Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Lysosome targeting fluorescence probe for imaging intracellular thiols†
Dnyaneshwar
Kand‡
a,
Tanmoy
Saha‡
a,
Mayurika
Lahiri
b and
Pinaki
Talukdar
*a
aDepartment of Chemistry, Indian Institute of Science Education and Research Pune, Pune 411008, India. E-mail: ptalukdar@iiserpune.ac.in
bDepartment of Biology, Indian Institute of Science Education and Research Pune, Pune 411008, India
First published on 25th June 2015
Abstract
A BODIPY-based fluorescence turn-on probe, exhibiting high selectivity and sensitivity towards intracellular thiols with excellent lysosomal localization is reported. The probe displayed fast response towards biothiols in aqueous solution. Localization of the probe in lysosome was demonstrated by intracellular colocalization studies with the aid of LysoSensor Green.
Cysteine (Cys), homocysteine (Hcy) and glutathione (GSH) are three important low molecular weight thiol biomolecules (biothiols) which perform various physiological functions essential for survival.1 Alterations in intracellular and plasma levels of biothiols are associated with various diseases and disorders,2e.g. abnormal levels of Cys result in hair depigmentation, edema, liver damage, slow growth in children etc.3 Hcy is known as a risk factor for cardiovascular and Alzheimer's diseases.4 GSH, a tripeptide is the most abundant intracellular nonprotein thiol which plays a critical role in controlling oxidative stress in order to maintain redox homeostasis,5 crucial for cell growth and function.6 Although, numerous fluorescent probes have been developed for selective detection of biothiols7–9 and cell imaging applications,7,10–13 probes specific for locating subcellular organelles in particular are rare.14–26 Fluorescent probes for detection of biothiols in mitochondria are reported.27,28
Lysosome is an important cell organelle that contains approximately 50 different degradative enzymes which are active at the acidic pH (pH = 4–6) of the compartment.29 The lysosomal membrane constitutes a physiological barrier between the lysosome matrix and the surrounding cytoplasm. The membrane's impermeability ensures the retention of both the lysosomal enzymes and their substrates within the lysosomes.30 It is believed that GSH may be involved in stabilizing lysosome membranes.31 Thiols facilitate intralysosomal proteolysis by reducing disulphide bonds.32 For example, Cys is an effective stimulator of albumin degradation in liver lysosomes.31 For better understanding of the role of lysosomal thiols it is important to develop thiol selective fluorescent probes capable of targeting lysosomes.
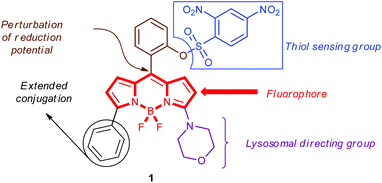
Herein, the design, synthesis and biothiol sensing properties of lysosome targeting fluorescence turn-on probe 1 are reported (Fig. 1). To obtain a photostable water soluble fluorescent thiol probe with excitation and emission wavelengths in the visible region, boron-dipyrromethene (BODIPY) was selected as the fluorophore.33 The necessary molecular decorations for thiol recognition and lysosome targeting were incorporated via the 2,4-dinitrobenzenesulfonyl (DNs) group and morpholine ring,34 respectively. As BODIPY-based chemosensors operate by perturbing the reduction potential of the meso-substituent,35 the DNs group was attached to the aryl group at the meso-position.36 Moreover, a phenyl ring at the 5-position provided extended conjugation resulting in excitation at longer wavelengths while maintaining a high quantum yield.37
Synthesis of probe 1 was carried out from salicylaldehyde in five steps (Scheme 1). Dipyrromethane 3 was synthesized from salicylaldehyde 2 and pyrrole in the presence of catalytic CF3COOH in 64% yield. Compound 3 was dibrominated with two equivalents of N-bromosuccinimide in tetrahydrofuran at −78 °C and oxidized with DDQ in dichloromethane followed by the addition of BF3·Et2O and Et3N which afforded the dibromo-BODIPY 4 in 60% yield. Reaction of 4 with morpholine resulted in BODIPY derivative 5 in 80% yield which on subsequent Suzuki coupling reaction with phenyl boronic acid provided compound 6 in 79% yield. Compound 6 upon treatment with 2,4-dinitrobenzenesulfonyl chloride provided probe 1 in 91% yield. Apart from the spectroscopic characterization of all compounds, single crystal X-ray diffraction structures for compound 5 and probe 1 were also recorded. Crystal structure analysis of probe 1 was useful to confirm the relative spatial arrangement between BODIPY and DNs moieties (Fig. S2†).
