Open Access Article
Open Access ArticleTryptophan prenyltransferases showing higher catalytic activities for Friedel–Crafts alkylation of o- and m-tyrosines than tyrosine prenyltransferases†
Aili
Fan
a,
Xiulan
Xie
b and
Shu-Ming
Li
*a
aInstitut für Pharmazeutische Biologie und Biotechnologie, Philipps-Universität Marburg, Deutschhausstrasse 17A, 35037 Marburg, Germany. E-mail: shuming.li@staff.uni-marburg.de
bFachbereich Chemie, Philipps-Universität Marburg, Hans-Meerwein-Strasse, 35032 Marburg, Germany
First published on 3rd June 2015
Abstract
Tryptophan prenyltransferases FgaPT2, 5-DMATS, 6-DMATSSv and 7-DMATS catalyse regiospecific C-prenylations on the indole ring, while tyrosine prenyltransferases SirD and TyrPT catalyse the O-prenylation of the phenolic hydroxyl group. In this study, we report the Friedel–Crafts alkylation of L-o-tyrosine by these enzymes. Surprisingly, no conversion was detected with SirD and three tryptophan prenyltransferases showed significantly higher activity than another tyrosine prenyltransferase TyrPT. C5-prenylated L-o-tyrosine was identified as a unique product of these enzymes. Using L-m-tyrosine as the prenylation substrate, product formation was only observed with the tryptophan prenyltransferases FgaPT2 and 7-DMATS. C4- and C6-prenylated derivatives were identified in the reaction mixture of FgaPT2. These results provided additional evidence for the similarities and differences between these two subgroups within the DMATS superfamily in their catalytic behaviours.
Introduction
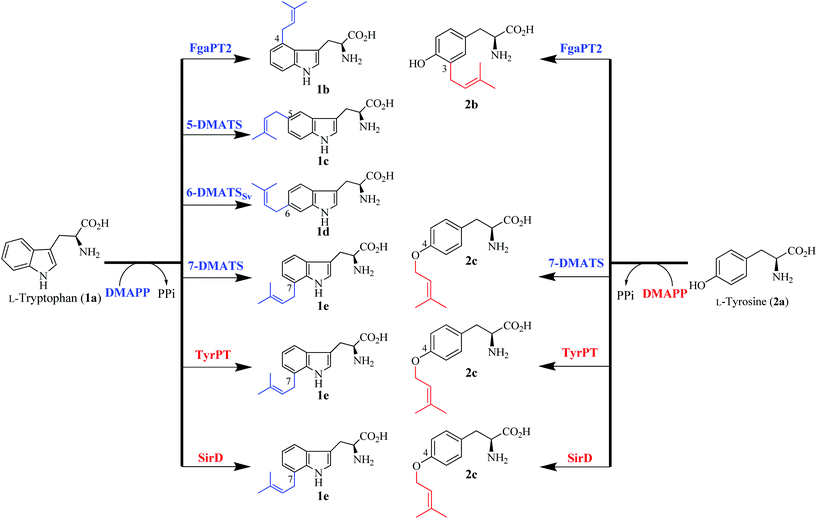
Prenylated secondary metabolites are widely distributed in nature. The biological and pharmacological activities of prenylated products are usually distinct from their non-prenylated precursors and therefore contribute largely to drug discovery and development programmes.1–3 Prenyltransferases are involved in the biosynthesis of these natural products and catalyse the regiospecific, in most cases Friedel–Crafts alkylations by transferring prenyl moieties from different prenyl donors to various acceptors. The prenyl moieties with different carbon chain lengths (C5, C10, C15 or C20 units) can be attached in the reverse or regular pattern and further modified by cyclization, oxidation and more. Therefore, prenyltransferases play an important role in the structural diversity of such products.4–6 These features have drawn considerable attention from scientists in different disciplines to prenyltransferases and prenylated derivatives. Based on their primary sequences, biochemical properties and structures, prenyltransferases can be divided into different subgroups including protein prenyltransferases, prenyl diphosphate synthases and aromatic prenyltransferases.4,7 In the last few years, significant progress has been achieved for aromatic prenyltransferases, especially for those from the dimethylallyl tryptophan synthase (DMATS) superfamily.6,8,9 So far, more than 40 such prenyltransferases have been identified and characterised biochemically from microorganisms, especially from fungi and bacteria.6,8,9 The majority of the DMATS superfamily uses indole derivatives including tryptophan and tryptophan-containing cyclic dipeptides as substrates. In the presence of dimethylallyl diphosphate (DMAPP), for example, FgaPT2,10 5-DMATS,11 6-DMATSSv![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 and 7-DMATS12 from this family catalyse regiospecific prenylations of L-tryptophan (1a) at C-4, C-5, C-6 and C-7 of the indole ring, respectively (Fig. 1). FgaPT2 and 7-DMATS from Aspergillus fumigatus are involved in the biosynthesis of fumigaclavine C13 and astechrome,14 respectively. 