Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Upgrading a microplate reader for photobiology and all-optical experiments†
Florian
Richter
*a,
Ulrike S.
Scheib
b,
Jennifer
Mehlhorn
b,
Roman
Schubert
a,
Jonas
Wietek
b,
Oliver
Gernetzki
b,
Peter
Hegemann
b,
Tilo
Mathes
bc and
Andreas
Möglich
*a
aHumboldt-Universität zu Berlin, Institut für Biologie, Biophysikalische Chemie, Berlin, Germany. E-mail: florian.richter.1@hu-berlin.de; andreas.moeglich@hu-berlin.de; Fax: +49-30-2093-8948; Tel: +49-30-2093-8850
bHumboldt-Universität zu Berlin, Institut für Biologie, Experimentelle Biophysik, Berlin, Germany
cDepartment of Physics, Faculty of Sciences, Vrije Universiteit Amsterdam, The Netherlands
First published on 24th October 2014
Abstract
Automation can vastly reduce the cost of experimental labor and thus facilitate high experimental throughput, but little off-the-shelf hardware for the automation of illumination experiments is commercially available. Here, we use inexpensive open-source electronics to add programmable illumination capabilities to a multimode microplate reader. We deploy this setup to characterize light-triggered phenomena in three different sensory photoreceptors. First, we study the photoactivation of Arabidopsis thaliana phytochrome B by light of different wavelengths. Second, we investigate the dark-state recovery kinetics of the Synechocystis sp. blue-light sensor Slr1694 at multiple temperatures and imidazole concentrations; while the kinetics of the W91F mutant of Slr1694 are strongly accelerated by imidazole, the wild-type protein is hardly affected. Third, we determine the light response of the Beggiatoa sp. photoactivatable adenylate cyclase bPAC in Chinese hamster ovary cells. bPAC is activated by blue light in dose-dependent manner with a half-maximal intensity of 0.58 mW cm−2; intracellular cAMP spikes generated upon bPAC activation decay with a half time of about 5 minutes after light switch-off. Taken together, we present a setup which is easily assembled and which thus offers a facile approach to conducting illumination experiments at high throughput, reproducibility and fidelity.
Introduction
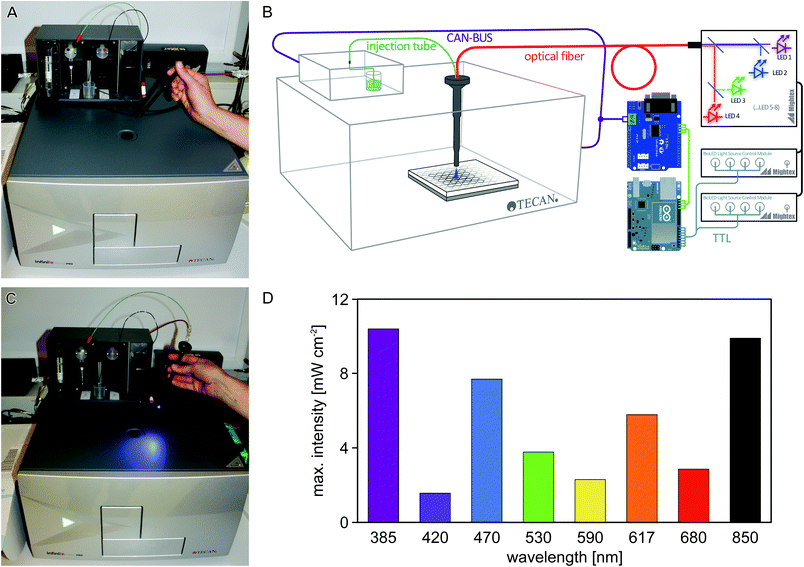
Laboratory automation and high-throughput experiments have a major impact on life-sciences research. As a core piece of laboratory automation equipment, multimode microplate readers can investigate multiple samples in parallel (usually in 96-, 384- or 1536-well format) with respect to absorption, fluorescence, and luminescence, while at the same time controlling the temperature and/or atmospheric composition of the sample environment. Such microplate readers can usually be programmed to automate measurements, so as to facilitate high sample throughput.In the photobiology field, numerous studies crucially require repetitive experimentation at varying parameters, e.g., the analysis of responses to different light qualities and quantities,1 or the screening of variant libraries of light-responsive systems.2 Usually, these studies mandate that sample illumination and measurement be conducted in quick succession. Using a microplate reader for such experiments therefore necessitates that individual wells are illuminated inside the instrument, and that illumination events are precisely programmed as part of a measurement protocol. Most commercially available microplate readers do not possess dedicated illumination capabilities, and the conventional probe light used for absorption and fluorescence measurements is generally of too low intensity to activate light-responsive systems to significant extent within reasonably short time intervals. To overcome this limitation, we used freely available open-source electronics and optical components to design and implement an illumination device for a widely used microplate reader. As illustrated in Fig. 1A, the Tecan Infinite M200 pro microplate reader can be equipped with an external injector module which allows for injection of liquids into individual wells at defined stages in a measurement protocol. This injector module consists of a hollow metal cylinder (referred to as ‘injection cylinder’) that houses two injection tubes, denoted A and B, which are connected to two pumps and two liquid reservoirs. The injection cylinder is inserted into the microplate reader through a hole in the top. When the microplate reader receives an injection command for either pump, the microplate is positioned such that the desired well is directly below the injection hole, and liquid from the respective reservoir (A or B) is injected into said well. We reasoned that by shining light through the injection hole, this setup could be repurposed for automatically ‘injecting’ light instead of liquid, and set out to implement an illumination setup where both the duration and wavelength of illumination can be specified through the standard iControl operating software of the Tecan microplate reader. The regular injection capabilities from line A are retained in the repurposed microplate reader, thus allowing experiments that rely on injection of both liquid and light. We demonstrate the utility of this setup for studies on three photoreceptor systems.
