, but in vitro neurite outgrowth activity similar to FK506 was maintained. These results demonstrate the potential of pathway engineering for the modification of structurally complex natural products, such as FK506, to create improved biological agents.
Introduction
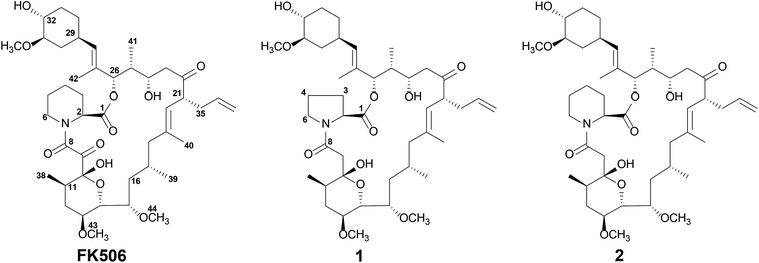
FK506 (also known as tacrolimus or fujimycin) is a 23-member macrolide originally isolated from Streptomyces tsukubaensis and is clinically used as an immunosuppressant drug for the prevention of allograft rejections.1–3 FK506 (Fig. 1) and similar drugs have been reported to alter several biochemical processes by interacting with cytoplasmic immunophilin proteins commonly referred to as FK506-binding proteins (FKBPs).4 FK506 blocks the action of calcineurin, a key enzyme in T-cell receptor signalling, preventing T-lymphocyte activation. According to a detailed study on the mechanism of FK506 immunosuppressive action, its chemical structure can be divided into two parts: an effector region which binds to calcineurin and a binding region which forms a complex with FKBPs.5–7 The FKBP-binding region containing a pipecolate moiety, a tricarbonyl group, and a cyclohexane ring is responsible for forming a binary complex with the FKBP12 protein. This leaves the remaining half (effector region) free to bind to calcineurin leading to a tripartite complex.6 In order to exert the immunosuppressive action, it is important that FK506 first binds to the FKBP12 protein. Once this binary complex is formed, it can then interact with calcineurin.5,6 In addition to its immunosuppressive action, FK506 has been reported to exhibit antifungal,8 anti-inflammatory,9 neuroprotective, and neuroregenerative activities.10,11 Attempting to reduce the immunosuppressive activity while at the same time maintaining these other impressive activities will allow the full therapeutic potential of FK506 to be harnessed.A hybrid polyketide synthase/nonribosomal peptide synthetase (PKS/NRPS) system is responsible for the biosynthesis of FK506. The chorismate-derived starter unit 4,5-dihydroxycyclohex-1-enecarboxylic acid (DHCHC)12 is extended in 10 condensation steps utilizing two malonyl-CoA, two methoxymalonyl-acyl carrier proteins (ACP), five methylmalonyl-CoA, and an allylmalonyl-CoA.13 Pipecolate derived from lysine by the action of FkbL is condensed with the linear polyketide chain by the NRPS FkbP and cyclized to yield a macrolide ring.14,15 This ring is further modified by post-PKS modifications such as O-methylation at the C-31 position catalyzed by FkbM and oxidation at the C-9 position by FkbD.16,17 Recently the detailed post-PKS modification route involving two independent parallel pathways was established via the characterization of all FK506 biosynthetic intermediates.18
During our efforts characterizing the functional role of enzymes FkbM and FkbD and identifying the biosynthetic intermediates present in the two parallel pathways for the post-PKS modification of FK506 biosynthesis, we encountered an additional peak in the HPLC-ESI-MS analysis of the organic extract of a Streptomyces sp. KCTC11604BP mutant strain in which the fkbD gene was inactivated by in-frame deletion (ΔfkbDin-frame). However, at that time we were only focusing on the identification of known biosynthetic intermediates of the post-PKS modification steps of FK506, so we did not try to isolate and identify this compound. Hence, in the present study, this compound was purified and identified as 9-deoxo-prolyl-FK506 (5-nor-9-deoxo-FK506) (1) on the basis of 1D- and 2D-NMR, HPLC-ESI-MS/MS, and HR-ESI-MS data. Additionally, the immunosuppressive and neurite outgrowth activities of 1 were evaluated.
