Gadolinium nicotinate clusters as potential MRI contrast agents†
Xinping Lina,
Qiongqiong Zhanga,
Jiahe Chend,
Xiangjian Kongc,
La-Sheng Longc,
Cheng Wangb and
Wenbin Lin*ab
aCollaborative Innovation Center of Chemistry for Energy Materials, College of Chemistry and Chemical Engineering, Xiamen University, Xiamen 361005, P. R. China. E-mail: wenbinlin@uchicago.edu
bDepartment of Chemistry, University of Chicago, 929 E 57th Street, Chicago, IL 60637, USA
cState Key Laboratory of Physical Chemistry of Solid Surfaces, Department of Chemistry, College of Chemistry and Chemical Engineering, Xiamen University, Xiamen 361005, China
dDepartment of Physics and Electronic Science, Fujian Key Laboratory of Plasma and Magnetic Resonance, Xiamen University, Xiamen 361005, China
First published on 28th November 2014
Abstract
Three multinuclear gadolinium(III) clusters containing the nicotinate (nic) ligand, [Gd2(nic)6(H2O)4] (1), [Gd4(μ3-OH)4(Hnic)5(H2O)12](ClO4)8·7H2O (2), and [Gd4(μ3-OH)4(nic)6(H2O)6]2(ClO4)4·4H2O (3) were synthesized and characterized. Potential applications of these Gd(III) clusters as high-field magnetic resonance imaging (MRI) contrast agents were evaluated. The longitudinal relaxivities (r1) of 1–3 in water at 7 T were determined to be 10.45 ± 0.16, 9.28 ± 0.06, and 2.04 ± 0.29 mM−1 s−1 on a per Gd ion basis, respectively. In 1% agarose solution, the r1 values increased slightly to 10.37 ± 0.31, 10.74 ± 0.11, and 4.60 ± 0.29 mM−1 s−1 for 1–3, respectively. The ability to tune the number of inner-sphere water molecules, cluster sizes, organic ligands, and Gd coordination modes in such Gd(III) clusters provide interesting opportunities to further enhance the MR relaxivities for potential MRI applications.
Introduction
Magnetic resonance imaging (MRI) has become one of the most important techniques in modern diagnostic medicine because of its ability to non-invasively produce three-dimensional images of biological specimens with a deep tissue penetration and at a high spatial resolution.1 In MRI, image contrasts result from differences of water relaxation time and proton density between adjacent tissues.2 To enhance the image contrast, Gd(III) chelates3 are typically employed as contrast agents to reduce the longitudinal relaxation times (T1) of water molecules. Gd(III) contrast agents effectively increase the sensitivity of MRI, allowing reliable diagnosis of diseased states that are typically not discernible in the non-contrast enhanced MR images. The effectiveness of a Gd(III) chelate as a contrast agent is determined by its ability to relax water protons, as expressed by the longitudinal relaxivity (r1) which is the slope of the linear dependence of 1/T1 vs. Gd concentration in mM.4 Clinically used Gd(III) contrast agents, such as Gd–DPTA,5,6 are mononuclear compounds with a tightly bound chelating ligand and one to two coordinated water molecules.7–10 These Gd contrast agents however tend to have modest r1 relaxivities, particularly at high magnetic field strengths. As a result, significant amounts of Gd chelates need to be administered in order to afford adequate image contrasts between normal and pathological tissues in the clinic.11 Significant research efforts have therefore been devoted to the development of more efficient MRI contrast agents, for example, based on dendrimers8,12–14 and fullerenes,15,16 Gd2O3 nanoparticles,17,18 nanoscale metal–organic frameworks,19–22 and other nanoparticles containing a high density of Gd chelates, in the past few years.Metal clusters have attracted considerable attention in coordination and materials chemistry, and can be prepared by modular and metal-directed self-assembly methods.23–29 Many of these large yet molecular species30–35 possess beautiful structures as well as interesting optical and magnetic properties. Some of these metal clusters have potential applications as MRI contrast agents. For example, Pan and co-workers described that Mn12–acetate [MnIII/IV12O12(CH3CO2)16(H2O)4] could be coated with a polymeric shell via ligand-exchange to afford contrast agents with large r1 and r2 relaxivities of 5.2 ± 0.12 mM−1 s−1 and 10.7 ± 0.24 at 1.5 T, respectively.36 Duan et al. reported the use of hexanuclear gadolinium-based octahedron (Gd–PT1) as selective MRI contrast agents for detecting glucosamine and observed enhanced r1 relativity (64.8 mM−1 s−1 on per a Gd ion basis, 400 MHz, 298 K) owing to the rigid facial bridging ligands.25 In this work, we report the synthesis and characterization of three Gd clusters based on the nicotinate (nic) ligand, Gd2(nic)6(H2O)4 (1), [Gd4(μ3-OH)4(Hnic)5(H2O)12](ClO4)8·7H2O (2)37 and [Gd4(μ3-OH)4(nic)6(H2O)6]2(ClO4)4·4H2O (3). We further examined their MR relaxivities using a 7 T scanner, and studied the influences of the number of Gd ions and coordinated water molecules as well as the size of the clusters on their MR relaxivities (Fig. 1).