Sensory photoreceptors are the biological agents that mediate vision and diverse light-driven adaptive responses in numerous organisms.3,4 Upon light absorption by its chromophore, the photoreceptor undergoes structural and dynamic changes that culminate in a light-dependent modulation of its biological activity. Based on the chromophore and the photochemistry employed for light detection, photoreceptors are grouped into distinct classes. One class, BLUF domains (sensors of blue light using flavin adenine dinucleotide) are widely distributed in prokaryotes and protists where they control the activity of diverse physiological processes in response to blue-light exposure.5,6 Photon absorption in the blue spectral region leads to the formation of the signaling state of the photoreceptor that then thermally reverts to the dark state over the course of seconds to minutes depending on BLUF domain. The molecular details of BLUF photochemistry are still under intense debate but both photoactivation and thermal inactivation are apparently accomplished by a rearrangement of the hydrogen-bonding network near the flavin-nucleotide cofactor.7 Another class, phytochromes (Phy) are biological red-/far-red-light receptors that occur in plants, (cyano)bacteria, fungi, and diatoms.8 Phys exist in intrinsic equilibrium between red-light-absorbing and far-red-light-absorbing forms, Pr and Pfr, which differ in the isomerization state of their bilin chromophore. Illumination with red and far-red light drives the Pr→Pfr and Pfr→Pr transitions, respectively; that is, Phys are photochromic and can be toggled between two states by exposure to different light colors. In higher plants, five phytochromes (PhyA through PhyE) control various aspects of physiology, e.g., photomorphogenesis and shade avoidance.8,9
In recent years, photoreceptors have garnered increased attention due to their use in optogenetics.10 For these applications, photoreceptors are introduced as DNA templates to target cells, tissues and organs which are thereby rendered light-sensitive. Application of light can hence be used to control cellular events with supreme spatiotemporal resolution, minimal invasiveness and full reversibility. As a case in point, the blue-light-activated adenylate cyclase bPAC from Beggiatoa sp. possesses a BLUF domain and catalyzes formation of the second messenger 3′,5′-cyclic adenosine monophosphate (cAMP) upon blue-light exposure.11,12 Since cAMP is involved in the regulation of manifold biological processes, bPAC represents a particularly versatile and widely used optogenetic tool.13–15 In a similar vein, the light-induced association of Arabidopsis thaliana PhyB with its interacting factors PIF16 has been exploited in the construction of light-responsive cellular circuits, e.g., to control by light gene expression17 or cell motility.18
Results and discussion
Design and calibration of automated illumination setup
To enable efficient and versatile studies on sensory photoreceptors and other light-responsive systems, we sought to equip the off-the-shelf Tecan Infinite M200 pro microplate reader with programmable illumination capabilities by using readily available optical and electrical components (Fig. 1A–C). Two separate aspects had to be realized: (i) light delivery through the injection hole; and (ii) control of an LED through the plate reader operating software. For (i), a fiber-optics waveguide (1.5 mm diameter) was threaded through an empty injection cylinder and was connected to a light source containing eight separate LEDs that are coupled into a single fiber output. For (ii), the communication between the microplate reader and its injection module was captured and processed using an Arduino microcontroller (Fig. 1B). We configured the setup such that injection commands issued to pump B of the injector are re-interpreted to control the LED fiber optics; in this way, illumination events can be conveniently implemented into measurement protocols using the standard iControl software (version 1.9) of the microplate reader. Notably, we also fitted an injection tube (connected to pump A) into the cylinder alongside the waveguide, thus enabling experiments that require both illumination and dispensing of liquid from line A. Details on the setup are provided in the Experimental section and as ESI.†We first measured the light power of the eight LEDs included in the light source (peak emission at 385, 420, 470, 530, 590, 617, 680, 850 nm, respectively) at maximum output and found that it ranged from about 2 to 10 mW cm−2 (Fig. 1D); lower intensities can be set in 0.1% increments via the control software of the LED light source.
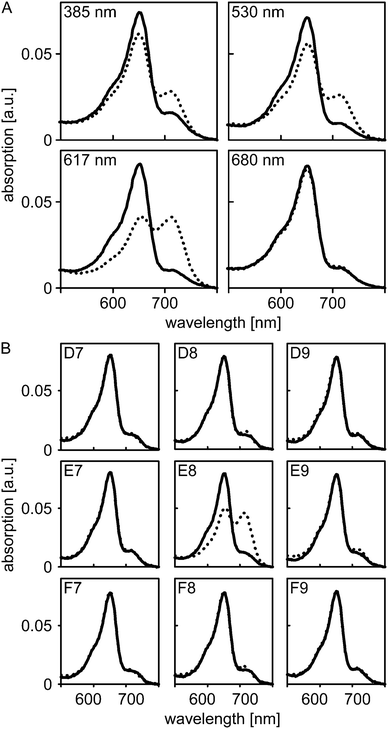
Next, we assessed whether light application via our new illumination setup can be implemented into standard iControl measurement protocols and whether the applied light intensities suffice to trigger photochemical reactions in sensory photoreceptors. To this end, we resorted to A. thaliana PhyB which absorbs light across the entire UV/visible spectral region and which can be reversibly switched between its Pr and Pfr states by illumination with different wavelengths. In its dark-adapted form, PhyB predominantly populates the Pr state which is characterized by an absorption maximum at 652 nm (Fig. 2A). Illumination with 385, 420, 470, 530, 590, 617 nm could be used to also populate the Pfr state, which maximally absorbs around 714 nm, to varying extent, where 590 and 617 nm were most efficient in driving the Pr→Pfr conversion. By contrast, illumination with 680 nm efficiently promoted the reverse Pfr→Pr conversion, and the resultant absorption spectrum largely corresponded to that of the dark-adapted state. Illumination with 850 nm elicited no spectral changes.