Results and discussion
The HPLC-ESI-MS/MS analysis of the organic extract of the ΔfkbDin-frame strain18 showed the presence of an ammonium adduct ion peak eluted at 13 min with m/z 793.1 along with a peak eluted at 28 min resulting from an ammonium adduct with m/z 807.1 which was previously identified as 9-deoxo-FK506 (2) (Fig. 2).The peak eluted at 13 min with m/z 793.1 was predicted to be 1 based on the fragment ions at 776.1, 758.1, 740.1, and 547.9 (Fig. S1†). The fragmentation pattern of 1 was similar to that of 2 with a difference of 14 Da between each of the fragment ions. The characteristic C-1–C-24 fragment ion appeared at m/z 561.9 in 2 whereas the same for 1 was denoted by an ion peak with m/z 547.9. This suggested that 1 had one less methylene group in the characteristic C-1–C-24 fragment than 2 and other 9-deoxo-FK506 analogues.18 Due to the relaxed substrate specificity of FkbP14 an FK506 analogue can be generated containing a proline19 rather than the common pipecoline moiety. Thus, 1 was assumed to contain a proline moiety. Although 1 was produced as a major compound by the ΔfkbDin-frame strain,18 we did not pay much attention to the peak corresponding to compound 1 in our previous study because our major interest focussed on identifying the known biosynthetic intermediates of FK506.
In order to confirm the structure of 1 by spectroscopic analysis (Table 1), the crude extract from a large cultivation of ΔfkbDin-frame strain (4 l) was subjected to repeated chromatographic separations to afford 7.4 mg of pure compound. HR-ESI-MS analysis of 1 yielded an [M + H]+ ion at m/z 776.4940, consistent with a molecular formula of C43H69NO11 (calcd m/z 776.4949). The 1H NMR spectrum (Fig. S2†) showed the characteristic signals of the FK506 skeleton:18 three doublets corresponding to methyl groups at δH 0.95/H3-38, 0.90/H3-41, and 0.75/H3-39; two methyl singlets at δH 1.67/H3-40 and 1.66/H3-42; three methoxy singlets at δH 3.40/H3-45, 3.37/H3-43, and 3.36/H3-44; and a multiplet of an olefinic proton at δH 5.70/H-36 (Table 1). Forty-two carbon signals from the 13C NMR spectrum (Fig. S3†) and one carbonyl carbon from the HMBC spectrum were identified accounting for all 43 expected carbons. However, careful comparison of the NMR data of 1 to those of 2 indicated that the signals in 1 corresponded to the pipecolate moiety (C-2–C-6) of 2 shifted. Further analysis of the obtained 1H, 13C, and HSQC spectra (Fig. S2–S4,† respectively) of 1 revealed only four carbon signals for this region at δC 58.9/C-2 (δH 4.35/H-2), 47.4/C-6 (δH 3.63/H-6a, 3.54/H-6b), 29.2/C-3 (δH 2.19/H-3a, 1.98/H-3b), and 24.7/C-4 (δH 1.96/H2-4) confirming the presence of the proline moiety in 1 rather than pipecolate. The proline moiety of 1 was further confirmed through COSY and HMBC analysis (Fig. 3). The COSY crosspeaks in the range δH 4.35 to δH 3.54 provided the H-2/H-3/H-4/H-6 connectivity (Fig. 3 and S5†). Furthermore, the key HMBC correlations (Fig. 3 and S6†) between the proton signals at δH 4.35/H-2, 2.19/H-3a, and 1.98/H-3b and the carbonyl ester carbon at δC 169.9/C-1, as well as protons δH 3.63/H-6a and 3.54/H-6b and the carbon signals at δC 58.9/C-2 and 29.2/C-3 were observed. The HSQC spectrum (Fig. S4†) showed that the proton signals at δH 2.64 (d, J = 15 Hz) and 2.56 (d, J = 15 Hz) were correlated with the carbon signal at δC 39.2. These proton signals at δH 2.64 and 2.56 showed HMBC correlations to both C-8 (δC 171.8) and C-10 (δC 98.6) (Fig. 3 and S6†) indicating 1 is a 9-deoxo-FK506 analogue similar to that of 2. The COSY spectrum (Fig. S5†) of 1 showed the remaining 4 spin systems (Fig. 3), which were connected on the basis of HMBC correlations (Fig. 3 and S6†). The positions of the methyl and methoxy groups were also assigned on the basis of the respective HMBC correlations (Fig. 3). Therefore, 1 was assigned as 9-deoxo-prolyl-FK506, and this is the first 9-deoxo-FK506 analogue containing a proline moiety instead of the common pipecolate ring. The stereochemistry of 1 was assumed to be the same as that of the parent compound FK506. In the wild-type Streptomyces strain, the prolyl analogue of FK506 was a minor metabolite compared to FK506.19 However, 1 was produced in a larger quantity than 2 in the ΔfkbDin-frame strain, suggesting that, for reasons unknown, proline can be incorporated more efficiently than pipecolate in the absence of the FkbD-catalyzed C9-oxidation.