To validate the fluorescence turn-on nature of sensing, the photophysical properties of compounds 1 and 6 were investigated in aqueous HEPES buffer solution (10 mM, pH 7.4, 1 mM CTAB). The absorption spectrum of probe 1 exhibited λmax = 510 nm with a molar extinction coefficient ε = 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 M−1 cm−1 (see Fig. S3†). The probe displayed very weak fluorescence (λex = 510 nm) and quantum yield, Φ = 0.0026 (standard: Rhodamine G, Φ = 0.76 in water).38 Compound 6 exhibited an absorption band centred at λmax = 515 nm with a molar extinction coefficient ε = 23
600 M−1 cm−1 (see Fig. S3†). The probe displayed very weak fluorescence (λex = 510 nm) and quantum yield, Φ = 0.0026 (standard: Rhodamine G, Φ = 0.76 in water).38 Compound 6 exhibited an absorption band centred at λmax = 515 nm with a molar extinction coefficient ε = 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 966 M−1 cm−1. The fluorescence spectrum acquired for 6 indicated a strong fluorescence emission centred at λem = 584 nm (λex = 510 nm) and Φ = 0.17 (standard: Rhodamine G, Φ = 0.76 in water). This photophysical data satisfies the criteria of probe 1 to act as an efficient fluorescent turn-on probe. Reactivity of the probe 1 (10 μM) towards n-BuNH2, Cys, GSH and Hcy (100 μM) was determined by fluorescence emission kinetics in HEPES buffer (10 mM, 1 mM CTAB, pH = 7.4). In each experiment, the emission intensity at λ = 585 nm (λex = 510 nm) was recorded with time (Fig. 2A). Addition of n-BuNH2 to probe 1 did not indicate the formation of fluorescent species during the reaction. A pronounced fluorescence intensity increase up to ∼64-fold was obtained within 1 min after the addition of Cys (t1/2 = 6.4 s with a pseudo first order rate constant, k = 0.108 s−1), Hcy (t1/2 = 14.49 s and k = 0.0478 s−1) and GSH (t1/2 = 7.47 s and k = 0.0928 s−1).
966 M−1 cm−1. The fluorescence spectrum acquired for 6 indicated a strong fluorescence emission centred at λem = 584 nm (λex = 510 nm) and Φ = 0.17 (standard: Rhodamine G, Φ = 0.76 in water). This photophysical data satisfies the criteria of probe 1 to act as an efficient fluorescent turn-on probe. Reactivity of the probe 1 (10 μM) towards n-BuNH2, Cys, GSH and Hcy (100 μM) was determined by fluorescence emission kinetics in HEPES buffer (10 mM, 1 mM CTAB, pH = 7.4). In each experiment, the emission intensity at λ = 585 nm (λex = 510 nm) was recorded with time (Fig. 2A). Addition of n-BuNH2 to probe 1 did not indicate the formation of fluorescent species during the reaction. A pronounced fluorescence intensity increase up to ∼64-fold was obtained within 1 min after the addition of Cys (t1/2 = 6.4 s with a pseudo first order rate constant, k = 0.108 s−1), Hcy (t1/2 = 14.49 s and k = 0.0478 s−1) and GSH (t1/2 = 7.47 s and k = 0.0928 s−1).
Quantitative turn-on response of probe 1 towards Cys was examined by fluorometric titrations. Sharp enhancements in the fluorescence intensity (at λem = 585 nm) were observed (Fig. 2B) when titrations were carried out by addition of increasing concentrations of Cys (0, 2, 4, 6, 8, 10, 20 and 40 μM) to the probe 1 (10 μM) in HEPES buffer (10 mM, 1 mM CTAB, pH = 7.4). An equilibration time of 1 min was given after each addition of Cys to ensure the completion of the reaction. When fluorescence intensities at 585 nm were plotted against concentrations of Cys, good linear correlation (regression factor, R = 0.9686) was observed up to one equivalent of the Cys added (Fig. S6A†). A detection limit of 8.2 nM was calculated for probe 1, based on the signal to noise ratio, S/N = 3. Sensing of Cys by 1 was also associated with the change in color from orchid to hot pink under ambient light (Fig. 2C) and excitation under a hand held UV-lamp (λex = 365 nm) resulted in the appearance of orange fluorescence (Fig. 2D).