To assess whether illumination via the fiber-optics setup can be confined to a single well without affecting adjacent wells on the microplate, we put nine equivalent aliquots of PhyB in individual wells in a 3-by-3 arrangement. Absorption spectra were recorded for all nine wells prior to and after illumination of the center well with 590 nm light at maximum intensity for 16 s (Fig. 2B). In the center well, conversion to the Pfr state occurred to the same extent as before; by contrast, in the surrounding wells the absorption spectra before and after illumination closely overlapped. These data show that individual wells can be addressed with minimal cross-talk, even when relatively high light doses are applied.
Temperature and imidazole effects on dark-state recovery kinetics in Slr1694
Having established the principal function of the customized microplate reader illumination setup, we analyzed the dark-state recovery kinetics after blue-light exposure of the Synechocystis sp. PCC 6803 Slr1694 BLUF protein at multiple, systematically varied experimental conditions (Fig. 3). While the response of these kinetics towards perturbations could well provide incisive insight into the still contentious mechanism of the BLUF photocycle, corresponding systematic studies have so far been hampered by the sizeable number of measurements required. However, using our newly implemented, automated illumination setup, only modest amounts of experimental labor are required to set up and conduct multiple measurements in reproducible and efficient fashion.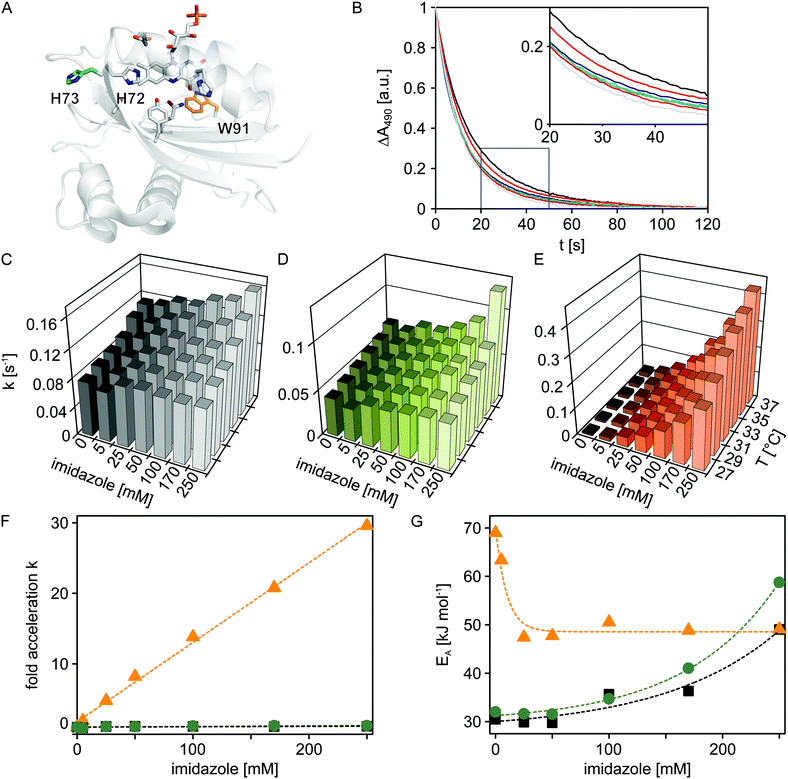 | ||
| Fig. 3 Temperature and imidazole effects on dark recovery in Slr1694 variants. (A) Structural model of Slr1694 in the W91in configuration according to Yuan et al.23 (B) Dark-state recovery kinetics monitored by absorption measurements at 490 nm as a function of imidazole concentration (from left to right: 0, 5, 25, 50, 100, 170 and 250 mM). The inset shows a close-up view of a portion of the data. (C–E) Temperature and imidazole dependence of rate constants for dark-state recovery in Slr1694 variants: wild-type (C), H73S (D), and W91F (E). (F) Factor acceleration of dark-state recovery as a function of imidazole concentration in Slr1694 wild-type (black), H73S (green) and W91F (orange). (G) Activation energies for dark-state recovery as a function of imidazole concentration in Slr1694 wild-type (black), H73S (green) and W91F (orange). | ||
It was previously reported that imidazole dramatically accelerates the dark recovery kinetics in the BLUF protein AppA from Rhodobacter sphaeroides,19 similar to observations for the related class of light-oxygen-voltage photoreceptors.20 Base catalysis thus might generally play an important role in the dark-state recovery mechanism, although the precise molecular basis remains unclear. To get a better understanding of this effect and to glean additional insight into the mechanism of BLUF photochemistry, we recorded dark-state recovery kinetics as a function of imidazole concentration (0, 5, 25, 50, 100, 170 and 250 mM) and temperature (27, 29, 31, 33, 35 and 37 °C). Surprisingly, the single-exponential dark-state recovery kinetics of Slr1694 (rate constant for dark recovery k = 0.08 s−1 at 27 °C) were largely insensitive towards imidazole addition up to concentrations of 250 mM (Fig. 3B and 3C). Evidently, the strongly accelerating effect of imidazole is not universally shared among all BLUF domains.