6-DMATSSv is likely to be involved in the biosynthesis of 6-dimethylallylindole-carbaldehyde in Streptomyces violaceusniger.8 A few members of the DMATS superfamily are responsible for the prenylation of non-indole substances. For example, SirD from Leptosphaeria maculans catalyses an O-prenylation of tyrosine (2a) (Fig. 1),15 the first specific step in the biosynthesis of sirodesmin PL.16 Last year, a new tyrosine O-prenyltransferase TyrPT (Fig. 1) was identified in Aspergillus niger, which was demonstrated to have similar functions to SirD.17 Further study showed that SirD and TyrPT also accepted L-tryptophan (1a) as a substrate in vitro and catalyse the same C7-prenylation as 7-DMATS (Fig. 1).15,17 Correspondingly, 7-DMATS catalyses the same O-prenylation of tyrosine as SirD and TyrPT (Fig. 1). Very recently, we demonstrated the C3-prenylation of L-tyrosine (2a) by the tryptophan C4-prenyltransferase FgaPT2 (Fig. 1).18 These results demonstrated the close relationship between tryptophan C-prenyltransferases and tyrosine O-prenyltransferases. The different regioselectivities of the mentioned enzymes with 2a encouraged us to test the acceptance of the 2a isomers, L-o-tyrosine (3a) and L-m-tyrosine (4a), by tryptophan and tyrosine prenyltransferases. 3a and 4a are important amino acid analogues found in metabolic pathways of human beings and have shown potential for treatments of different diseases.19–21 Prenylations of these two compounds have not yet been reported and their prenylated derivatives could be interesting candidates for further biological and pharmacological investigations.
8 and 7-DMATS12 from this family catalyse regiospecific prenylations of L-tryptophan (1a) at C-4, C-5, C-6 and C-7 of the indole ring, respectively (Fig. 1). FgaPT2 and 7-DMATS from Aspergillus fumigatus are involved in the biosynthesis of fumigaclavine C13 and astechrome,14 respectively. 6-DMATSSv is likely to be involved in the biosynthesis of 6-dimethylallylindole-carbaldehyde in Streptomyces violaceusniger.8 A few members of the DMATS superfamily are responsible for the prenylation of non-indole substances. For example, SirD from Leptosphaeria maculans catalyses an O-prenylation of tyrosine (2a) (Fig. 1),15 the first specific step in the biosynthesis of sirodesmin PL.16 Last year, a new tyrosine O-prenyltransferase TyrPT (Fig. 1) was identified in Aspergillus niger, which was demonstrated to have similar functions to SirD.17 Further study showed that SirD and TyrPT also accepted L-tryptophan (1a) as a substrate in vitro and catalyse the same C7-prenylation as 7-DMATS (Fig. 1).15,17 Correspondingly, 7-DMATS catalyses the same O-prenylation of tyrosine as SirD and TyrPT (Fig. 1). Very recently, we demonstrated the C3-prenylation of L-tyrosine (2a) by the tryptophan C4-prenyltransferase FgaPT2 (Fig. 1).18 These results demonstrated the close relationship between tryptophan C-prenyltransferases and tyrosine O-prenyltransferases. The different regioselectivities of the mentioned enzymes with 2a encouraged us to test the acceptance of the 2a isomers, L-o-tyrosine (3a) and L-m-tyrosine (4a), by tryptophan and tyrosine prenyltransferases. 3a and 4a are important amino acid analogues found in metabolic pathways of human beings and have shown potential for treatments of different diseases.19–21 Prenylations of these two compounds have not yet been reported and their prenylated derivatives could be interesting candidates for further biological and pharmacological investigations.