We have already stated that the lysosomal pH ranges between 4 and 6.39 As a lysosome targeting probe for biothiols, the molecule should also respond to biothiols in the pH range of lysosome. To verify this, fluorescence intensities (at 585 nm) of probe 1 (10 μM) and compound 6 (10 μM) were individually recorded in phosphate buffer (10 mM, 1 mM CTAB) at pH values 4, 5, 6, 7 and 8 (Fig. 3A). Simultaneously the sensing activity of 1 (10 μM) was monitored at pH = 4–8 by treating with Cys (100 μM) in phosphate buffer (10 mM, 1 mM CTAB) for 10 min. From this study, response of the probe towards Cys at pH lower than the physiological pH was observed, and fluorescence enhancements in the pH range of 4–8 were also comparable. Encouraged by these results, reactivity of the probe 1 (10 μM) towards Cys (100 μM) at pH 5 was monitored in phosphate buffer (10 mM, 1 mM CTAB, pH = 5). For the free probe, no fluorescence intensity enhancement λ = 585 nm (λex = 510 nm) was observed indicating the stability of the compound under acidic pH (Fig. 3B). However, a sharp enhancement of fluorescence intensity up to ∼95-fold was obtained within 8 min after the addition of Cys. From this reaction kinetics, k = 0.00553 s−1 and t1/2 = 125.3 s were calculated. The slower rate in comparison with pH 7.4 can be rationalized with the lower nucleophilicity of the thiol group of Cys. Moreover, quantitative turn-on response of probe 1 towards Cys in phosphate buffer (10 mM, 1 mM CTAB, pH = 5) was also examined by fluorometric titrations. Significant enhancements in fluorescence intensity (at λem = 585 nm) were observed when titrations were carried out by adding increasing concentrations of Cys (0, 2, 4, 6, 8, 10, 20, 40 and 100 μM) into the probe 1 (10 μM) at this pH (Fig. 3C). An equilibration time of 8 min was given after each addition of Cys to ensure the completion of the reaction. When fluorescence intensities at 585 nm were plotted against concentrations of Cys, a linear correlation (regression factor, R2 = 0.9787) was observed up to one equivalent of the Cys added (Fig. S6B†). A detection limit of 95.7 nM was calculated for the probe 1, based on the signal to noise ratio, S/N = 3.
To prove the formation of compound 6 during the thiol sensing, HPLC studies were carried out using a gradient method using CH3CN and H2O as eluents (for detailed information see the ESI†). A HPLC chromatograph of pure probe 1 provided the retention time, tR = 17.56 min, while tR = 16.34 min was obtained for compound 6 (Fig. 4). Compound 1 upon treatment with Cys (0.5 and 1.0 equiv.) clearly showed consumption of the probe and formation of 6 (Fig. 4). MALDI-TOF analysis of the isolated compound corresponding to tR = 16.34 min showed m/z = 445.13 confirming the formation of compound 6 (Fig. S9†).
 | ||
| Fig. 4 HPLC chromatographs of probe 1 (10 μM) upon reaction with Cys (0.5 and 1.0 equivalent) recorded in a gradient solvent system of CH3CN and H2O. | ||
In the next stage, the selectivity of probe 1 towards biological thiols was examined under physiological pH. In each case, probe 1 (10 μM) was treated separately with 100 μM of each analyte (either of Ala, Arg, His, NaCl, Na2SO4, NaSCN, NaNO3, BuNH2, Ser, GSH, Cys and Hcy) in HEPES buffer (10 mM, 1 mM CTAB, pH = 7.4) and fluorescence spectra (λex = 510 nm) were recorded after 5 minutes at room temperature. No significant fluorescence enhancement was observed for non-thiol analytes (Fig. 5). Treatment of probe 1 with GSH, Cys and Hcy under identical conditions provided strong fluorescence enhancements in the range of 54–63-fold. Comparable fluorescence enhancements were observed upon addition of Cys to the solutions pre-treated with non-thiol analytes. This study demonstrates the effectiveness of compound 1 as a selective probe for biothiols under competitive environments.
