Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A structure activity-relationship study of the bacterial signal molecule HHQ reveals swarming motility inhibition in Bacillus atrophaeus†
F. Jerry
Reen
a,
Rachel
Shanahan
b,
Rafael
Cano
b,
Fergal
O'Gara
*ac and
Gerard P.
McGlacken
*b
aBIOMERIT Research Centre, Department of Microbiology, University College Cork, Ireland. E-mail: f.ogara@ucc.ie; Fax: +353 21 4903101
bDepartment of Chemistry and Analytical & Biological Chemistry Research Facility (ABCRF), University College Cork, Ireland. E-mail: g.mcglacken@ucc.ie; Fax: +353 21 4274097
cSchool of Biomedical Sciences, Curtin University, Perth, WA 6845, Australia
First published on 16th April 2015
Abstract
The sharp rise in antimicrobial resistance has been matched by a decline in the identification and clinical introduction of new classes of drugs to target microbial infections. Thus new approaches are being sought to counter the pending threat of a post-antibiotic era. In that context, the use of non-growth limiting small molecules, that target virulence behaviour in pathogens, has emerged as a solution with real clinical potential. We have previously shown that two signal molecules (HHQ and PQS) from the nosocomial pathogen Pseudomonas aeruginosa have modulatory activity towards other microorganisms. This current study involves the synthesis and evaluation of analogues of HHQ towards swarming and biofilm virulence behaviour in Bacillus atrophaeus, a soil bacterium and co-inhibitor with P. aeruginosa. Compounds with altered C6–C8 positions on the anthranilate-derived ring of HHQ, display a surprising degree of biological specificity, with certain candidates displaying complete motility inhibition. In contrast, anti-biofilm activity of the parent molecule was completely lost upon alteration at any position indicating a remarkable degree of specificity and delineation of phenotype.
Introduction
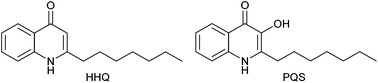
Quinolones are fused heterocyclic compounds that form the active structure of a wide range of potent broad-spectrum antimicrobial agents.1 In fact, fluoroquinolones are the most successful non-natural product class of antibiotics.2 Another important class of quinolones are the 2-alkyl-4-quinolones (AHQs) which are used as quorum sensing (QS) signaling molecules by pathogenic Gram-negative bacteria, including Pseudomonas aeruginosa,3,4 and certain Burkholderia,5 and Alteromonas6 strains. AHQ signaling pathways in these species have been shown to control production of multiple virulence determinants.7–9 This includes biofilm formation10,11 which is a structured community of bacterial cells enclosed in a self-produced polymeric matrix adhering to an inert or living surface.12,13 This mode of growth is particularly resistant to phagocytosis by white blood cells and to antibodies and antibiotics compared to planktonic cells.13 In multi-drug resistant bacteria, biofilms play a key role in allowing the pathogen to overcome host defences and contribute to its virulence. The primary autoinducers of the P. aeruginosa AHQ signalling pathway are 2-heptyl-4-quinolone (referred to as the Pseudomonas quinolone signal, or PQS) and its biological precursor 2-heptyl-4-quinolone (HHQ) (Fig. 1)11 and PQS has been shown to mediate biofilm formation in P. aeruginosa.7Bacillus atrophaeus (also named B. globigii and previously B. subtilis) is a Gram positive bacterium which co-inhabits the soil environment with P. aeruginosa.14 It is indistinguishable from the typed B. subtilis strain except for the production of pigment, a feature that marks the organism as particularly useful in the phenotypic analysis described here. Bacillus subtilis species are an excellent and well-utilised model system for Gram positive bacteria, and a cross-species influence with P. aeruginosa has been shown.15–17 In light of the co-existence of both B. atrophaeus and P. aeruginosa in the soil, it is perhaps unsurprising that communication mechanisms between both organisms would exist and this type of community dynamic and microbiome has received considerable attention.18 However, the extent of these networks and molecular mechanisms remain to be ascertained. From a microbial control point of view, suppression of key virulence phenotypes in bacteria, independent of growth inhibition, should be less susceptible to the build-up of resistance than with traditional antibiotic treatment as metabolic processes and bacterial growth are not directly targeted.
The SAR analysis presented in this study represents the preliminary steps in delineating the action of AHQs towards Gram positive bacteria, potentially providing a platform for future therapeutic developments.