 | ||
| Fig. 1 Molecular parameters that influence relaxivity: rotation (τR), water exchange (τM) and hydration number (q). | ||
Experimental
Materials and general methods
Nicotinic acid, nitric acid, gadolinium oxide, perchloric acid, sodium hydroxide, and gadolinium oxide were obtained from commercial sources. Other solvents were of reagent grade and used without further purification. Elemental analyses were performed on an Elemental Vario EL analyzer. Thermogravimetric analysis (TGA) results were obtained on a SDT Q600 Thermal Analyzer. HR-MS spectra were recorded on a Bruker En Apex ultra 7.0T FT-MS apparatus. X-Ray powder diffraction patterns were obtained on a Rigaku Ultima IV XRD.Synthesis of Gd2(nic)6(H2O)4 (1)
The cluster 1 was prepared by a method that was modified from the procedure reported by Hong et al.38 10 mL of H2O was added to a mixture of GdCl3·6H2O (750 mg, 2 mmol) and nicotinic acid (490 mg, 4 mmol). 650 mg of NaN3 (10.0 mmol) was added to the solution, followed by the addition of 300 μL of 2 M HNO3. The mixture was transferred to a Teflon-lined high pressure reactor and then heated at 180 °C for three days. The reactor was slowly cooled at 1 °C per 10 min to room temperature, and the mixture was filtered. Colorless crystals of 1 were obtained after the solution was allowed to stand at room temperature for two days. Yield: 0.1 g (13.4%, based on nicotinic acid). Anal. calcd for C36H32Gd2N6O16: C, 38.6%; H, 2.86%; N, 7.50%. Found: C, 37.43%; H, 3.14%; N, 7.27%. IR (KBr): 3505 (m), 1645 (s), 1597 (s), 1573 (w), 1547 (m), 1476 (m), 1433 (s), 1405 (s), 1195 (m), 1166 (w), 1114 (w), 1093 (m), 1028 (m), 976 (w), 864 (m), 853 (m), 759 (vs.), 702 (vs.), 634 (m), 566 (m), 567 (m) cm−1. F.W. = 1119.18.Synthesis of [Gd4(μ3-OH)4(Hnic)5(H2O)12](ClO4)8·7H2O (2)
The cluster 2 was synthesized in the same way as that reported by Kong et al.39 Block-shaped colorless crystals of 2 were obtained. Yield: 0.2 g (8.2% based on nicotinic acid). Anal. calcd for C30Cl8H67Gd4N5O65: C, 14.28%; H, 2.34%; N, 2.53%. Found: C, 14.47%; H, 2.22%; N, 2.91%. IR (KBr): 3388 (m), 2861 (w), 2025 (w), 1643 (s), 1593 (s), 1417 (s), 1352 (w), 1305 (w), 1089 (s), 940 (w), 835 (w), 750 (s), 688 (s), 672 (m), 627 (s), 560 (m) cm−1. F.W. = 2450.47.Synthesis of [Gd4(μ3-OH)4(nic)6(H2O)6]2(ClO4)4 ·4H2O (3)
The cluster 3 was synthesized by a method that was modified from the procedure reported by Kong et al.39 Nicotinic acid (0.62 g, 5 mmol) in 5 mL of deionized water was titrated with NaOH (1.0 M) to reach a pH of 8, which was then added dropwise to the solution of 10 mL of Gd(ClO4)3 (1.0 M) that have been dropwise added a fresh NaOH (1.0 M) solution to the point of incipient but permanent precipitation at 90 °C. After stirring at 90 °C for one additional hour, the resulting white epinephelos mixture was quickly filtered while hot. The mixture was allowed to stand at room temperature for two days to afford block-shaped colorless crystals. Yield: 0.13 g (8.8% based on nicotinic acid). Anal. calcd for C72H88Gd8Cl4N12O64: C, 24.4%; H, 2.48%; N, 4.74%. Found: C, 24.90%; H, 2.32%; N, 4.84%. IR (KBr): 3388 (m), 1617 (s), 1566 (s), 1476 (w), 1409 (s), 1195 (m), 1158 (m), 1097 (m), 1030 (m), 973 (w), 842 (m), 759 (s), 698 (vs.), 636 (m), 563 (w) cm−1. F.W. = 3545.34.X-Ray crystal structure determination