Inspection of the structures of AppA21,22 and Slr169423 pinpoints the histidine residue 73 in Slr1694 which is absent in AppA (Fig. 3A). Interestingly, His73 is situated in immediate vicinity of the strictly conserved histidine 72 which probably plays a crucial role in BLUF photochemistry because of its involvement in a proton-relay pathway between the exterior solvent and the flavin chromophore.24 We reasoned that His73 might serve as an imidazole proxy which would account both for the intrinsically faster recovery kinetics of Slr1694 compared to AppA and for its low sensitivity towards imidazole addition. To test this notion, we generated the H73S mutant of Slr1694 which exhibited slightly slower recovery kinetics than wild-type (k = 0.04 s−1 at 27 °C). However, the sensitivity towards imidazole was only marginally enhanced (Fig. 3D). By contrast, we discovered that the previously generated Slr1694 mutant W91F25,26 displays a very strong effect of imidazole on the lifetime of the signaling state (Fig. 3E). The intrinsic dark recovery in Slr1694 W91F is around 10-fold slower than in wild-type (k = 8 × 10−3 s−1 at 27 °C) but is accelerated by about 30-fold in the presence of 250 mM imidazole (Fig. 3F).
To better characterize the imidazole effects among the different Slr1694 variants, we recorded recovery kinetics at several temperatures and calculated activation energies (Fig. 3G). In the absence of imidazole, wild-type Slr1694 and the H73S mutant showed closely similar activation energies of EA = 30 kJ mol−1 and EA = 32 kJ mol−1, respectively. By contrast, the recovery kinetics in the W91F mutant displayed stronger temperature dependence with EA amounting to 69 kJ mol−1. At elevated imidazole concentrations ≥ 25 mM, the activation energy for dark recovery in W91F reached an asymptotic value of 49 kJ mol−1. These data can be explained by assuming two parallel pathways for dark recovery, one that is independent of imidazole, and one that is accelerated by imidazole. In the absence of imidazole, only the first pathway contributes to the observable kinetics, whereas at higher concentrations the kinetics are increasingly dominated by the imidazole-dependent pathway. Related observations were also made for the recovery kinetics of Slr1694 wild-type and H73S. However, in these cases, imidazole addition led to an increase of EA to about 50 kJ mol−1 and 60 kJ mol−1, respectively, at 250 mM imidazole. Due to the lower imidazole sensitivity of these variants, the accelerating effect cannot be saturated, and both recovery pathways contribute to the observable kinetics at all tested imidazole concentrations.
In the absence of structural information on the Slr1694 mutants and on the signaling state, a molecular interpretation of these findings is difficult. However, Fourier transform infrared spectroscopy showed that mutation of tryptophan 91 to phenylalanine increases the flexibility of the β5 strand of the BLUF domain which has been centrally implicated in signal transduction.24,26 Resultant changes in protein dynamics could govern solvent accessibility to the flavin chromophore and could thereby affect the stability and lifetime of the signaling state. More experiments are needed to clarify the mechanism of dark reversion in BLUF photoreceptors. Doubtless, the automated microplate reader illumination setup we present here will greatly facilitate pertinent measurements.
Light-activated adenylate cyclase activity in eukaryotic cell culture
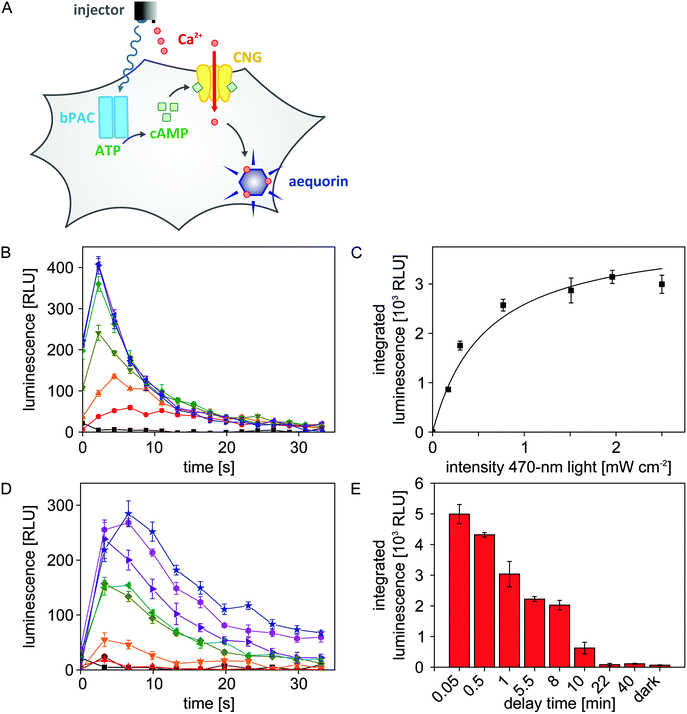
As detailed above, we configured the microplate reader such that both light and liquid can be dispensed in a single experiment. We exploited this previously unavailable setup to efficiently map the blue-light-dependent activity of bPAC in eukaryotic cells. To readily visualize adenylate cyclase activity, we resorted to a previously constructed Chinese hamster ovary (CHO) reporter cell line27,28 (Fig. 4A). Briefly, this cell line stably expresses a cyclic-nucleotide-gated (CNG) ion channel and the aequorin luciferase from Aequorea victoria. An increase of the cAMP concentration inside these cells triggers opening of the CNG channel which allows influx of Ca2+ ions from the extracellular medium. Once aequorin binds Ca2+, it rapidly oxidizes its cofactor coelenterazine to coelenteramide, with concomitant release of CO2 and emission of a photon in the blue-green spectral region. The resultant, transient luminescence signal can be measured with great sensitivity via the dedicated luminescence channel of the microplate reader.To assess the light-regulated adenylate cyclase activity of bPAC, the CHO reporter cell line was transiently transfected with bPAC-encoding DNA. Following seeding into 96-well microplates and incubation in the dark with the cofactor