Results and discussion
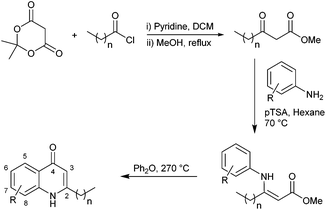
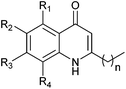
In light of the important roles played by PQS and HHQ in virulence, both molecules have been the subject of a number of structure–activity relationship (SAR) analyses in recent years.9,15,16,19 A study carried out by Hodgkinson's group showed evidence that the PQS regulator protein (PqsR), iron-chelating ability (specifically PqsR-independent production of pyoverdine) and membrane vesicle formation are altered upon changes to the PQS framework.19 For example, shortening or lengthening the C2 alkyl chain decreased activation of PqsR and substitution of the anthranilate ring compromised either the specific signalling, or membrane vesicle promotion. This indicates that there are multiple signalling mechanisms by which HHQ and PQS may possibly act. Antagonism of biofilm formation or any cross-species effects were not reported. A structure–activity investigation carried out by Reen et al. focused on substitution of the C-3 position.16 HHQ was shown to inhibit biofilm formation in species that inhabit the same environment as P. aeruginosa whereas PQS and 3-halo-analogues did not. Very recently Steinbach and Hartmann also reported ‘blocking’ the C-3 position thus preventing hydroxylation to a PQS analogue.20 Following on from their initial investigations, a simple quinolone was elegantly designed to target PqsR, and subsequently displayed antivirulence activity in vivo.In the current work, we generated a diverse range of novel HHQ analogues by using derivatized anthranilates and, in some cases, by extension of the alkyl chain at C-2 from seven to nine carbons (Table 1). HHQ analogues were prepared using a procedure similar to that described by Somanathan et al.,21 and us,22 involving a Conrad–Limpach type cyclisation. Thus β-keto-esters were prepared by acylation of Meldrum's acid (2,2-dimethyl-1,3-dioxane-4,6-dione) with octanoyl chloride or decanoyl chloride, followed by methanolysis (Scheme 1). Condensation with a variety of anilines in the presence of acid using a Dean–Stark apparatus and subsequent cyclisation of the enamine at reflux in diphenyl ether gave the corresponding 2-heptyl- (1–8) or 2-nonyl-4-quinolone (9–13). During the course of this work, an alternative method for accessing the enamine intermediate was also used.23 On occasion, this method provided a more efficient route. As mentioned earlier, B. atrophaeus was selected due to is co-existence with P. aeruginosa in the soil. Additionally, the organism is widely used as a surrogate for B. anthracis investigations in biodefense research,24 as well as being used as a sterilisation control strain in industry.25B. atrophaeus also exhibits strong swarming and biofilm phenotypes that are characteristic of the multicellular behaviour underpinning virulence in many pathogens.26 Firstly, growth of B. atrophaeus was investigated in the presence of HHQ, PQS and the suite of analogues. Prepared in honeycomb 100-well plates, in TSB supplemented with 10 μM of compound or equivalent volume of methanol as control, all readings were taken on a BioScreen Analyser.
| Compound | n | R1 | R2 | R3 | R4 | Yielda [%] |
|---|---|---|---|---|---|---|
| a % yields are isolated yields over two steps. | ||||||
| 1 | 6 | H | Me | H | H | 31 |
| 2 | 6 | H | n-Hex | H | H | 35 |
| 3 | 6 | H | H | –CH–CH–CH–CH– | 46 | |
| 4 | 6 | H | H | H | OMe | 12 |
| 5 | 6 | H | H | OMe | H | 33 |
| 6 | 6 | H | OMe | H | H | 34 |
| 7 | 6 | H | Cl | H | H | 21 |
| 8 | 6 | H | H | H | Cl | 19 |
| 9 | 8 | H | H | H | H | 23 |
| 10 | 8 | H | OMe | H | H | 28 |
| 11 | 8 | H | F | H | H | 16 |
| 12 | 8 | H | H | –CH–CH–CH–CH– | 51 | |
| 13 | 8 | H | n-Hex | H | H | 16 |
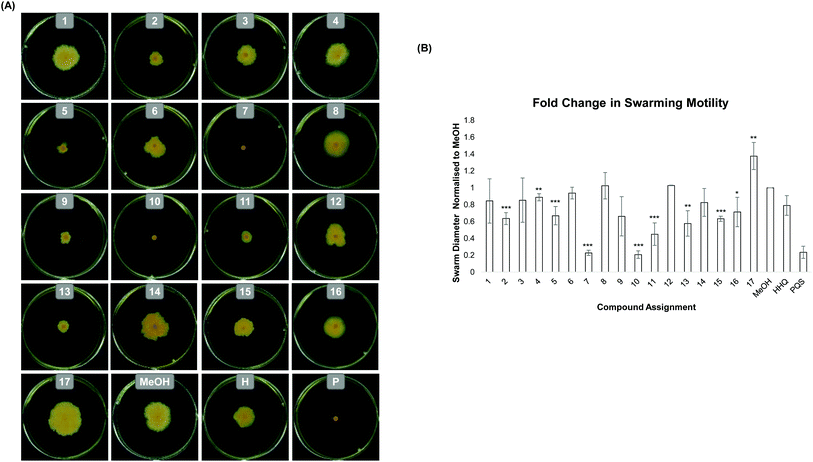