Based on the aforementioned outcome, localization of probe 1 in the lysosome and its ability to sense biological thiols in living cells were examined. First, the cell permeability and intracellular thiol sensing ability of probe 1 were evaluated by live-cell imaging of the human cervical cancer cell line (HeLa). Strong fluorescence was observed when HeLa cells were incubated with probe 1 (10 μM in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 DMSO–DNEM v/v, pH = 7.4) at 37 °C for 10 min (Fig. 6B and E). These cells then incubated with commercially available lysosome specific dye-LysoSensor Green (1.0 μM) showed green fluorescence (Fig. 6D). A colocalization image of green and red channels shows the localization of probe 1 in lysosomes (Fig. 6F). In the control experiment, cells were pre-treated with an excess (5 mM) of the thiol-reactive N-phenylmaleimide and then incubated with probe 1. The confocal microscopic studies did not show the fluorescence signal (Fig. 6C). This confirms the specificity of probe 1 for thiols over other analytes in living cells.
100 DMSO–DNEM v/v, pH = 7.4) at 37 °C for 10 min (Fig. 6B and E). These cells then incubated with commercially available lysosome specific dye-LysoSensor Green (1.0 μM) showed green fluorescence (Fig. 6D). A colocalization image of green and red channels shows the localization of probe 1 in lysosomes (Fig. 6F). In the control experiment, cells were pre-treated with an excess (5 mM) of the thiol-reactive N-phenylmaleimide and then incubated with probe 1. The confocal microscopic studies did not show the fluorescence signal (Fig. 6C). This confirms the specificity of probe 1 for thiols over other analytes in living cells.
Next, the colocalization experiments were performed by co-staining HeLa cells with LysoSensor Green and probe 1 to determine the location of fluorescence emission. When HeLa cells were incubated with probe 1 (5 μM in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 DMSO–DNEM v/v, pH = 7.4) at 37 °C for 10 min, a strong red fluorescence was observed (Fig. 7A). These cells were then incubated with LysoSensor Green (1 μM in 1
100 DMSO–DNEM v/v, pH = 7.4) at 37 °C for 10 min, a strong red fluorescence was observed (Fig. 7A). These cells were then incubated with LysoSensor Green (1 μM in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 DMSO–DNEM v/v, pH = 7.4) at 37 °C for 10 min and showed green fluorescence (Fig. 7B). As seen in Fig. 7C the fluorescence image of probe 1 was mainly overlapped with that of LysoSensor Green indicating the ability of probe 1 to target lysosomes. The intensity profiles of the linear regions of interest (ROI) across HeLa cells stained with probe 1 and LysoSensor Green vary in close synchrony (Fig. 7D). Pearson's coefficient and overlap coefficient are 0.963 and 0.984, respectively; evaluated using the conventional dye overlay method. Overlap coefficients k1 and k2 were found to be 0.9 and 1.1 respectively. Colocalization coefficients (Manders’ coefficients) M1 = 0.77 (fraction of LysoSensor Green overlapping probe) and M2 = 0.818 (fraction of probe overlapping LysoSensor Green) also confirm an excellent overlap. An intensity correlation analysis (ICA) is employed to assess the intensity distribution of the two co-existing dyes. The pixel intensity of the LysoSensor Green was plotted against that of the probe 1 (Fig. 7E). The dependent staining results in a highly correlated plot, and the ICA plots for the two stains generate an unsymmetrical hourglass-shaped scatter plot that is markedly skewed toward positive values (Fig. 7F and G). Li's intensity correlation quotient (ICQ) for the two dyes is 0.459, very close to 0.5, suggesting that the staining intensities are dependent on each other.
100 DMSO–DNEM v/v, pH = 7.4) at 37 °C for 10 min and showed green fluorescence (Fig. 7B). As seen in Fig. 7C the fluorescence image of probe 1 was mainly overlapped with that of LysoSensor Green indicating the ability of probe 1 to target lysosomes. The intensity profiles of the linear regions of interest (ROI) across HeLa cells stained with probe 1 and LysoSensor Green vary in close synchrony (Fig. 7D). Pearson's coefficient and overlap coefficient are 0.963 and 0.984, respectively; evaluated using the conventional dye overlay method. Overlap coefficients k1 and k2 were found to be 0.9 and 1.1 respectively. Colocalization coefficients (Manders’ coefficients) M1 = 0.77 (fraction of LysoSensor Green overlapping probe) and M2 = 0.818 (fraction of probe overlapping LysoSensor Green) also confirm an excellent overlap. An intensity correlation analysis (ICA) is employed to assess the intensity distribution of the two co-existing dyes. The pixel intensity of the LysoSensor Green was plotted against that of the probe 1 (Fig. 7E). The dependent staining results in a highly correlated plot, and the ICA plots for the two stains generate an unsymmetrical hourglass-shaped scatter plot that is markedly skewed toward positive values (Fig. 7F and G). Li's intensity correlation quotient (ICQ) for the two dyes is 0.459, very close to 0.5, suggesting that the staining intensities are dependent on each other.