coelenterazine, the transfected cells were exposed for 8 s to different intensities of 470 nm light (0, 0.17, 0.30, 0.77, 1.51, 1.96, 2.50 mW cm−2) to stimulate bPAC activity.11,12 Immediately after light exposure, CaCl2 was added to a final concentration of 3 mM, and resultant luminescence signals were recorded over time (Fig. 4B). The integrated luminescence signal, reflective of intracellular adenylate cyclase activity, increases with applied light intensity but approaches saturation for intensities ≥2 mW cm−2. A hyperbolic fit was used to determine the half-maximal light dose for bPAC activation of E50 = (0.58 ± 0.12) mW cm−2 (Fig. 4C). This number corresponds to a previously reported value of (0.40 ± 0.04) mW cm−2 for the half-maximal light dose although the underlying experiments were performed in vitro on purified bPAC, and high-performance liquid chromatography was used for detection.12
Empowered by the efficient microplate reader illumination setup, we sought to investigate for how long cAMP spikes persist in the CHO cells after blue-light exposure ceases (470 nm, 2.50 mW cm−2). To this end, we successively increased the time lag between light application and CaCl2 injection from 3 seconds to 40 minutes. With increasing delay time, the observable luminescence rapidly decayed, and at delay times larger than 20 minutes, luminescence had returned to baseline values (Fig. 4D and E). Based on these data, we estimate that intracellular cAMP spikes that result from blue-light-triggered bPAC activity decay with a half life of approximately 5 minutes.
Taken together, our experiments provide valuable benchmarks for optogenetic applications of bPAC. More broadly, the newly developed illumination system allows to assess the light sensitivity of bPAC and intracellular cAMP persistence in a time-resolved and automated way, thereby complementing single-cell electrophysiological activity assays.12 The ability to dispense both light and liquid within the same measurement protocol proved of decisive advantage, as it enables multiplexing and ensures reproducibility.
Conclusion
In summary, we have presented an easily implementable strategy for adding illumination capabilities to a widely used, commercially available multimode microplate reader. All optical and electronic components are readily obtainable; notably, the LED light source used presently can easily be substituted by any other light source that can be controlled by TTL signals. The newly established, programmable illumination setup offers several decisive benefits: samples can be illuminated with multiple wavelengths within one measurement (cf. Fig. 2); data acquisition is greatly facilitated thus enabling high throughput, reproducibility and fidelity (cf. Fig. 3); experiments that rely on precisely synchronized injections of both liquid and light are feasible (cf. Fig. 4).We expect our setup to be broadly applicable to studies on sensory photoreceptors as demonstrated here. Of particular interest, the setup is suitable for optogenetic studies as it supports all-optical experiments in which a system is both perturbed and probed by light. For example, light-gated ion channels34 can be combined with fluorescent voltage sensors to achieve continuous and precise online control over membrane voltage.35 In a similar vein, bPAC11,12 (or, other light-activated nucleotide cyclases36,37) can be combined with a newly available, red-light-activated cAMP/cGMP-specific phosphodiesterase38 and suitable cNMP indicator dyes to establish corresponding, tight control over intracellular cNMP levels.
More generally, the approach we present here not only applies to photosensory signal transduction but also to any other biological, physical or chemical reaction that can be triggered by light and whose progress can be monitored spectroscopically.
Experimental
Design of the automated illumination setup
The rejigging of the Tecan Infinite M200 pro microplate reader (Tecan Group Ltd, Männedorf) for illumination comprised two principal steps: (i) delivering light into the microplate reader through the injection hole; and (ii) turning an LED on and off in response to an injection signal sent by the microplate reader iControl 1.9 software.For (i), an empty injection cylinder was generously provided by Tecan, into which a 1.5 mm waveguide (Thorlabs, Dachau) and an injection tube were fitted. The injection tube was connected to pump A of the injector. The waveguide was hooked up to an LED light source (Mightex Systems, Toronto) containing eight different LEDs (385 [full-width at half maximum: 10 nm], 420 [13], 470 [20], 530 [32], 590 [17], 617 [18], 680 [21], 850 [32] nm), thus allowing simultaneous delivery of light of several wavelengths through the injection hole into the microplate reader.