Growth of B. atrophaeus was not significantly altered in the presence of any of the modified analogues (SM Fig. 1) and thus any virulence effects were likely to be growth independent. Swarming motility is a key multicellular behaviour in bacteria, considered a virulence phenotype in many organisms27 and underpinned by both intra and intercellular signalling. In the case of B. atrophaeus, swarming cells are joined in chain formations and the phenotype is promoted by the secretion of surfactant ahead of the swarm front.28,29 Previously, we showed that addition of PQS to swarm plates completely abolished swarming motility in B. atrophaeus, while addition of HHQ led to little reduction in this key virulence phenotype.15 However in the study reported herein, addition of HHQ analogues (1–13) to swarming media led to a significant alteration in activity compared to the parent compound HHQ. Remarkably, some analogues exhibited PQS like inhibition, while others had the effect of promoting swarming activity in the model organism (Fig. 2). Of the compounds tested, compounds 1, 3, 4, 6, 8, 12 and 14 led to a phenotype statistically similar to that observed with the carrier control MeOH. Quinolones 2, 5, 9, 11 and 13 gave a moderate decrease in swarming, whereas quinolones 7 and 10 completely abrogated swarming activity in B. atrophaeus as shown in Fig. 2. Overall substitution at the 6-position afforded the most response. In compounds with a C-2 heptyl chain, an electron withdrawing group (7) gave a dramatic decrease in swarming. Interestingly, this is consistent with Hartmann's observation, where a nitro group at the 6-position gave antagonistic behaviour in P. aeruginosa.20 When we prepared an analogue possessing an electron releasing group (6) no decrease in swarming was observed. Remarkably, extension of the C-2 alkyl chain in the non-inhibitory analogue, afforded a molecule (10) capable of returning anti-motility behaviour.30
4-Quinolones such as HHQ exist as two interconverting tautomeric forms (a 4-quinolone and a 4-hydroxyquinoline) and equilibrium tends towards the quinolone form under physiological conditions.31 We were intrigued to delineate these two structural isomers and elucidate if either structural form maintained activity if ‘frozen out’ by methylation at oxygen or nitrogen.
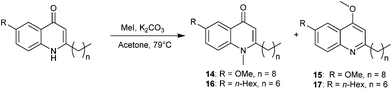
Preparation of the N- and O-methylated compounds was carried out by reaction of quinolone substrate 10 with methyl iodide in acetone and K2CO3. The resulting isomeric mixture was purified by column chromatography to afford the desired products 14 (quinolone) and 15 (quinoline) in 14 and 42% respectively (Scheme 2). When tested on B. atrophaeus, both compounds had lost anti-swarming activity (Fig. 2) relative to the parent quinolone (10) but the quinoline form 15 did show a slight enhancement of anti-swarming activity relative to quinolone 14. We also chose to carry out a similar investigation using quinolone 2, which displayed enhanced suppression of swarming motility relative to MeOH (and HHQ itself). In this case the N–Me derivative 16 (isolated in 10%) again showed little or no variation. However, addition of the O–Me variant 17 (isolated in 37%) led to significant enhancement of swarming motility, in direct contrast to the parent molecule.
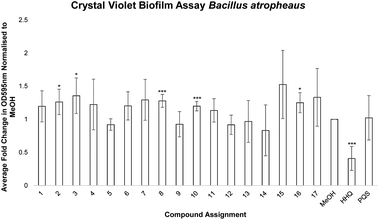
Having established the influence of the analogues on swarming motility, it was expected that a similar profile would emerge from an investigation of their impact on biofilm formation. Swarming motility is associated with biofilm formation in many pathogens, sometimes in what is an inverse relationship. Therefore, each analogue was tested for its impact on pellicle formation and attachment in B. atrophaeus. Surprisingly, all analogues had lost their anti-pellicle and anti-biofilm formation activity with respect to the parent molecule HHQ. Modification of any part of the quinolone framework led to loss of activity, even for those where suppression of swarming motility was enhanced (Fig. 3).