| Position | δC | δH, m (J in Hz) | 1H–1H COSY | HMBC |
|---|---|---|---|---|
| a Signal was assigned from HMBC spectrum.b Signals with similar values could be interchanged. | ||||
| 1 | 169.9 | |||
| 2 | 58.9 | 4.35, dd (8.0, 3.0) | H-3a, H-3b | C-1, C-3, C-4, C-6 |
| 3 | 29.2 | 2.19, m | H-2, H-4 | C-1, C-2, C-4, C-6 |
| 1.98, m | H-2 | C-1, C-2, C-6 | ||
| 4 | 24.7 | 1.96, m | H-6a, H-6b, H-3a | C-3, C-6 |
| 5 | ||||
| 6 | 47.4 | 3.63, m | H-4 | C-2, C-3, C-4 |
| 3.54, m | H-4 | C-2, C-3, C-4 | ||
| 7 | ||||
| 8 | 171.8 | |||
| 9 | 39.2 | 2.64, d (15.0) | H-9b | C-8, C-10 |
| 2.56, d (15.0) | H-9a | C-8, C-10 | ||
| 10 | 98.6 | |||
| 11 | 38.6 | 1.61, m | H-12a, H-38 | C-10 |
| 12 | 32.7 | 1.99, m | H-11, H-13 | C-10, C-13, C-14, C-38 |
| 1.55, m | H-13 | C-10, C-13, C-38 | ||
| 13 | 74.6 | 3.40, (overlapped) | H-12a, H-12b, H-14 | C-14, C-43 |
| 14 | 71.0 | 3.84, dd (10.0, 2.5) | H-13, H-15 | C-10, C-12, C-13 |
| 15 | 77.1 | 3.53, m | H-14, H-16a, H-16b | |
| 16 | 36.4 | 1.46, m | H-15 | C-17, C-19 |
| 1.35, m | H-15, H-17 | C-17, C-19 | ||
| 17 | 25.6 | 1.61, (overlapped) | H-39, H-16b, H-18a | |
| 18 | 49.0 | 2.33, m | H-17 | C-17, C-19, C-20, C-39 |
| 1.69, m | ||||
| 19 | 141.1 | |||
| 20 | 121.9 | 5.01, overlapped | H-21 | C-18, C-21, C-22, C-40 |
| 21 | 53.4 | 3.36, overlapped | H-20, H-35a, H-35b | C-22, C-20, C-35, C-36 |
| 22 | 214.0a | |||
| 23 | 44.0 | 2.69, dd (17.0, 3.0) | H-23b | C-22, C-24 |
| 2.34, dd (17.0, 7.0) | H-23a, H-24 | C-22, C-24, C-25 | ||
| 24 | 69.1 | 4.02, dd (7.0, 3.0) | H-23b, H-25 | C-22, C-26, C-41 |
| 25 | 41.2 | 1.81, m (3.0) | H-24, H-26, H-41 | |
| 26 | 78.0 | 5.17, d (3.0) | H-25 | C-1, C-24, C-25, C-27, C-28, C-41, C-42 |
| 27 | 132.4 | |||
| 28 | 129.7 | 4.98, (overlapped) | H-29 | C-26, C-27, C-29, C-30, C-34, C-42 |
| 29 | 35.0 | 2.27, m | H-28, H-30a, H-30b, H-34a, H-34b | C-30 |
| 30 | 34.9 | 2.05, m | H-29, H-30b, H-31 | C-31 |
| 0.97, (overlapped) | H-29, H-30a, H-31 | C-31 | ||
| 31 | 84.4 | 2.99, ddd (8.5, 4.5, 2.5) | H-30a, H-30b, H-32 | C-32, C-45 |
| 32 | 73.7 | 3.40, (overlapped) | H-31, H-33a, H-33b | |
| 33 | 31.4 | 1.98, m | H-32, H-33b, H-34a, H-34b | |
| 1.35, m | H-32, H-33a, H-34a, H-34b | |||
| 34 | 30.8 | 1.61, m | H-29, H-33a, H-33b, H-34b | |
| 1.04, m | H-29, H-33a, H-33b, H-34a | |||
| 35 | 35.7 | 2.45, m (7.0) | H-36, H-21 | C-20, C-21, C-22, C-36, C-37 |
| 2.25, m | H-36, H-21 | C-20, C-21, C-22, C-36, C-37 | ||