For (ii), we exploited the fact that in the Tecan Infinite series, the injection module is connected to the microplate reader through an external cable. Through this cable, the control software and the injector module pass messages back and forth using a so-called ‘Controller-Area-Network’, or CAN-bus protocol (http://en.wikipedia.org/wiki/CAN_bus). The graphical user interface of the iControl software allows users to specify the volume (in μL), the speed (in μL s−1) and the source reservoir/pump (A or B) of an injection event. We used an Arduino-Mega microcontroller (EXP GmbH, Saarbrücken) in combination with a Seeed-Studio CAN Bus shield (EXP GmbH, Saarbrücken) to intercept and record message traffic on the CAN-Bus in response to an injection command. Analysis of these recorded messages indicated that the control software passes the three user-specified parameters of pump identity, injection volume and injection speed to the injector which then performs a corresponding injection and reports back to the control software. The quotient of injection volume over injection speed yields the injection duration, i.e. the time that the injection target well is situated below the injection hole, and consequently the time that this well would be illuminated if light were delivered through the injection hole. We next connected the Arduino microcontroller to the TTL (transistor-transistor logic) trigger port of the Mightex light source, thus enabling the LEDs to be controlled through signals generated by the Arduino controller. Finally, we programmed the Arduino controller to, in response to a pump-B injection command passed by the iControl software, turn on the LED for the injection duration. As an added feature, the Arduino controller was programmed to turn on different LEDs depending on the last digit of the user-specified injection volume, meaning that both the duration and color of illumination can be encoded in a single injection command. The Arduino source code for decoding microplate-reader communications and switching LEDs in response has been deposited at https://gist.github.com/flosopher/6fd3ff661df9db708632. A list of parts used for the illumination upgrade as well as photographs that illustrate the setup are provided as ESI.†
In summary, by fitting a waveguide through the injection hole, and splicing a microcontroller into the data connection between microplate reader and injector, we were able to illuminate individual microplate wells in response to injection commands, thus enabling the variable programming of illumination events during measurement protocols through the regular microplate reader iControl software. Since the injector is equipped with two pumps that are separately addressable by the control software, measurement protocols that feature decoupled illumination and injection events are also feasible. All experiments reported presently were conducted with 96-well microplates; while the plate reader also handles 384-well plates, we have not tested the illumination setup for such plates.
Experiments on A. thaliana phytochrome B
The PAS-GAF-PHY photosensory core module of A. thaliana PhyB (residues 1–651) was co-expressed in Escherichia coli BL21 with heme oxygenase and PcyA from Synechocystis sp. which together provide the phycocyanobilin chromophore.29 After protein expression for 20 h at 18 °C, cells were lysed by sonication, and PhyB was purified by immobilized metal ion affinity chromatography (Protino Ni-NTA, Macherey-Nagel, Düren) using an Äkta pure system (GE Healthcare, Freiburg). Fractions containing PhyB were pooled and further purified via anion exchange (HiTrap Q, GE Healthcare). The PhyB sample was concentrated and dialyzed into storage buffer (25 mM Tris pH 8.0, 200 mM NaCl, 10% (v/v) glycerol); sample purity and chromophore incorporation were confirmed by gel electrophoresis and zinc-acetate staining.30 Concentration of PhyB was determined by absorption spectroscopy using an extinction coefficient of 93![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1 at 652 nm. The ratio of the absorption at 652 nm and at 280 nm amounted to 1.12 which is closely similar to the previously reported value of 1.05 for a corresponding PhyB construct.31 Microplate reader measurements were conducted at 30 °C in 96-well μClear microplates (Greiner BioOne, Frickenhausen) on 100 μL solution containing 5 μM PhyB. Absorption spectra were recorded prior to and following illumination for 16 s at maximum intensity of the respective LED in the light source.
000 M−1 cm−1 at 652 nm. The ratio of the absorption at 652 nm and at 280 nm amounted to 1.12 which is closely similar to the previously reported value of 1.05 for a corresponding PhyB construct.31 Microplate reader measurements were conducted at 30 °C in 96-well μClear microplates (Greiner BioOne, Frickenhausen) on 100 μL solution containing 5 μM PhyB. Absorption spectra were recorded prior to and following illumination for 16 s at maximum intensity of the respective LED in the light source.