The X-ray diffraction data of 1 and 3 were collected on Agilent Supernova with Mo-Kα radiation (λ = 0.71073 Å) at 100 K. Absorption corrections were applied by using the multiscan program CrysAlis Red. The structures were solved by direct and Fourier methods and refined by full-matrix least squares based on F2 using the SHELX and Olex2 software. Detailed crystal data and refinement parameters are listed in Table S1.†Measurement of r1 relaxivities
To measure the r1 relaxivities of 1–3, samples with different Gd(III) concentrations were dispersed in Mill-Q water or in 1% agarose solution. The Gd(III) concentrations were measured by inductively coupled plasma-mass spectrometry (ICP-MS). The samples were scanned (at 300![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K) on both 0.5 T and 7 T MRI scanners. T1-weighted MRI phantom images of the three samples are performed on a 7 T MRI scanner.
K) on both 0.5 T and 7 T MRI scanners. T1-weighted MRI phantom images of the three samples are performed on a 7 T MRI scanner.
Results and discussion
Synthesis
Cluster 1 was synthesized in 13.4% yield by first hydrothermally treating a mixture of nicotinic acid, GdCl3, and NaN3 in the presence of a small amount of HNO3 followed by crystallization at room temperature over a period of two days.38 Clusters 2 and 3 were prepared in low yield (about 8%) by first heating appropriate mixtures at 90 °C, followed by crystallization at room temperature. Such a ligand-controlled hydrolytic approach has been previously established for Gd cluster synthesis.40,41 The structures and compositions of 1–3 have been established by single-crystal X-ray crystallography, powder X-ray diffraction, and elemental analysis. Cluster 2 was prepared by a one-pot synthesis method whereby an aqueous solution of NaOH was added dropwise into the mixture containing both the nicotinic acid ligand and the Gd salt. In contrast, cluster 3 was obtained by a two-step procedure. The pH of the ligand aqueous solution and the lanthanide perchlorate was adjusted individually to appropriate values before they were mixed. The one-pot synthesis produced the tetranuclear cluster of [Gd4(μ3-OH)4(Hnic)5(H2O)12](ClO4)8·7H2O, while the two-step synthesis afforded [Gd4(μ3-OH)4(nic)6(H2O)6]2(ClO4)4·4H2O. Cluster 3 slightly differs from [Gd4(μ3-OH)4(nic)6(H2O)8]2(ClO4)4·3H2O which was previously described by Long et al. The pyridyl nitrogen atoms of the nic ligands in cluster 2 was protonated, while the nic ligands in cluster 3 were completely deprotonated.X-Ray single crystal structures
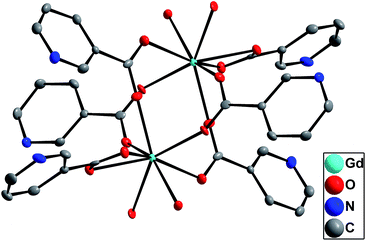
Single-crystal X-ray diffraction analyses reveal that 1 is a neutral dinuclear compound that crystallizes in the monoclinic space group P21/c. In 1, each Gd3+ ion is coordinated to eight oxygen atoms (Fig. 2), with two oxygen atoms coming from a chelating nicotinic acid ligand, four oxygen atoms coming from four bridging nicotinic acid ligands, and two oxygen atoms from two water molecules.42 Each Gd ion is thus coordinated to two water molecules in 1. In addition, there are two crystallization water molecules for each molecule of 1 in the crystals.The structure of 2 was previously reported by Kong et al.37 Briefly, the [Gd4(μ3-OH)4]8+ cationic core adopts a distorted cubane structure (Fig. 3). Two Gd ions (Gd1 and Gd2) have two coordinated water molecules whereas two other Gd ions (Gd3 and Gd4) have four coordinated water molecules. All of the pyridyl nitrogen atoms of the nicotinate ligands in 2 are protonated to maintain charge balance.