| 36 | 135.6 | 5.70, ddt (17.0, 10.0, 7.0) | H-35a, H-35b, H-37 | C-21, C-35 |
| 37 | 116.7 | 5.00, br s | H-36 | C-35, C-36 |
| 38 | 17.1 | 0.95, d (6.5) | H-11 | C-10, C-11, C-12 |
| 39 | 19.0 | 0.75, d (6.5) | H-17 | C-16, C-17, C-18 |
| 40 | 15.7 | 1.67b, s | C-19, C-20 | |
| 41 | 10.0 | 0.90, d (6.5) | H-25 | C-24, C-25, C-26 |
| 42 | 14.4 | 1.66b, s | C-27, C-28 | |
| 43 | 56.3 | 3.37b, s | C-13 | |
| 44 | 57.8 | 3.36b, s | C-15 | |
| 45 | 56.7 | 3.40, s | C-31 | |
| 10-OH | 6.76, s | C-8, C-9 | ||
The evaluation of IL-2 secretions from T-lymphocytes activated by CD3/CD28 antibodies treated with 1 and FK506 indicated a loss of immunosuppressive activity for 1 (Fig. 4A). This reduced immunosuppression by 1 corroborates with previous reports wherein prolyl-FK506 and 9-deoxo-FK506 also displayed decreased immunosuppressive activity when compared to the parent molecule, FK506.5,19 Similarly, the relative nerve regeneration activity of 1 compared to FK506 was assessed using rat pheochromocytoma (PC12) cells (Fig. 4B and 5). Compound 1 exhibited only slightly reduced (∼10%) in vitro neurite outgrowth activity compared to FK506. Because the immunosuppression and neuroregeneration actions of FK506 arise via different mechanisms (complexes formed with FKBP12 and FKBP52, respectively),10,11 the loss of immunosuppressive activity due to its reduced ability to form a binary complex with FKBP12 should not prevent the binding to FKBP52. Thus, neuroregenerative activity can be maintained while immunosuppression is lost. A detailed study needs to be undertaken, however, to identify the underlying mechanistic differences between the immunosuppressive and neurite outgrowth activities of 1 and FK506. Because it has been known that weak cytotoxic compounds also induce neurite-outgrowth with NGF,20 the cytotoxicity of 1 was tested on primary cortical neurons using MTT and cell count assays. As a result, compound 1 showed no cytotoxicity up to the concentration of 40 μg ml−1 (∼50 μM) (Fig. S7†) suggesting that its neurite-outgrowth activity is not related to its cytotoxicity. This significant reduction of immunosuppressive activity, while maintaining neurite outgrowth activity, suggests the potential of 1 for the use in both the treatment of neurodegenerative diseases and as a new molecular probe to investigate the detailed mechanism underlying neuroregeneration by neuroimmunophilin ligands.