Experiments on Synechocystis sp. PCC 6803 Slr1694
Expression constructs for Slr1694 wild-type and the W91F variant were prepared as previously described;32,33 the corresponding construct for the H73S variant was obtained by site-directed mutagenesis (QuikChange, Stratagene, Agilent, Waldbronn). Protein expression in a fermenter and subsequent purification were carried out as described.32,33 Spectroscopic measurements were performed in 96-well μClear microplates with each well containing 100 μL of 100 μM Slr1694 protein and varying imidazole concentrations (0, 5, 25, 50, 100, 170 and 250 mM). Samples were exposed to 470 nm light (7.7 mW cm−2) for 8 s, and dark-state recovery kinetics were followed by measuring absorption at 490 nm. Measurements were performed at temperatures of 27, 29, 31, 33, 35 and 37 °C; temperature fluctuations over time were on the order of ± 0.2 °C or less. For data analysis, all obtained curves were fitted to a single-exponential decay using the ‘leastsq’ optimization algorithm implemented in the Python module SciPy (http://scipy.org/). The reported rate constants for dark recovery, k, are mean ± s.d. of triplicate measurements.Experiments on Beggiatoa sp. bPAC
Light-triggered alterations of intracellular cAMP levels mediated by bPAC were detected in a CHO cell line that stably expresses a cAMP-sensitive heterotetrameric CNG channel and the luminescence reporter aequorin (provided by Dr F. Wunder, Bayer Pharma).28 Cells were cultured at 37 °C and 5% (v/v) CO2 in Dulbecco's modified Eagle's medium/NUT mix F-12 with L-glutamine, supplemented with 10% (v/v) inactivated fetal calf serum, 50 U mL−1 penicillin, 50 μg mL−1 streptomycin, 2.5 μg mL−1 amphotericin B, 0.6 mg mL−1 hygromycin B, and 0.25 mg mL−1 zeocin. In preparation of activity measurements, 2.5–3.0 × 104 cells were seeded into single wells of 96-well μClear microplates containing 100 μL medium. One day after seeding, wells were supplied with 60 μL medium-transfection mix, containing 400 ng DNA encoding bPAC-mCherry12 and 1.4 μL FuGENE HD transfection reagent (Promega, Mannheim). After 48 h, cells were washed once with Ca2+-free Tyrode solution (130 mM NaCl, 5 mM KCl, 20 mM HEPES, 1 mM MgCl2, 4.8 mM NaHCO3 pH 7.4) and incubated at 37 °C with 0.6 μg μL−1 coelenterazine (Promega). After 1–1.5 hours, cells were washed and supplemented with 100 μL Ca2+-free Tyrode buffer per well.To analyze light-activated adenylate cyclase activity, wells were illuminated for 8 s with different intensities of 470 nm light provided by the fiber-optics light source. After a defined delay, CaCl2 was injected to a final concentration of 3 mM; for experiments in Fig. 4B this delay was held constant at 3 s, for experiments in Fig. 4D this delay was varied between 3 s and 40 min. Resultant luminescence was monitored over time using a (520 ± 20) nm bandpass emission filter. Luminescence values were integrated over 33 s and corrected for background; reported values represent mean ± s.e.m. of four independent measurements. The light-dose dependency of the luminescence signals was fitted to a hyperbolic function using the non-linear least squares algorithm in Origin Pro 8.0.
Acknowledgements
We thank members of the Hegemann and Möglich groups for discussion. Helpful advice and support by H. W. Stotz and Dr D. Truckenbrodt (Tecan Deutschland GmbH) are greatly appreciated, as is stellar support by G. Schmitt of the mechanical workshop at Humboldt-Universität. Dr M. Stierl provided valuable advice on plate reader luminescence measurements; the CHO reporter cell line was generously provided by Dr F. Wunder (Bayer Pharma); and plasmids used for expression of PhyB were received from Dr W. Weber (Universität Freiburg). Funding through a Sofja-Kovalevskaya Award by the Alexander-von-Humboldt Foundation (A.M.), by the Einstein Foundation grant IPF-2012-148 (F.R. & A.M.), by Deutsche Forschungsgemeinschaft grants RI2468/1-1 (F.R.) and HE3824/24-1 (P.H. & T.M.), and by the Leibniz Graduate School of Molecular Biophysics (R.S. & A.M.) is gratefully acknowledged.Notes and references
- N. C. Rockwell, D. Duanmu, S. S. Martin, C. Bachy, D. C. Price, D. Bhattacharya, A. Z. Worden and J. C. Lagarias, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 3871–3876 CrossRef CAS PubMed.
- A. Möglich and K. Moffat, Photochem. Photobiol. Sci., 2010, 9, 1286–1300 Search PubMed.
- P. Hegemann, Annu. Rev. Plant Biol., 2008, 59, 167–189 CrossRef CAS PubMed.
- A. Möglich, X. Yang, R. A. Ayers and K. Moffat, Annu. Rev. Plant Biol., 2010, 61, 21–47 CrossRef PubMed.
- M. Iseki, S. Matsunaga, A. Murakami, K. Ohno, K. Shiga, K. Yoshida, M. Sugai, T. Takahashi, T. Hori and M. Watanabe, Nature, 2002, 415, 1047–1051 CrossRef CAS PubMed.
- M. Gomelsky and G. Klug, Trends Biochem. Sci., 2002, 27, 497–500 CrossRef CAS.
- M. Gauden, I. H. van Stokkum, J. M. Key, D. C. Lührs, R. van Grondelle, P. Hegemann and J. T. Kennis, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 10895–10900 CrossRef CAS PubMed.
- N. C. Rockwell, Y. S. Su and J. C. Lagarias, Annu. Rev. Plant Biol., 2006, 57, 837–858 CrossRef CAS PubMed.
- J. Hughes, Annu. Rev. Plant Biol., 2013, 64, 377–402 CrossRef CAS PubMed.
- K. Deisseroth, G. Feng, A. K. Majewska, G. Miesenböck, A. Ting and M. J. Schnitzer, J. Neurosci., 2006, 26, 10380–10386 CrossRef CAS PubMed.
- M.-H. Ryu, O. V. Moskvin, J. Siltberg-Liberles and M. Gomelsky, J. Biol. Chem., 2010, 285, 41501–41508 CrossRef CAS PubMed.
- M. Stierl, P. Stumpf, D. Udwari, R. Gueta, R. Hagedorn, A. Losi, W. Gärtner, L. Petereit, M. Efetova, M. Schwarzel, T. G. Oertner, G. Nagel and P. Hegemann, J. Biol. Chem., 2011, 286, 1181–1188 CrossRef CAS PubMed.
- R. J. De Marco, A. H. Groneberg, C.-M. Yeh, L. A. Castillo Ramírez and S. Ryu, Front. Neural Circuits, 2013, 7, 82 Search PubMed.