 | ||
| Fig. 1 Selected examples of prenyl transfer reactions catalysed by tryptophan and tyrosine prenyltransferases. | ||
Results and discussion
Comparison of the enzyme activities of tryptophan and tyrosine prenyltransferases towards tryptophan (1a) and tyrosine (2a)
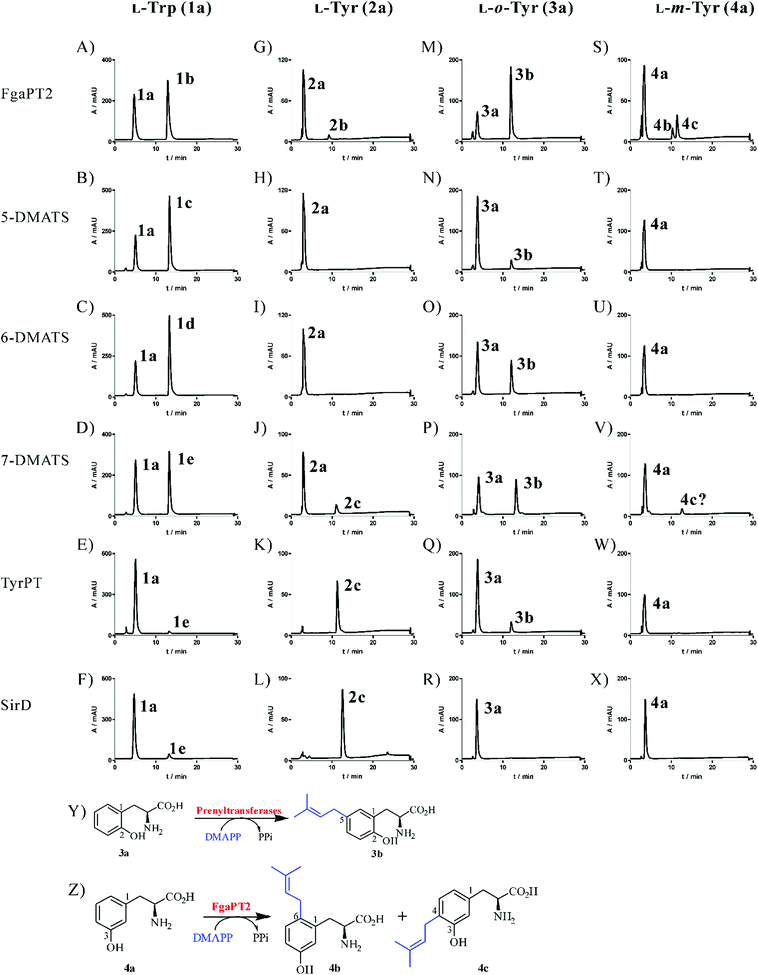
For better comparison of their catalytic activities, we carried out incubation of 1a and 2a with FgaPT2, 5-DMATS, 6-DMATSSv, 7-DMATS, SirD and TyrPT under the same conditions, i.e. 2 μg enzyme for 1a and 20 μg for 2a, 1 mM 1a or 2a, 5 mM CaCl2 and 1 mM DMAPP in 100 μL reaction mixtures. With the exception of 2a with 5-DMATS and 6-DMATSSv, HPLC analysis of the reaction mixtures (Fig. 2A–L) revealed the formation of one product each, confirming the results published previously.17,18,22 Product yields of these reactions are summarized in Table 1. It is obvious that tryptophan prenyltransferases FgaPT2, 5-DMATS, 6-DMATSSv and 7-DMATS accepted 1a much better than 2a. Over 50% conversion yields were observed with 1a as the substrate by tryptophan prenyltransferases after incubation at 37 °C for 1.5 h (Table 1; Fig. 2A–D). With 2a as the substrate, product formation was detected only in the reaction mixtures of FgaPT2 and 7-DMATS for tryptophan prenyltransferases, with product yields of 6.2 ± 1.0 and 13.8 ± 1.3%, respectively (Table 1). As expected, 2a was much better accepted by tyrosine than by tryptophan prenyltransferases. Almost total conversion of 2a was observed for TyrPT and SirD (Table 1; Fig. 2K and L). Product yields of 1a with TyrPT and SirD were found to be 5.2 ± 0.40 and 5.9 ± 1.0% (Table 1, Fig. 2), respectively.| 1a | 2a | 3a | 4a | |||||
|---|---|---|---|---|---|---|---|---|
| Prenylated positiona | Product yield [%] | Prenylated positiona | Product yield [%] | Prenylated positiona | Product yield [%] | Prenylated positiona | Product yield [%] | |
| a For structures see Fig. 1; “—” not determined. | ||||||||
| FgaPT2 | C-4 | 51.2 ± 4.6 | C-3 | 6.2 ± 1.0 | C-5 | 73.6 ± 3.6 | C-4; C-6 | 33.4 ± 1.4 |
| 5-DMATS | C-5 | 62.1 ± 2.8 | — | ≤0.5 | C-5 | 10.9 ± 2.5 | — | ≤0.5 |
| 6-DMATSSv | C-6 | 63.6 ± 2.9 | — | ≤0.5 | C-5 | 29.9 ± 2.5 | — | ≤0.5 |
| 7-DMATS | C-7 | 52.1 ± 1.9 | O-4 | 13.8 ± 1.3 | C-5 | 40.2 ± 3.8 | — | 4.8 ± 1.6 |
| TyrPT | C-7 | 5.2 ± 0.40 | O-4 | 100 | C-5 | 10.8 ± 0.7 | — | ≤0.5 |
| SirD | C-7 | 5.9 ± 1.0 | O-4 | 100 | C-5 | ≤0.5 | — | ≤0.5 |
Acceptance of L-o-tyrosine (3a) by all the tested tryptophan prenyltransferases, but not by the tyrosine prenyltransferase SirD
In the presence of 1 mM DMAPP, L-o-tyrosine (3a) was incubated with the six prenyltransferases (20 μg in 100 μL assay) mentioned in Fig. 2 at 37 °C for 1.5 h. Subsequent HPLC analysis revealed product formation in five reaction mixtures (Fig. 2M–Q). Surprisingly, 3a was accepted by all the tested tryptophan prenyltransferases, but not by the tyrosine prenyltransferase SirD (Fig. 2R), although TyrPT and SirD shared nearly the same behaviour towards the diverse substrates tested before.17 Furthermore, the tryptophan prenyltransferases FgaPT2, 6-DMATSSv and 7-DMATS with product yields of 73.6 ± 3.6, 29.9 ± 2.5 and 40.2 ± 3.8%, respectively, showed significantly higher activities than TyrPT with a product yield of 10.8 ± 0.7%, which is comparable to that of 5-DMATS. The product peaks detected in these assays display the same retention time, indicating the formation of an identical enzyme product by different prenyltransferases.C5-prenylated o-tyrosine (3b) as the unique product of tyrosine and tryptophan prenyltransferases