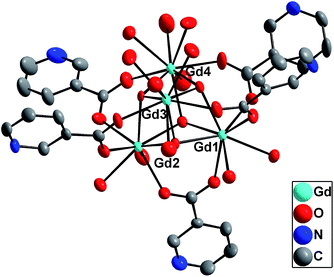
The molecular structure of 3 consists of a dimer of [Gd4(μ3-OH)4(nic)6(H2O)6]2+ as shown in Fig. 4. There is an inversion center residing at the center of the dimer. The Gd4(μ3-OH)4 cubane core is encapsulated by six nicotinate ligands. These ligands display two different coordination modes: five of them use their carboxylate groups to bridge two Gd3+ ions whereas the sixth one uses its carboxylate group to bridge two Gd3+ ions of one Gd4(μ3-OH)4 cubane core and uses its pyridyl N atom to coordinate a Gd3+ ion in the other Gd4(μ3-OH)4 cubane core (as shown for Gd4 and Gd3 in Fig. 4).
Gd1, Gd2, and Gd3 centers in 3 are thus 8-coordinate, by binding to three oxygen atoms from the bridging nicotinate ligands, three μ3-OH groups, and two water molecules. The Gd4 center is seven-coordinate and binds to three oxygen atoms from the bridging nicotinate ligands, three μ3-OH groups, and one pyridyl atom of the bridging nicotinate ligand. The structure of 3 is related to, but distinct from an octanuclear Gd complex with the formula of [Gd4(μ3-OH)4(nic)6(H2O)8]2(ClO4)4·3H2O that was reported earlier.37
Characterization of clusters 1–3 by PXRD, TGA and ESI-MS
We have used PXRD to verify the phase purity of the clusters. As shown in Fig. 5a–c, the experimental PXRD patterns of 1–3 generally match the PXRD patterns that were simulated from their single crystal structures well, indicating that the bulk phase of these clusters are identical to those of the single crystals chosen for X-ray diffraction studies. We note a slight peak shift in PXRD pattern of 3 from the simulated pattern, which can be attributed to different temperature between the single crystal X-ray and PXRD experiment as well as possible partial loss of lattice solvent molecules in the powdery samples for PXRD. Indexing the PXRD pattern of 3 gives similar lattice parameters to those determined from single crystal data (Table S3 and Fig. S7†).Thermogravimetric analyses (TGA) were also performed on clusters 1–3. As shown in Fig. 5d, the TGA curves for 1–3 display an initial weight loss 6.4% (calcd 7.9%) for 1, 12.5% (calcd 13.9%) for 2, and 8.1% (calcd 9.0%) for 3 at the room temperature to 300 °C temperature range, which correspond to the loss of guest and coordinated water molecules. When the temperature is higher than 300 °C, clusters of 1–3 were rapidly decomposed to form Gd2O3 with remaining weight of 32.5% (calcd 32.4%) for 1, 30.1% (calcd 29.6%) for 2, and 40.0% (calcd 40.9%) for 3 at 900 °C.
We have also tried to characterize the clusters using electrospray ionization-mass spectrometry to ensure that the clusters remain intact when they are dissolved in aqueous solutions. As shown in Fig. S1,† the ESI-MS spectrum (positive ion mode) of 1 gave the most intense peak at m/z = 1049.00, which corresponding to the fragment of [Gd2(nic)6 + H]+. This assignment was further confirmed by the isotopic distribution pattern of the [Gd2(nic)6 + H]+ fragment. As shown in Fig. 6a and b, the experimental and theoretical isotopic distribution patterns match extremely well.
The ESI-MS spectra of 2 and 3 indicate that the clusters have undergone extensive fragmentation during the electrospray ionization process. We have not been able to unambiguously assign the major peaks in their ESI-MS spectra. Instead, we resorted to dynamic light scattering (DLS) to probe the species resulted from the dissolution of 2 and 3 in aqueous solutions.