Conclusions
In this study we prepared a number of analogues of known P. aeruginosa signalling molecule HHQ, present in complex multi-bacterial and multi-kingdom environments, and tested them for anti-swarming and anti-biofilm activity towards B. atrophaeus. Several of the novel analogues show potent suppression of swarming motility, which suggests some degree of freedom regarding the structural interaction between the quinolone core molecule and the receiving organism. However, the lack of anti-biofilm activity exhibited by all the analogues would suggest that the anti-biofilm activity exhibited by HHQ is highly sensitive to structural modification and may reflect a highly specific protein–ligand interaction.Previously, HHQ itself has shown very minor anti-swarming activity in a range of organisms in contrast to PQS, which completely abolished swarming activity.15 Strikingly, modification of the quinolone backbone structure described here, resulted in quinolones possessing potent suppression of swarming, similar to that with PQS. While there is no evidence of a pqsH type-activity encoded in a genome outside of P. aeruginosa, and thus no known mechanism for the hydroxylation of our HHQ analogues to PQS analogues,15 other biosynthetic steps could be at play and our investigations in this area will be published in due course.
Acknowledgements
The authors would like to thank David Woods for technical assistance. This research was supported in part by grants awarded to GMG by Science Foundation of Ireland (SFI) (SFI/12/TIDA/B2405, SFI/12/IP/1315, SFI/09/RFP/CHS2353 and SSPC2 12/RC/2275) and the Irish Research Council and to FOG (SFI: SSPC2 12/RC/2275; 13-TIDA-B2625; 07/ IN.1/B948; 12/TIDA/B2411; 12/TIDA/B2405; 09/RFP/BMT 2350); the Department of Agriculture, Fisheries and Food (DAFF11/F/009 MabS; FIRM/RSF/CoFoRD; FIRM 08/RDC/629); the Environmental Protection Agency (EPA 2008-PhD/S-2); the Irish Research Council for Science, Engineering and Technology (GOIPG/2014/647; PD/2011/2414; RS/2010/2413); the European Commission (H20/20 EU-634486; FP7-PEOPLE-2013-ITN, 607786; OCEAN2012, 287589; FP7-KBBE-2012-6, CP-TP 311975; FP7-KBBE-2012-6, CP-TP-312184; Marie Curie 256596); the Marine Institute (Beaufort award C2CRA 2007/ 082); Teagasc (Walsh Fellowship 2013), the Health Research Board (HRA/2009/146) and the Irish Thoracic Society via the 2014 MRCG/HRB scheme (MRCG/2014/6).Notes and references
- D. C. Hooper and E. Rubinstein, Quinolone Antimicrobial Agents, ASM Press, Washington D.C., 2003 Search PubMed.
- D. O'Hagan, J. Fluorine Chem., 2010, 131, 1071 CrossRef.
- S. Swift, J. A. Downie, N. A. Whitehead, A. M. Barnard, G. P. Salmond and P. Williams, Adv. Microb. Physiol., 2001, 45, 199 CrossRef CAS PubMed.
- C. Fuqua, M. R. Parsek and E. P. Greenberg, Ann. Rev. Genet., 2001, 35, 439 CrossRef CAS PubMed.
- S. P. Diggle, P. Cornelis, P. Williams and M. Camara, Int. J. Med. Microbiol., 2006, 296, 83 CrossRef CAS PubMed.
- R. A. Long, A. Qureshi, D. J. Faulkner and F. Azam, Appl. Environ. Microbiol., 2003, 69, 568 CrossRef CAS PubMed.
- S. P. Diggle, K. Winzer, S. R. Chhabra, K. E. Worrall, M. Camara and P. Williams, Mol. Microbiol., 2003, 50, 29 CrossRef CAS PubMed.
- E. C. Pesci, J. B. J. Milbank, J. P. Pearson, S. McKnight, A. S. Kende, E. P. Greenberg and B. H. Iglewski, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 11229 CrossRef CAS.
- L. Mashburn-Warren, J. Howe, K. Brandenburg and M. Whiteley, J. Bacteriol., 2009, 191, 3411 CrossRef CAS PubMed.