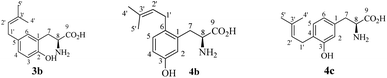
To elucidate their structures, the enzyme products of 3a with FgaPT2, 5-DMATS, 6-DMATSSv, 7-DMATS and TyrPT were isolated on HPLC from 5–10 ml of incubation mixtures and subjected to NMR (Table 2, Fig. S1 in the ESI†) and HR-MS analyses (Experimental section). HR-MS data confirmed the monoprenylation of the isolated products by detection of molecular masses, which are 68 Da larger than that of 3a. 1H-NMR data proved that FgaPT2, 5-DMATS, 6-DMATSSv, 7-DMATS and TyrPT converted indeed 3a to the same product 3b. Inspection of the 1H-NMR spectra (Fig. S1 in the ESI†) of 3b revealed the presence of signals for a regular prenyl moiety at δH 3.21 (d, H-1′), 5.25 (tsept, H-2′), 1.70 (d, H-4′) and 1.69 ppm (br s, H-5′). Furthermore, signals for three aromatic protons indicated that the prenylation took place on the aromatic ring, rather than at the hydroxyl group. The signals of the aromatic protons at δH 6.76 (d, 8.0 Hz), 6.91 (dd, 8.0, 2.0 Hz) and 6.99 (d, 2.0 Hz) of 3b proved the prenylation at C-4 or C-5 of 3a (Table 2). For the C5-prenylated 3a, it is expected that the signal of H-3 should be found in the high-field with a coupling constant of approximately 8.0 Hz and the signal of H-6 in the low-field with a coupling constant of 2.0 Hz. The obtained data corresponded very well to this prediction and therefore confirmed 3b to be the C5-prenylated product (Fig. 2Y). These results sound somewhat surprising, because an O-prenylated derivative was identified in the reaction mixtures of L-tyrosine (2a) with 7-DMATS and TyrPT.17,22 However, the C3-prenylated derivative was the enzyme product of FgaPT2 with 2a (Fig. 1).18| Pos | δ H, multi, J | δ H, multi, J | δ C | δ H, multi, J |
|---|---|---|---|---|
| a Signals overlapping with those of solvent. | ||||
| 1 | — | — | 134.9 | — |
| 2 | — | 6.69, d, 2.5 | 116.4 | 6.69, s |
| 3 | 6.76, d, 8.0 | — | 155.4 | — |
| 4 | 6.91, dd, 8.0, 2.0 | 6.64, dd, 8.0, 2.5 | 114.1 | — |
| 5 | — | 7.00, d, 8.0 | 130.6 | 7.00, d, 7.5 |
| 6 | 6.99, d, 2.0 | — | 131.2 | 6.67, d, 7.5 |
| 7 | Approx. 3.30a | 3.42, dd, 15.0, 4.5 | 34.4 | 3.19, dd, 14.0, 3.0 |
| 2.95, dd, 14.3, 8.5 | 2.80, dd, 15.0, 10.5 | 2.86, dd, 14.0, 9.3 | ||
| 8 | 3.85, dd, 8.5, 4.0 | 3.72, dd, 10.5, 4.5 | 55.5 | 3.69, m |
| 9 | — | — | 172.6 | — |
| 1′ | 3.21, d, 7.0 | 3.33, d, 7.0 | 30.0 | 3.24, d, 7.5 |
| 2′ | 5.25, tsept, 7.5, 1.5 | 5.19, br t, 7.0 | 123.4 | 5.28, br t, 7.5 |
| 3′ | — | — | 131.3 | — |
| 4′ | 1.70, d, 1.0 | 1.73, s | 16.6 | 1.70, d, 1.0 |
| 5′ | 1.69, br s | 1.72, s | 24.4 | 1.69, s |
| MeOH-d4 | MeOH-d4 | MeOH-d4 | MeOH-d4 | |
Acceptance of L-m-tyrosine (4a) by two tryptophan prenyltransferases and identification of C4- and C6-prenylated derivatives as products of the FgaPT2 reaction
After the identification of the C5-prenylated 3a from reaction mixtures of tyrosine and tryptophan prenyltransferases, we tested the behaviour of these enzymes towards L-m-tyrosine (4a) under the same conditions as for 3a. As shown in Fig. 2S–X, product formation was only observed in the reaction mixtures of FgaPT2 and 7-DMATS, with product yields of 33.4 ± 1.4 and 4.8 ± 1.6% (Table 1), respectively. Unexpectedly, 4a was not accepted by the two tyrosine prenyltransferases SirD and TyrPT (Fig. 2W and X). Another surprise is the much better acceptance of 4a by FgaPT2 than by 7-DMATS (Fig. 2S and V), which differs clearly from the preference of 2a towards these two enzymes (Fig. 2G and J). It seems that the positions of the hydroxyl groups at the benzene ring of phenylalanine have critical influence on their acceptance by tryptophan and tyrosine prenyltransferases.Inspection of the HPLC chromatograms in Fig. 2S and V revealed the presence of two product peaks 4b and 4c with a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