The cytotoxicity of probe 1 was determined by the MTT assay.
Various concentrations of probe 1 (5, 10, 20 and 50 μM) were used to determine the toxicity level of the probe towards HeLa cells. The result revealed that cells were not affected by incubation with probe 1 (up to 10 μM) for 2 h as about 95% cell viability was determined at 10 μM concentration of probe 1 (Fig. S8†).
Conclusions
In summary, we have developed a lysosome targeting BODIPY-based fluorescence turn-on probe for rapid, selective and sensitive detection of biothiols. At pH 7.4, the probe displayed a very fast reaction with biothiols such as Cys, Hcy and GSH. Reaction of the probe with Cys provided the pseudo first order rate constant, k = 0.108 s−1 and t1/2 = 6.4 s. The reaction also provided an ∼64-fold fluorescence enhancement within 1 min of reaction. The probe was also reactive towards the biothiol at pH 5 however, with a lower value of k = 0.00553 s−1 and longer t1/2 = 125.3 s. At this pH, a sharp enhancement of fluorescence intensity up to ∼95-fold was obtained after 8 min of Cys addition. Live-cell imaging studies of HeLa cells confirmed the cell permeability, lysosome specificity and intracellular biothiol detection ability of the probe. The MTT assay disclosed about 95% cell viability at 10 μM concentration of the probe.Acknowledgements
We acknowledge IISER Pune and DST-SERB (Grant No. SR/S1/OC-65/2012) for financial support. D.K. thanks CSIR and T.S. thanks UGC for research fellowship.Notes and references
- S. Zhang, C.-N. Ong and H.-M. Shen, Cancer Lett., 2004, 208, 143–153 CrossRef CAS PubMed.
- J. Nourooz-Zadeh, in Biothiols in Health and Disease, ed. L. Packer and E. Cadenas, 1996 Search PubMed.
- S. Shahrokhian, Anal. Chem., 2001, 73, 5972–5978 CrossRef CAS.
- H. Refsum, P. M. Ueland, O. Nygard and S. E. Vollset, Annu. Rev. Med., 1998, 49, 31–62 CrossRef CAS PubMed.
- Z. A. Wood, E. Schroder, J. Robin Harris and L. B. Poole, Trends Biochem. Sci., 2003, 28, 32–40 CrossRef CAS.
- T. P. Dalton, H. G. Shertzer and A. Puga, Annu. Rev. Pharmacol. Toxicol., 1999, 39, 67–101 CrossRef CAS PubMed.
- H. S. Jung, X. Chen, J. S. Kim and J. Yoon, Chem. Soc. Rev., 2013, 42, 6019–6031 RSC.
- S. Sreejith, K. P. Divya and A. Ajayaghosh, Angew. Chem., Int. Ed., 2008, 47, 7883–7887 CrossRef CAS PubMed.
- X. Zhang, X. Ren, Q.-H. Xu, K. P. Loh and Z.-K. Chen, Org. Lett., 2009, 11, 1257–1260 CrossRef CAS PubMed.
- X. Chen, Y. Zhou, X. Peng and J. Yoon, Chem. Soc. Rev., 2010, 39, 2120–2135 RSC.
- L.-L. Yin, Z.-Z. Chen, L.-L. Tong, K.-H. Xu and B. Tang, Fenxi Huaxue, 2009, 37, 1073–1081 CAS.
- L. Zhu, Z. Yuan, J. T. Simmons and K. Sreenath, RSC Adv., 2014, 4, 20398–20440 RSC.
- H. Li, J. Fan and X. Peng, Chem. Soc. Rev., 2013, 42, 7943–7962 RSC.
- X. Chen, S.-K. Ko, M. J. Kim, I. Shin and J. Yoon, Chem. Commun., 2010, 46, 2751–2753 RSC.