- J. F. Dubern and S. P. Diggle, Mol. Biosyst., 2008, 4, 882 RSC.
- S. P. Diggle, S. Matthijs, V. J. Wright, M. P. Fletcher, S. R. Chhabra, I. L. Lamont, X. Kong, R. C. Hider, P. Cornelis, M. Camara and P. Williams, Chem. Biol., 2007, 14, 87 CrossRef CAS PubMed.
- J. W. Costerton, Int. J. Antimicrob. Agents, 1999, 11, 217 CrossRef CAS PubMed.
- J. W. Costerton, P. S. Stewart and E. P. Greenberg, Science, 1999, 284, 1318 CrossRef CAS PubMed.
- S. A. Burke, J. D. Wright, M. K. Robinson, B. V. Bronk and R. L. Warren, Appl. Environ. Microbiol., 2004, 70, 2786 CrossRef CAS PubMed.
- F. J. Reen, M. J. Mooij, L. J. Holcombe, C. M. McSweeney, G. P. McGlacken, J. P. Morrissey and F. O'Gara, FEMS Microbiol. Ecol., 2011, 77, 413 CrossRef CAS PubMed.
- F. J. Reen, S. L. Clarke, C. Legendre, C. M. McSweeney, K. S. Eccles, S. E. Lawrence, F. O'Gara and G. P. McGlacken, Org. Biomol. Chem., 2012, 10, 8903 CAS.
- Y. Tashiro, S. Ichikawa, T. Nakajima-Kambe, H. Uchiyama and N. Nomura, Microbes Environ., 2010, 25, 120 CrossRef PubMed.
- R. East, Nature, 2013, S18–S19, DOI:10.1038/501S18a.
- J. T. Hodgkinson, S. D. Bowden, W. R. Galloway, D. R. Spring and M. Welch, J. Bacteriol., 2010, 192, 3833 CrossRef CAS PubMed.
- C. Lu, C. K. Maurer, B. Kirsch, A. Steinbach and R. W. Hartmann, Angew. Chem., Int. Ed., 2014, 53, 1109 CrossRef CAS PubMed.
- R. Somanathan and K. M. Smith, J. Heterocycl. Chem., 1981, 18, 1077 CrossRef CAS.
- G. P. McGlacken, C. M. McSweeney, T. O'Brien, S. E. Lawrence, C. J. Elcoate, F. J. Reen and F. O'Gara, Tetrahedron Lett., 2010, 51, 5919 CrossRef CAS.
- A. V. Narsaiah, A. R. Reddy, B. V. S. Reddy and J. S. Yadav, Open Catal. J., 2011, 4, 43 CrossRef.
- S. R. Kane, S. E. Létant, G. A. Murphy, T. M. Alfaro, P. W. Krauter, R. Mahnke, T. C. Legler and E. Raber, J. Microbiol. Methods, 2009, 76, 278 CrossRef CAS PubMed.
- S. E. Létant, G. A. Murphy, T. M. Alfaro, J. R. Avila, S. R. Kane, E. Raber, T. M. Bunt and S. R. Shah, Appl. Environ. Microbiol., 2011, 77, 6570 CrossRef PubMed.
- S. T. Rutherford and B. T. Bassler, Cold Spring Harbor Perspect. Med., 2012, 2(11), 1, DOI:10.1101/cshperspect.a012427 . pii: a012427.
- D. E. Kearns, Nat. Rev. Microbiol., 2010, 8, 634 CrossRef CAS PubMed.
- K. Hamze, S. Autret, K. Hinc, S. Laalami, D. Julkowska, R. Briandet, M. Renault, C. Absalon, I. B. Holland, H. Putzer and S. J. Séror, Microbiol., 2011, 157, 2456 CrossRef CAS PubMed.
- S. Heeb, M. P. Fletcher, S. R. Chhabra, S. P. Diggle, P. Williams and M. Cámara, FEMS Microbiol. Rev., 2011, 35, 247 CrossRef CAS PubMed.
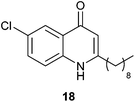
- One Referee suggested the synthesis and biological testing of compound 18 to delineate the requirement for alteration at the C6 position and the length of the alkyl chain. No anti-swarming activity was observed, further demonstrating the exquisite specificity of the quinolone structure. No anti-biofilm activity was observed.
. - P. K. Singh, A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. Welch and E. P. Greenberg, Nature, 2000, 407, 762 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ob00315f |
| This journal is © The Royal Society of Chemistry 2015 |