- M. Efetova, L. Petereit, K. Rosiewicz, G. Overend, F. Haußig, B. T. Hovemann, P. Cabrero, J. A. T. Dow and M. Schwärzel, J. Cell Sci., 2013, 126, 778–788 CrossRef CAS PubMed.
- A. Hartmann, R. D. Arroyo-Olarte, K. Imkeller, P. Hegemann, R. Lucius and N. Gupta, J. Biol. Chem., 2013, 288, 13705–13717 CrossRef CAS PubMed.
- M. Ni, J. M. Tepperman and P. H. Quail, Cell, 1998, 95, 657–667 CrossRef CAS.
- S. Shimizu-Sato, E. Huq, J. M. Tepperman and P. H. Quail, Nat. Biotechnol., 2002, 20, 1041–1044 CrossRef CAS PubMed.
- A. Levskaya, O. D. Weiner, W. A. Lim and C. A. Voigt, Nature, 2009, 461, 997–1001 CrossRef CAS PubMed.
- W. Laan, M. Gauden, S. Yeremenko, R. van Grondelle, J. T. M. Kennis and K. J. Hellingwerf, Biochemistry, 2006, 45, 51–60 CrossRef CAS PubMed.
- M. T. Alexandre, J. C. Arents, R. van Grondelle, K. J. Hellingwerf and J. T. Kennis, Biochemistry, 2007, 46, 3129–3137 CrossRef CAS PubMed.
- S. Anderson, V. Dragnea, S. Masuda, J. Ybe, K. Moffat and C. Bauer, Biochemistry, 2005, 44, 7998–8005 CrossRef CAS PubMed.
- A. Jung, T. Domratcheva, M. Tarutina, Q. Wu, W. H. Ko, R. L. Shoeman, M. Gomelsky, K. H. Gardner and I. Schlichting, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 12350–12355 CrossRef CAS PubMed.
- H. Yuan, S. Anderson, S. Masuda, V. Dragnea, K. Moffat and C. Bauer, Biochemistry, 2006, 45, 12687–12694 CrossRef CAS PubMed.
- T. Majerus, T. Kottke, W. Laan, K. Hellingwerf and J. Heberle, ChemPhysChem, 2007, 8, 1787–1789 CrossRef CAS PubMed.
- C. Bonetti, M. Stierl, T. Mathes, I. H. M. van Stokkum, K. M. Mullen, T. A. Cohen-Stuart, R. van Grondelle, P. Hegemann and J. T. M. Kennis, Biochemistry, 2009, 48, 11458–11469 CrossRef CAS PubMed.
- S. Masuda, K. Hasegawa and T. Ono, Plant Cell Physiol., 2005, 46, 1894–1901 CrossRef CAS PubMed.
- F. Wunder, J.-P. Stasch, J. Hütter, C. Alonso-Alija, J. Hüser and E. Lohrmann, Anal. Biochem., 2005, 339, 104–112 CrossRef CAS PubMed.
- F. Wunder, A. Rebmann, A. Geerts and B. Kalthof, Mol. Pharmacol., 2008, 73, 1235–1243 CrossRef CAS PubMed.
- L. O. Essen, J. Mailliet and J. Hughes, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 14709–14714 CrossRef CAS PubMed.
- T. R. Berkelman and J. C. Lagarias, Anal. Biochem., 1986, 156, 194–201 CrossRef CAS.
- K. Mukougawa, H. Kanamoto, T. Kobayashi, A. Yokota and T. Kohchi, FEBS Lett., 2006, 580, 1333–1338 CrossRef CAS PubMed.
- T. Mathes, C. Vogl, J. Stolz and P. Hegemann, J. Mol. Biol., 2009, 385, 1511–1518 CrossRef CAS PubMed.
- J. Mehlhorn, H. Steinocher, S. Beck, J. T. M. Kennis, P. Hegemann and T. Mathes, PLoS One, 2013, 8, e79006 CAS.
- G. Nagel, D. Ollig, M. Fuhrmann, S. Kateriya, A. M. Musti, E. Bamberg and P. Hegemann, Science, 2002, 296, 2395–2398 CrossRef CAS PubMed.
- D. R. Hochbaum, Y. Zhao, S. L. Farhi, N. Klapoetke, C. A. Werley, V. Kapoor, P. Zou, J. M. Kralj, D. Maclaurin, N. Smedemark-Margulies, J. L. Saulnier, G. L. Boulting, C. Straub, Y. K. Cho, M. Melkonian, G. K.-S. Wong, D. J. Harrison, V. N. Murthy, B. L. Sabatini, E. S. Boyden, R. E. Campbell and A. E. Cohen, Nat. Methods, 2014, 11, 825–833 CrossRef CAS PubMed.
- G. M. Avelar, R. I. Schumacher, P. A. Zaini, G. Leonard, T. A. Richards and S. L. Gomes, Curr. Biol., 2014, 24, 1234–1240 CrossRef CAS PubMed.
- M.-H. Ryu, I.-H. Kang, M. D. Nelson, T. M. Jensen, A. I. Lyuksyutova, J. Siltberg-Liberles, D. M. Raizen and M. Gomelsky, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 10167–10172 CrossRef CAS PubMed.
- C. F. Gasser, S. Taiber, C.-M. Yeh, C. H. Wittig, P. Hegemann, S. Ryu, F. Wunder and A. Möglich, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 8803–8808 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4pp00361f |
| This journal is © The Royal Society of Chemistry and Owner Societies 2015 |