Experimental
General experimental procedures
The optical rotation was measured with a Jasco P-1010 polarimeter using a 0.1 dm path length cell. The UV spectrum was recorded with a Scinco S-3100 spectrophotometer, and the IR spectrum was obtained with Varian FTS-800 FTIR spectrophotometer. NMR spectra were acquired using a Varian INOVA 500 spectrometer operating at 500 MHz for 1H and 125 MHz for 13C nuclei. Chemical shifts were given in ppm using tetramethylsilane (TMS) as an internal reference. All NMR data processing was done using Mnova software (Mestrelab Research S.L.). Samples for NMR analysis were prepared by dissolving the pure compound in 250 μl of CDCl3 (Sigma), and placing the solutions in 5 mm Shigemi advanced NMR microtubes (Sigma) matched to the solvent. HPLC-ESI-MS/MS spectra using an ACQUITY UPLC BEH C18 column (50 × 2.1 mm, 1.7 μm; Waters) were recorded on a Waters/Micromass Quattro micro MS interface consisting of a Waters 2695 separation module connected directly to a Micromass Quattro micro MS. Tracing was done by MS/MS operated in multiple reactions monitoring mode choosing mass pairs specific for the selected analytes to detect the transition from parent ion as an ammonium adduct to product ion: 793 > 547 for 1 and 807 > 561 for 2. The HR-ESI-MS data were obtained using Waters SYNAPT G2 mass spectrometer coupled with UPLC. HPLC purification was performed using semipreparative Watchers 120 ODS-BP (250 × 10 mm, 5 μm) column on an Acme 9000 HPLC system (YL Instrument Co. Ltd., Korea) consisting of a SP930D gradient pump coupled with a UV730D UV detector set to 205 nm and a CTS30 column oven set to 50 °C. HPLC grade solvents used in the experiments were purchased from J. T. Baker.Construction of mutant and culture conditions
The inactivation of the fkbD gene in the FK506-producing strain Streptomyces sp. KCTC11604BP by in-frame deletion was previously described.18 Spores of Streptomyces sp. KCTC11604BP and its fkbD deletion mutant ΔfkbDin-frame were generated on ISP4 agar plates, and a seed culture was prepared in R2YE21 broth. Vegetative cells (50 mg) grown in the seed culture were inoculated into 250 ml baffled flasks containing 50 ml of R2YE medium and cultivated on an orbital shaker (set at 180 rpm) for 6 days at 28 °C.Extraction and isolation
The culture broth (4 l) of ΔfkbDin-frame strain was centrifuged and the supernatant was subjected to solvent–solvent partition with ethyl acetate (twice). The ethyl acetate-soluble layers were combined and evaporated under reduced pressure to yield a dark reddish extract. This dark extract was subjected to fractionation by preparative reversed-phase HPLC using 60% aqueous methanol as the mobile phase with a flow rate of 2 ml min−1. Resulting fractions were monitored for the presence of target compound 1 using HPLC-ESI-MS analysis. The fraction containing 1 was further separated by semipreparative reversed-phase HPLC employing 50% aqueous acetonitrile as the mobile phase with a flow rate of 2 ml min−1 to obtain two subfractions. Subfraction 2 containing 1 was purified using the same column and the same HPLC conditions to afford a pure amorphous white solid (7.4 mg, tR 90 min).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 227 nm (2.0); IR (film) νmax 3450, 2960, 1750, 1640, 1170, 1050 cm−1; 1H and 13C NMR, see Table 1; (+)-ESI-MS m/z 793.1 [M + NH4]+; (+)-MS/MS: m/z 776.1, 758.1, 740.1, 547.9; (+)-HR-ESI-MS m/z 776.4940 [M + H]+ (calcd for C43H70NO11, 776.4949).