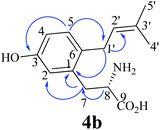
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 in the reaction mixture of FgaPT2 at 10.3 and 11.4 min and only one in that of 7-DMATS at 11.4 min. The UV maxima of 4b at 220 and 283 nm differed slightly from those of 4c at 220 and 278 nm. For structure elucidation, 4b and 4c were isolated on HPLC from the incubation mixture of 4a with FgaPT2 and subjected to NMR (Table 2; Fig. S2–S5 in the ESI†) and HR-MS analyses (Experimental section). HR-MS data confirmed the monoprenylation of the isolated products. Inspection of the 1H-NMR spectra (Table 2) of 4b and 4c revealed the presence of signals for a regular prenyl moiety each at δH 3.33 or 3.24 (d, H-1′), 5.19 or 5.28 (br t H-2′), 1.73 or 1.70 (s H-4′) and 1.72 or 1.69 ppm (s, H-5′). The signals of the aromatic protons at 6.69 (d, 2.5 Hz, H-2), 6.64 (dd, 8.0, 2.5 Hz, H-4), and 7.00 (d, 8.0 Hz, H-5) of 4b and 6.69 (s, H-2), 7.00 (d, 7.5 Hz, H-5), and 6.67 (d, 7.5 Hz, H-6) of 4c indicated the prenylation at C-4 in one case and C-6 in another case. To determine the structures of 4b and 4c, we recorded HSQC and HMBC spectra for 4b (Fig. 3, Fig. S3 and S4 in the ESI†). The important correlation between H-1′ and C-1 prove unequivocally that 4b is the C6-prenylated derivative, i.e. prenylation at the para-position to the phenolic hydroxyl group (Fig. 2Z). Consequently, 4c is the C4-prenylated product (Fig. 2Z). Due to the low quality, the structure of the enzyme product of 4a with 7-DMATS could not be elucidated in this study. However, from its same retention time and UV absorption maxima with those of 4c, it could be speculated that this substance has the same structure as 4c.
2 in the reaction mixture of FgaPT2 at 10.3 and 11.4 min and only one in that of 7-DMATS at 11.4 min. The UV maxima of 4b at 220 and 283 nm differed slightly from those of 4c at 220 and 278 nm. For structure elucidation, 4b and 4c were isolated on HPLC from the incubation mixture of 4a with FgaPT2 and subjected to NMR (Table 2; Fig. S2–S5 in the ESI†) and HR-MS analyses (Experimental section). HR-MS data confirmed the monoprenylation of the isolated products. Inspection of the 1H-NMR spectra (Table 2) of 4b and 4c revealed the presence of signals for a regular prenyl moiety each at δH 3.33 or 3.24 (d, H-1′), 5.19 or 5.28 (br t H-2′), 1.73 or 1.70 (s H-4′) and 1.72 or 1.69 ppm (s, H-5′). The signals of the aromatic protons at 6.69 (d, 2.5 Hz, H-2), 6.64 (dd, 8.0, 2.5 Hz, H-4), and 7.00 (d, 8.0 Hz, H-5) of 4b and 6.69 (s, H-2), 7.00 (d, 7.5 Hz, H-5), and 6.67 (d, 7.5 Hz, H-6) of 4c indicated the prenylation at C-4 in one case and C-6 in another case. To determine the structures of 4b and 4c, we recorded HSQC and HMBC spectra for 4b (Fig. 3, Fig. S3 and S4 in the ESI†). The important correlation between H-1′ and C-1 prove unequivocally that 4b is the C6-prenylated derivative, i.e. prenylation at the para-position to the phenolic hydroxyl group (Fig. 2Z). Consequently, 4c is the C4-prenylated product (Fig. 2Z). Due to the low quality, the structure of the enzyme product of 4a with 7-DMATS could not be elucidated in this study. However, from its same retention time and UV absorption maxima with those of 4c, it could be speculated that this substance has the same structure as 4c.