- D. Kand, A. M. Kalle, S. J. Varma and P. Talukdar, Chem. Commun., 2012, 48, 2722–2724 RSC.
- W. Lin, L. Long and W. Tan, Chem. Commun., 2010, 46, 1503–1505 RSC.
- Y.-Q. Sun, M. Chen, J. Liu, X. Lv, J.-f. Li and W. Guo, Chem. Commun., 2011, 47, 11029–11031 RSC.
- B. Tang, L. Yin, X. Wang, Z. Chen, L. Tong and K. Xu, Chem. Commun., 2009, 5293–5295 RSC.
- M. Zhang, M. Yu, F. Li, M. Zhu, M. Li, Y. Gao, L. Li, Z. Liu, J. Zhang, D. Zhang, T. Yi and C. Huang, J. Am. Chem. Soc., 2007, 129, 10322–10323 CrossRef CAS PubMed.
- J. Bouffard, Y. Kim, T. M. Swager, R. Weissleder and S. A. Hilderbrand, Org. Lett., 2008, 10, 37–40 CrossRef CAS PubMed.
- W. Sun, W. Li, J. Li, J. Zhang, L. Du and M. Li, Tetrahedron Lett., 2012, 53, 2332–2335 CrossRef CAS PubMed.
- D. Kand, A. M. Kalle and P. Talukdar, Org. Biomol. Chem., 2013, 11, 1691–1701 CAS.
- B. Zhu, X. Zhang, Y. Li, P. Wang, H. Zhang and X. Zhuang, Chem. Commun., 2010, 46, 5710–5712 RSC.
- J. Shao, H. Sun, H. Guo, S. Ji, J. Zhao, W. Wu, X. Yuan, C. Zhang and T. D. James, Chem. Sci., 2012, 3, 1049–1061 RSC.
- K. Sreenath, Z. Yuan, J. R. Allen, M. W. Davidson and L. Zhu, Chem. – Eur. J., 2015, 21, 867–874 CrossRef PubMed.
- D. Kim, G. Kim, S.-J. Nam, J. Yin and J. Yoon, Sci. Rep., 2015, 5, 1–8 Search PubMed.
- S. Singha, D. Kim, A. S. Rao, T. Wang, K. H. Kim, K.-H. Lee, K.-T. Kim and K. H. Ahn, Dyes Pigm., 2013, 99, 308–315 CrossRef CAS PubMed.
- C. S. Lim, G. Masanta, H. J. Kim, J. H. Han, H. M. Kim and B. R. Cho, J. Am. Chem. Soc., 2011, 133, 11132–11135 CrossRef CAS PubMed.
- H. Zhu, J. Fan, Q. Xu, H. Li, J. Wang, P. Gao and X. Peng, Chem. Commun., 2012, 48, 11766–11768 RSC.
- C. L. Andrew, A. R. Klemm and J. B. Lloyd, Biochim. Biophys. Acta, 1997, 1330, 71–82 CrossRef CAS.
- J. L. Mego, Biochem. J., 1984, 218, 775–783 CAS.
- B. Arunachalam, U. T. Phan, H. J. Geuze and P. Cresswell, Proc. Nat. Acad. Sci. U. S. A., 2000, 97, 745–750 CrossRef CAS.
- N. Boens, V. Leen and W. Dehaen, Chem. Soc. Rev., 2012, 41, 1130–1172 RSC.
- T. Liu, Z. Xu, D. R. Spring and J. Cui, Org. Lett., 2013, 15, 2310–2313 CrossRef CAS PubMed.
- A. Loudet and K. Burgess, Chem. Rev., 2007, 107, 4891–4932 CrossRef CAS PubMed.
- H. Guo, Y. Jing, X. Yuan, S. Ji, J. Zhao, X. Li and Y. Kan, Org. Biomol. Chem., 2011, 9, 3844–3853 CAS.
- C. Thivierge, R. Bandichhor and K. Burgess, Org. Lett., 2007, 9, 2135–2138 CrossRef CAS PubMed.
- J. Olmsted, III, J. Phys. Chem., 1979, 83, 2581–2584 CrossRef.
- K. A. Christensen, J. T. Myers and J. A. Swanson, J. Cell Sci., 2002, 115, 599–607 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental procedures, crystal structure parameters, analytical and spectroscopic data. CCDC 980654 and 980655. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c5ob00889a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2015 |