ε) 227 nm (2.0); IR (film) νmax 3450, 2960, 1750, 1640, 1170, 1050 cm−1; 1H and 13C NMR, see Table 1; (+)-ESI-MS m/z 793.1 [M + NH4]+; (+)-MS/MS: m/z 776.1, 758.1, 740.1, 547.9; (+)-HR-ESI-MS m/z 776.4940 [M + H]+ (calcd for C43H70NO11, 776.4949).In vitro T-cell activation assay
The relative immunosuppressive property of 1, compared to authentic FK506, was determined using T-lymphocytes as described previously.13 Levels of interleukin-2 secretion were quantified after treatment of CD3/CD28-activated human T-cells with the above compounds (0.1 nM concentration) for 16 to 20 h (see Fig. 4A).In vitro neurite outgrowth assay
The relative nerve regeneration activity of 1, compared to authentic FK506, was determined using rat pheochromocytoma cells (PC12) as described in the literature.13 The PC12 cells were treated with nerve growth factor (NGF; KOMA Biotech; 10 ng ml−1) in the presence or absence of 10 nM FK506 and 1. The neurite lengths were measured on photographic prints as previously described (Fig. 4B and 5).22Cytotoxicity assay on primary cortical neurons
Cortical tissue from neuronal mice was dissected and trypsinized to obtain isolated cells. For neuronal culture, cells were seeded at the density of 7 × 105 cells per ml on 96-well plate and maintained in Neurobasal A media (Gibco) supplemented with 1% L-glutamine (Invitrogen), 1% B27 supplement (Gibco), and 100 U ml−1 penicillin–streptomycin. Media were renewed every 2–3 days until use.The cytotoxicity of compound 1 at 1, 10, and 40 μg ml−1 concentration was tested in cultured neurons. After 48 h of treatment, number of trypan blue-stained living cells in each well was counted with haemocytometer. MTT assay (ATCC) was also performed to determine the cytotoxicity.
Conclusions
In conclusion, we have isolated and identified from the fkbD deletion mutant of Streptomyces sp. KCTC11604BP a new FK506 analogue, 9-deoxo-prolyl-FK506 (1), lacking the C9-keto group and containing a proline moiety instead of the common pipecoline ring. Compound 1 displayed reduced in vitro immunosuppressive activity but similar neuroregenerative activity in comparison to FK506. This research has successfully demonstrated the potential of biosynthetic engineering approaches, such as the deletion of a biosynthetic gene from a gene cluster, to generate new bioactive molecules where the structural complexity of the natural product discourages the use of chemical synthesis. Overall, this study demonstrates that neuroregenerative drugs can be developed from the FK506 macrolide class of natural products without the side effects attributed to immunosuppression.Acknowledgements
We thank Dr Kris Rathwell for critically reading this manuscript. This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (MISP) (2009-0083522, 2010-0017984, and 2013R1A2A1A01014230) and the Intelligent Synthetic Biology Center of the Global Frontier Project funded by MISP (2012054879). We would like to thank Dr Young Hye Kim from the Division of Mass Spectrometry Research, KBSI, Ochang for recording HR-ESI-MS data.Notes and references
- H. Kino, H. Hatanaka, M. Hashimoto, M. Nishiyama, T. Goto, M. Okuhara, M. Kohsaka, H. Aoki and H. Imanaka, J. Antibiot., 1987, 40, 1249 CrossRef.
- H. Kino, H. Hatanaka, S. Miyata, N. Inamura, M. Nishiyama, T. Yajima, T. Goto, M. Okuhara, M. Kohsaka, H. Aoki and T. Ochiai, J. Antibiot., 1987, 40, 1256 CrossRef.
- J. J. Fung, Transplantation, 2004, 77, S41 CrossRef CAS.
- C. B. Kang, H. Ye, S. Dhe-Paganon and H. S. Yoon, Neurosignals, 2008, 16, 318 CrossRef CAS PubMed.
- M. T. Goulet, K. M. Rupprecht, P. J. Sinclair, M. J. Wyvratt and W. H. Parsons, Perspect. Drug Discovery Des., 1994, 2, 145 CrossRef CAS.
- W. H. Parsons, N. H. Sigal and M. J. Wyvratt, Ann. N. Y. Acad. Sci., 1993, 685, 22 CrossRef CAS PubMed.
- J. P. Griffith, J. L. Kim, E. E. Kim, M. D. Sintchak, J. A. Thomson, M. J. Fitzgibbon, M. A. Fleming, P. R. Caron, K. Hsiao and M. A. Navia, Cell, 1995, 82, 507 CrossRef CAS.