Our results mentioned above suggest that the acceptance of a substrate by tryptophan or tyrosine prenyltransferases would not strictly depend on the chemical category of the aromatic nucleus, but strongly rely on the position of the functional groups on the aromatic nucleus, which perform the direct interaction with the enzyme active sites. Furthermore, these results reveal that tryptophan and tyrosine prenyltransferases possess very likely similar active sites and strengthen their close relationship in the evolution.
Kinetic study of the enzymatic Friedel–Crafts reactions
To get insights into the catalytic efficiency, kinetic parameters were determined for FgaPT2, 5-DMATS, 6-DMATSSv, 7-DMATS and TyrPT towards 3a and FgaPT2 towards 4a (Table 3; Fig. S6–S11 in the ESI†). As expected for unnatural substrates, KM values between 0.17 ± 0.0076 and 0.58 ± 0.081 mM were determined for 3a, much higher than for their natural substrates.8,10–12,17 The KM value of FgaPT2 towards 4a was found to be 0.90 ± 0.013 mM, higher than that with 3a (Table 3). In agreement with the product yields given in Table 1, FgaPT2 showed the highest turnover number and catalytic efficiency toward 3a, followed by 7-DMATS and 6-DMATSSv. TyrPT and 5-DMATS demonstrated comparable kinetic parameters for their reactions with 3a (Table 3).| K M (mM) | k cat (s−1) | k cat/KM (s−1 M−1) | |
|---|---|---|---|
| L-o-Tyrosine (3a) | |||
| FgaPT2 | 0.47 ± 0.0024 | 0.29 ± 0.029 | 600 ± 58 |
| 5-DMATS | 0.49 ± 0.0031 | 0.013 ± 0.00011 | 27 ± 0.40 |
| 6-DMATSSv | 0.58 ± 0.081 | 0.034 ± 0.00038 | 59 ± 3.6 |
| 7-DMATS | 0.17 ± 0.0076 | 0.045 ± 0.036 | 270 ± 8.9 |
| TyrPT | 0.58 ± 0.014 | 0.014 ± 0.00066 | 24 ± 1.7 |
| L-m-Tyrosine (4a) | |||
| FgaPT2 | 0.90 ± 0.013 | 0.0065 ± 0.00044 | 6.8 ± 0.093 |
Proposed reaction mechanisms of Friedel–Crafts alkylation of o- and m-tyrosine
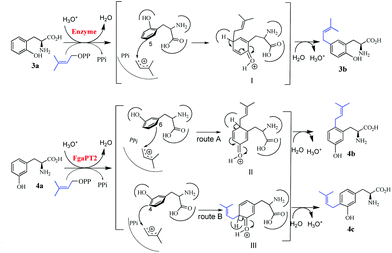
Previous studies on structures of several prenyltransferases and mutagenesis experiments have shown that the prenyl transfer reactions catalysed by the prenyltransferases of the DMATS superfamily are initiated by the formation of a dimethylallyl carbocation.23–26 Nucleophilic attack of this ion by an electron-rich aromatic ring leads to the formation of non-aromatic intermediates, which will be rearomatized by elimination of a proton and result in the formation of the prenylated derivatives. A similar mechanism could also be proposed for reactions observed in this study. As shown in Fig. 4, attack of the dimethylallyl carbocation by C-5 of 3a would result in the formation of the intermediate I, which would undergo proton elimination to form the final product 3b. For the FgaPT2 reaction with 4a, attack from two positions, C-6 and C-4, would be possible. It can be expected that two different intermediates II and III will be formed via routes A and B, respectively, and then converted to 4b and 4c after proton elimination (Fig. 4).Experimental section
Chemicals
DMAPP was synthesized according to the method described for geranyl diphosphate (GPP) reported previously.27L-o-Tyrosine and L-m-tyrosine used for the enzyme assays were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) and Alfa Aesar (Karlsruhe, Germany), respectively.Overexpression and purification of recombinant proteins
Protein overproduction and purification were carried out for FgaPT2,10 5-DMATS,11 6-DMATSSv,8 7-DMATS,12 SirD15 and TyrPT17 as described previously.Assays for determination of enzyme activities
Reaction mixtures (100 μl) for determination of the enzyme activities contained an aromatic substrate (1 mM), CaCl2 (5 mM), DMAPP (1 mM), glycerol (1.0–6.0% v/v), dimethyl sulfoxide (DMSO, 0–5.0% v/v), 50 mM Tris-HCl (pH 7.5) and a purified recombinant protein (2 or 20 μg). The reaction mixtures were incubated at 37 °C for 1.5 h and then terminated by the addition of 100 μl MeOH. Protein was removed by centrifugation at 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 20 min.
000 rpm for 20 min.