As shown in Fig. S3 and S4,† DLS data indicate the presence of hydrodynamic diameter peaks centering at 0.9 nm and 1.8 nm for 2 and 3, respectively. These DLS hydrodynamic diameters are consistent with the molecular dimensions of 2 and 3: the short and long dimensions of 2 are 0.6 and 1.7 nm, respectively, and the short and long dimensions of 3 are 0.9 and 2.7 nm, respectively. The DLS data thus support the notion that clusters 2 and 3 remain intact upon dissolution in aqueous solutions.
r1 Measurements and T1-weighted MRI phantom images
With seven unpaired electrons in each Gd ion, we expect that the synthesized Gd clusters to exhibit strong ability to relax water protons. We determined the longitudinal relaxivity (r1) values of 1–3 at both 0.5 T and 7 T. Each type of the clusters was prepared by dissolving them in 1% agarose solution with total Gd concentrations of 0.025, 0.1, 0.2, and 0.4 mM. As shown in Fig. 7a, the T1-weighted signal intensities in a 7 T scanner increase as the Gd concentration of the clusters increase, indicating that all of the clusters have the ability to enhance MRI contrasts with T1-weighted pulse sequences. At 7 T, the r1 values for 1–3 are 10.45 ± 0.16, 9.28 ± 0.06, and 2.04 ± 0.29 mM−1 s−1 in water through the linear fit in Fig. 7b, respectively. In 1% agarose solution, the r1 values increased slightly to 10.37 ± 0.31, 10.74 ± 0.11, and 4.60 ± 0.29 mM−1 s−1 for 1–3, respectively. The r1 values of the cluster 1 and 2 in 1% agarose are significantly higher than that of Gd–DTPA (r1 = 3.30 ± 0.25 mM−1 s−1 at 7 T) which we have determined under the same conditions. As expected, the r1 values for 1–3 decrease as the field strength increases; at 0.5 T, the r1 values are 11.00 ± 0.25, 10.08 ± 0.22, and 2.09 ± 0.32 mM−1 s−1 for 1–3 in water, respectively (Fig. S5†). The two Gd clusters of the 1 and 2 show high r1 relaxivities at 7 T, and they exhibit interesting relaxivity trends due to their different coordination environments. As described in the Solomon–Bloembergen–Morgan equations, the key factors contributing to the inner-sphere relaxivity include the rotational tumbling time (τR), the number of inner-sphere water molecules (q), and the residence lifetime of inner-sphere water molecules (τM). A larger relaxivity value will result when inner-sphere water have greater access to the paramagnetic metal center (i.e., shorter τM) and the inner-sphere relaxivity is linearly proportional to the number of inner-sphere water molecules (q).43 Since the molecular weights of the cluster ions range from 1119 to 3075 for 1–3, their tumbling rates in water should be relatively fast. We do not expect that the differences in tumbling rates of these clusters will significantly influence their r1 values. Instead, the number of coordinated water molecules and the steric hindrance around the water coordination sites for 1–3 should have significant impacts on their r1 values. Cluster 2 has the largest average number of coordinated water molecules per Gd (qave = 3), whereas cluster 3 has the smallest average number of coordinated water molecules per Gd (qave = 1.5). As a result, the r1 value for 2 is much higher than that of 3. Although 2 has a larger average number of coordinated water molecules per Gd than 1 (qave = 2), their r1 values are essentially identical. X-Ray structure comparisons indicate that the water coordination sites in 1 are more open than those in 2, and as a result, the τM value for 2 should be smaller, which compensate for its smaller qave value. | ||
| Fig. 7 The phantom study (a) and relaxivity plots (b) of the three clusters performed on a 7 T MRI scanner. | ||
Conclusions
We have prepared and characterized three gadolinium nicotinate clusters of different sizes. These Gd clusters of the cluster 1 and 2 have very high relaxivities at 7 T when compared to the clinically used Gd–DTPA. Because of the ability to tune the number of inner-sphere water molecules, cluster sizes, organic ligands, and Gd coordination modes in such Gd(III) clusters, we believe that Gd clusters provide interesting opportunities for designing new multinuclear MRI contrast agents for potential biological and biomedical imaging.Acknowledgements
The work was supported by the National Natural Science Foundation of P. R. China (21471126), the National Thousand Talents Program of P.R. China, and the 985 Program of the Chemistry and Chemical Engineering disciplines of Xiamen University. We acknowledge Ms Zhiwei Lin for the FT-MS measurement and Ms Ruiyun Huang for administrative help.Notes and references
- X. Zhang, X. Jing, T. Liu, G. Han, H. Li and C. Duan, Inorg. Chem., 2012, 51, 2325 CrossRef CAS PubMed.