- H. Nakagawa, T. Etoh, Y. Yokota, F. Ikeda, K. Hatano, N. Teratani, K. Shimomura, Y. Mine and T. Amaya, Clin. Drug Invest., 1996, 12, 244 CrossRef CAS PubMed.
- K. Migita and K. Eguchi, Curr. Med. Chem.: Anti-Inflammatory Anti-Allergy Agents, 2003, 2, 260 CrossRef CAS.
- B. G. Gold, Expert Opin. Invest. Drugs, 2000, 9, 2331 CrossRef CAS PubMed.
- B. G. Gold, V. Densmore, W. Shou, M. M. Matzuk and H. S. Gordon, J. Pharmacol. Exp. Ther., 1999, 289, 1202 CAS.
- J. N. Andexer, S. G. Kendrew, M. Nur-e-Alam, O. Lazos, T. A. Foster, A. S. Zimmermann, T. D. Warneck, D. Suthar, N. J. Coates, F. E. Koehn, J. S. Skotnicki, G. T. Carter, M. A. Gregory, C. J. Martin, S. J. Moss, P. F. Leadlay and B. Wilkinson, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 4776 CrossRef CAS PubMed.
- S. J. Mo, D. H. Kim, J. H. Lee, J. W. Park, D. B. Basnet, Y. H. Ban, Y. J. Yoo, S. Chen, S. R. Park, E. A. Choi, E. Kim, Y. Jin, S. Lee, J. Y. Park, Y. Liu, M. O. Lee, K. S. Lee, S. J. Kim, D. Kim, B. C. Park, S. Lee, H. J. Kwon, J. Suh, B. S. Moore, S. Lim and Y. J. Yoon, J. Am. Chem. Soc., 2011, 133, 976 CrossRef CAS PubMed.
- G. J. Gatto, S. M. McLoughlin, N. L. Kelleher and C. T. Walsh, Biochemistry, 2005, 44, 5993 CrossRef CAS PubMed.
- H. Motamedi and A. Shafiee, Eur. J. Biochem., 1998, 256, 528 CAS.
- H. Motamedi, A. Shafiee, S. J. Cai, S. L. Streicher, B. H. Arison and R. R. Miller, J. Bacteriol., 1996, 178, 5243 CAS.
- A. Shafiee, H. Motamedi, F. J. Dumont, B. H. Arison and R. R. Miller, J. Antibiot., 1997, 50, 418 CrossRef CAS.
- Y. H. Ban, P. B. Shinde, J.-Y. Hwang, M.-C. Song, D. H. Kim, S.-K. Lim, J. K. Sohng and Y. J. Yoon, J. Nat. Prod., 2013, 76, 1091 CrossRef CAS PubMed.
- H. Hatanaka, H. Kino, M. Asano, T. Goto, H. Tanaka and M. Okuhara, J. Antibiot., 1989, 42, 620 CrossRef CAS.
- Y. Obara, N. Nakahata, Y. Mizushina, F. Sugawara, K. Sakaguchi and Y. Ohizumi, Life Sci., 2000, 67, 1659 CrossRef CAS.
- T. Kieser, M. J. Bibb, M. J. Buttner, K. F. Chater and D. A. Hopwood, in Practical Streptomyces Genetics, John Innes Centre, Norwich, England, 2000, p. 408 Search PubMed.
- W. P. Revill, J. Voda, C. R. Reeves, L. Chung, A. Schirmer, G. Ashley, J. R. Carney, M. Fardis, C. W. Carreras, Y. Zhou, L. Feng, E. Tucker, D. Robinson and B. G. Gold, J. Pharmacol. Exp. Ther., 2002, 302, 1278 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: ESI-MS/MS, 1D- and 2D-NMR spectra of 1. See DOI: 10.1039/c4ra11907j |
| ‡ These authors contributed equally. |
| § Present address: Institute of Bioinformatics and Biotechnology, Savitribai Phule Pune University, Pune 411-007, India. |
| This journal is © The Royal Society of Chemistry 2015 |