Enzyme assays for product isolation and structure elucidation
Assays for isolation of the enzyme products were carried out in large scales (5–10 ml) containing an aromatic substrate (1 mM), DMAPP (1.5 mM), CaCl2 (5 mM), glycerol (1.0–9.9% v/v), DMSO (0–5.0% v/v), 50 mM Tris-HCl (pH 7.5) and a recombinant protein (0.2–0.6 mg per ml assay). After incubation for 16 h at 37 °C, the reaction mixtures were terminated by the addition of 5–10 ml methanol. After removal of the precipitated protein by centrifugation at 6000 rpm for 30 min, the reaction mixtures were concentrated in a rotating vacuum evaporator at 35 °C to a final volume of 1 ml before injection into HPLC.Enzyme assays for determination of the kinetic parameters
Assays for determination of the kinetic parameters contained CaCl2 (5 mM), glycerol (1.0–9.9% v/v), DMSO (0–5.0% v/v), 50 mM Tris HCl (pH 7.5), DMAPP (1 mM) and an aromatic substrate at final concentrations up to 2.0 mM. The reactions were then terminated with 100 μl MeOH. Protein was removed by centrifugation at 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 20 min.
000 rpm for 20 min.
HPLC analysis and isolation of enzyme products for structure elucidation
The enzyme products were routinely analysed by HPLC on an Agilent series 1200 by using a Multospher 120 RP-18 column (250 × 4 mm, 5 μm C + S Chromatographie Service, Langerwehe, Germany) at a flow rate of 1 ml min−1. Water (solvent A) and methanol (solvent B) were used as solvents for analysis and isolation of the enzyme products. A linear gradient of 30–100% (v/v) solvent B in 20 min was used for analysis of the enzymatic products. The column was then washed with 100% solvent B for 5 min and equilibrated with 30% solvent B for another 5 min. Detection was carried out on a photodiode array detector.For isolation of the enzyme products, the same HPLC equipment with a Multospher 120 RP-18 column (250 × 10 mm, 5 μm, C + S Chromatographie Service, Langerwehe, Germany) was used. A linear gradient was run with 50–100% (v/v) of methanol (solvent B) in water (solvent A) in 50–80 min and a flow rate at 2.5 ml min−1. The column was then washed with 100% (v/v) solvent B for 10 min and equilibrated with 50% (v/v) solvent B for 10 min.
HR-ESI-MS and NMR analysis of the enzyme products
The obtained products were analysed by NMR (in CD3OD) and MS analyses. NMR spectra were recorded at room temperature on a Bruker Avance 600 MHz or a JEOL ECA-500 spectrometer. The heteronuclear single quantum correlation (HSQC) and heteronuclear multiple bond correlation (HMBC) spectra were recorded with standard methods28 and processed with MestReNova.5.2.2. The positive HR-ESI-MS was carried out on an AutoSpec instrument (Micromass Co. UK Ltd) with the following results:3b: m/z 249.1374 [M]+ (C14H19NO3, cal.: 249.1365); 4b: m/z 249.1373 [M]+ (C14H19NO3, cal.: 249.1365); 4c: m/z 249.1348 [M]+ (C14H19NO3, cal.: 249.1365).
Conclusions
Prenyltransferases of the DMATS superfamily are known for their broad aromatic substrates.6 As shown in Fig. 2, tyrosine prenyltransferases TyrPT and SirD also accepted tryptophan as the substrate, but with lower activity than with tyrosine. Correspondingly, tryptophan prenyltransferases FgaPT2 and 7-DMATS also prenylated tyrosine, but with lower activity than for tryptophan. In this study, we demonstrated surprisingly that L-o-tyrosine (3a) was accepted by all of the tested tryptophan prenyltransferases for prenylations at C-4, C-5, C-6 and C-7 of the indole ring, but not by the tyrosine prenyltransferase SirD. The unique product C5-prenylated o-tyrosine was identified in all the reaction mixtures of FgaPT2, 5-DMATS, 6-DMATSSv, 7-DMATS and TyrPT, although these enzymes utilized different natural substrates and catalysed diverse regioselective reactions.
L-m-Tyrosine (4a) was only converted by two tryptophan prenyltransferases, but by none of the tyrosine prenyltransferases. Structure elucidation of the enzyme products of 4a with FgaPT2 revealed a low regioselectivity. C4- and C6-prenylated m-tyrosines with a ratio of approximately 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 were identified as the reaction products.
1 were identified as the reaction products.
Comparison studies in the future on the structure level of these two groups of prenyltransferases could provide detailed insights into the difference and similarity of their substrate and catalytic promiscuities. Meanwhile, it could even provide suggestions for protein engineering to create new biocatalysts in order to meet a given target, such as broader substrate spectra and/or different regioselectivity.29,30
Acknowledgements
This work was supported in part by a grant from DFG (Li844/4-1 to S.-M. Li). Aili Fan is a recipient of a scholarship from CSC. We thank Lena Ludwig for synthesis of DMAPP, Nina Zitzer and Stefan Newel, all from Philipps-Universität Marburg, for recording MS and NMR spectra, respectively.References
- S.-M. Li, Nat. Prod. Rep., 2010, 27, 57–78 RSC.
- P. Ruiz-Sanchis, S. A. Savina, F. Albericio and M. Alvarez, Chemistry, 2011, 17, 1388–1408 CrossRef CAS PubMed.