- L. N. Goswami, L. Ma, S. Chakravarty, Q. Cai, S. S. Jalisatgi and M. F. Hawthorne, Inorg. Chem., 2013, 52, 1694 CrossRef CAS PubMed.
- T. Courant, V. G. Roullin, C. Cadiou, M. Callewaert, M. C. Andry, C. Portefaix, C. Hoeffel, M. C. de Goltstein, M. Port, S. Laurent, L. V. Elst, R. Muller, M. Molinari and F. Chuburu, Angew. Chem., Int. Ed., 2012, 51, 9119 CrossRef CAS PubMed.
- E. J. Werner, A. Datta, C. J. Jocher and K. N. Raymond, Angew. Chem., Int. Ed., 2008, 47, 8568 CrossRef CAS PubMed.
- H. Gries, U. Speck, H.-J. Weinmann, H. P. Niendorf and W. Seifert, DE3640708A1, 1988.
- C. T. Williams, J. P. Stack, B. Loveday, Y. Watson and I. Isherwood, Br. J. Radiol., 1988, 61, 596 CrossRef CAS PubMed.
- D. T. Puerta, M. Botta, C. J. Jocher, E. J. Werner, S. Avedano, K. N. Raymond and S. M. Cohen, J. Am. Chem. Soc., 2006, 128, 2222 CrossRef CAS PubMed.
- P. Caravan, J. J. Ellison, T. J. McMurry and R. B. Lauffer, Chem. Rev., 1999, 99, 2293 CrossRef CAS PubMed.
- Z. Zhang, A. F. Kolodziej, M. T. Greenfield and P. Caravan, Angew. Chem., Int. Ed., 2011, 50, 2621 CrossRef CAS PubMed.
- J. Xu, D. G. Churchill, M. Botta and K. N. Raymond, Inorg. Chem., 2004, 43, 5492 CrossRef CAS PubMed.
- H. Ersoy and F. J. Rybicki, J. Magn. Reson. Imaging, 2007, 26, 1190 CrossRef PubMed.
- E. Boros, M. Polasek, Z. Zhang and P. Caravan, J. Am. Chem. Soc., 2012, 134, 19858 CrossRef CAS PubMed.
- P. Caravan, G. Parigi, J. M. Chasse, N. J. Cloutier, J. J. Ellison, R. B. Lauffer, C. Luchinat, S. A. McDermid, M. Spiller and T. J. McMurry, Inorg. Chem., 2007, 46, 6632 CrossRef CAS PubMed.
- E. Garanger, S. A. Hilderbrand, J. T. Blois, D. E. Sosnovik, R. Weissleder and L. Josephson, Chem. Commun., 2009, 4444 RSC.
- K. B. Ghiassi, M. M. Olmstead and A. L. Balch, Dalton Trans., 2014, 43, 7346 RSC.
- M. Mikawa, H. Kato, M. Okumura, M. Narazaki, Y. Kanazawa, N. Miwa and H. Shinohara, Bioconjugate Chem., 2001, 12, 510 CrossRef CAS PubMed.
- M. L. Viger, J. Sankaranarayanan, C. de Gracia Lux, M. Chan and A. Almutairi, J. Am. Chem. Soc., 2013, 135, 7847 CrossRef CAS PubMed.
- A.-A. Guay-Bégin, P. Chevallier, L. Faucher, S. Turgeon and M.-A. Fortin, Langmuir, 2011, 28, 774 CrossRef PubMed.
- K. M. L. Taylor, A. Jin and W. Lin, Angew. Chem., Int. Ed., 2008, 47, 7722 CrossRef CAS PubMed.
- J. Della Rocca, D. Liu and W. Lin, Acc. Chem. Res., 2011, 44, 957 CrossRef CAS PubMed.
- M. D. Rowe, C.-C. Chang, D. H. Thamm, S. L. Kraft, J. F. Harmon, A. P. Vogt, B. S. Sumerlin and S. G. Boyes, Langmuir, 2009, 25, 9487 CrossRef CAS PubMed.