- C. Wallwey and S.-M. Li, Nat. Prod. Rep., 2011, 28, 496–510 RSC.
- L. Heide, Curr. Opin. Chem. Biol., 2009, 13, 171–179 CrossRef CAS PubMed.
- K. Yazaki, K. Sasaki and Y. Tsurumaru, Phytochemistry, 2009, 70, 1739–1745 CrossRef CAS PubMed.
- X. Yu and S.-M. Li, Methods Enzymol., 2012, 516, 259–278 CAS.
- S.-M. Li, Appl. Microbiol. Biotechnol., 2009, 84, 631–639 CrossRef CAS PubMed.
- J. Winkelblech and S.-M. Li, ChemBioChem, 2014, 15, 1030–1039 CrossRef CAS PubMed.
- K. Miyamoto, F. Ishikawa, S. Nakamura, Y. Hayashi, I. Nakanishi and H. Kakeya, Bioorg. Med. Chem., 2014, 22, 2517–2528 CrossRef CAS PubMed.
- I. A. Unsöld and S.-M. Li, Microbiology, 2005, 151, 1499–1505 CrossRef PubMed.
- X. Yu, Y. Liu, X. Xie, X.-D. Zheng and S.-M. Li, J. Biol. Chem., 2012, 287, 1371–1380 CrossRef CAS PubMed.
- A. Kremer, L. Westrich and S.-M. Li, Microbiology, 2007, 153, 3409–3416 CrossRef CAS PubMed.
- S.-M. Li and I. A. Unsöld, Planta Med., 2006, 72, 1117–1120 CrossRef CAS PubMed.
- W.-B. Yin, J. A. Baccile, J. W. Bok, Y. Chen, N. P. Keller and F. C. Schroeder, J. Am. Chem. Soc., 2013, 135, 2064–2067 CrossRef CAS PubMed.
- A. Kremer and S.-M. Li, Microbiology, 2010, 156, 278–286 CrossRef CAS PubMed.
- D. M. Gardiner, A. J. Cozijnsen, L. M. Wilson, M. S. Pedras and B. J. Howlett, Mol. Microbiol., 2004, 53, 1307–1318 CrossRef CAS PubMed.
- A. Fan, H. Chen, R. Wu, H. Xu and S.-M. Li, Appl. Microbiol. Biotechnol., 2014, 98, 10119–10129 CrossRef CAS PubMed.
- A. Fan, G. Zocher, E. Stec, T. Stehle and S.-M. Li, J. Biol. Chem., 2015, 290, 1364–1373 CrossRef PubMed.
- G. A. Molnar, V. Nemes, Z. Biro, A. Ludany, Z. Wagner and I. Wittmann, Free Radical Res., 2005, 39, 1359–1366 CrossRef CAS PubMed.
- G. A. Molnar, Z. Wagner, L. Marko, S. T. Ko, M. Mohas, B. Kocsis, Z. Matus, L. Wagner, M. Tamasko, I. Mazak, B. Laczy, J. Nagy and I. Wittmann, Kidney Int., 2005, 68, 2281–2287 CrossRef CAS PubMed.
- C. E. Humphrey, M. Furegati, K. Laumen, L. La Vecchia, T. Leutert, J. C. D. Müller-Hartwieg and M. Vögtle, Org. Process Res. Dev., 2007, 11, 1069–1075 CrossRef CAS.
- A. Fan and S.-M. Li, Tetrahedron Lett., 2014, 55, 5199–5202 CrossRef CAS PubMed.
- L. Y. P. Luk and M. E. Tanner, J. Am. Chem. Soc., 2009, 131, 13932–13933 CrossRef CAS PubMed.
- M. Jost, G. Zocher, S. Tarcz, M. Matuschek, X. Xie, S.-M. Li and T. Stehle, J. Am. Chem. Soc., 2010, 132, 17849–17858 CrossRef CAS PubMed.
- U. Metzger, C. Schall, G. Zocher, I. Unsöld, E. Stec, S.-M. Li, L. Heide and T. Stehle, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 14309–14314 CrossRef CAS PubMed.
- E. Stec, N. Steffan, A. Kremer, H. Zou, X. Zheng and S.-M. Li, ChemBioChem, 2008, 9, 2055–2058 CrossRef CAS PubMed.
- A. B. Woodside, Z. Huang and C. D. Poulter, Org. Synth., 1988, 66, 211–215 CrossRef CAS.
- S. Berger and S. Braun, 200 and More NMR Experiments. A Practical Course, Wiley-VCH, Weinheim, Germany, 2004 Search PubMed.
- U. T. Bornscheuer and R. J. Kazlauskas, Angew. Chem., Int. Ed., 2004, 43, 6032–6040 CrossRef CAS PubMed.
- M. Hohne and U. T. Bornscheuer, Angew. Chem., Int. Ed., 2014, 53, 1200–1202 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ob01040c |
| This journal is © The Royal Society of Chemistry 2015 |