- W. Hatakeyama, T. J. Sanchez, M. D. Rowe, N. J. Serkova, M. W. Liberatore and S. G. Boyes, ACS Appl. Mater. Interfaces, 2011, 3, 1502 CAS.
- M. W. Cooke and G. S. Hanan, Chem. Soc. Rev., 2007, 36, 1466 RSC.
- X.-J. Kong, Y. Wu, L.-S. Long, L.-S. Zheng and Z. Zheng, J. Am. Chem. Soc., 2009, 131, 6918 CrossRef CAS PubMed.
- C. He, X. Wu, J. Kong, T. liu, X. Zhang and C. Duan, Chem. Commun., 2012, 48, 9290 RSC.
- D. T. Thielemann, A. T. Wagner, E. Rösch, D. K. Kölmel, J. G. Heck, B. Rudat, M. Neumaier, C. Feldmann, U. Schepers, S. Bräse and P. W. Roesky, J. Am. Chem. Soc., 2013, 135, 7454 CrossRef CAS PubMed.
- Y.-L. Hou, G. Xiong, P.-F. Shi, R.-R. Cheng, J.-Z. Cui and B. Zhao, Chem. Commun., 2013, 49, 6066 RSC.
- L.-X. Chang, G. Xiong, L. Wang, P. Cheng and B. Zhao, Chem. Commun., 2013, 49, 1055 RSC.
- S. K. Langley, B. Moubaraki and K. S. Murray, Inorg. Chem., 2012, 51, 3947 CrossRef CAS PubMed.
- J. Bai, A. V. Virovets and M. Scheer, Science, 2003, 300, 781 CrossRef CAS PubMed.
- T. C. Stamatatos, K. A. Abboud, W. Wernsdorfer and G. Christou, Angew. Chem., Int. Ed., 2007, 46, 884 CrossRef CAS PubMed.
- H.-H. Zou, R. Wang, Z.-L. Chen, D.-C. Liu and F.-P. Liang, Dalton Trans., 2014, 43, 2581 RSC.
- N. M. Randell, M. U. Anwar, M. W. Drover, L. N. Dawe and L. K. Thompson, Inorg. Chem., 2013, 52, 6731 CrossRef CAS PubMed.
- J.-M. Jia, S.-J. Liu, Y. Cui, S.-D. Han, T.-L. Hu and X.-H. Bu, Cryst. Growth Des., 2013, 13, 4631 CAS.
- F.-S. Guo, Y.-C. Chen, L.-L. Mao, W.-Q. Lin, J.-D. Leng, R. Tarasenko, M. Orendáč, J. Prokleška, V. Sechovský and M.-L. Tong, Chem.–Eur. J., 2013, 19, 14876 CrossRef CAS PubMed.
- B. Kim, A. H. Schmieder, A. J. Stacy, T. A. Williams and D. Pan, J. Am. Chem. Soc., 2012, 134, 10377 CrossRef CAS PubMed.
- X.-J. Kong, L.-S. Long, L.-S. Zheng, R. Wang and Z. Zheng, Inorg. Chem., 2009, 48, 3268 CrossRef CAS PubMed.
- M. Wu, F. Jiang, X. Kong, D. Yuan, L. Long, S. A. Al-Thabaiti and M. Hong, Chem. Sci., 2013, 4, 3104 RSC.
- X.-J. Kong, L.-S. Long, L.-S. Zheng, R. Wang and Z. Zheng, Inorg. Chem., 2009, 48, 3268 CrossRef CAS PubMed.
- R. Wang, Z. Zheng, T. Jin and R. J. Staples, Angew. Chem., Int. Ed., 1999, 38, 1813 CrossRef CAS.
- R. Wang, H. Liu, M. D. Carducci, T. Jin, C. Zheng and Z. Zheng, Inorg. Chem., 2001, 40, 2743 CrossRef CAS PubMed.
- W. Chen and S. Fukuzumi, Inorg. Chem., 2009, 48, 3800 CrossRef CAS PubMed.
- E. L. Que and C. J. Chang, Chem. Soc. Rev., 2010, 39, 51 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: X-Ray crystallography, electrospray mass spectrum, IR spectra and DLS spectra. CCDC 1014973 and 1014974. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c4ra07853e |
| This journal is © The Royal Society of Chemistry 2015 |