Open Access Article
Open Access ArticleA galactose-functionalized dendritic siRNA-nanovector to potentiate hepatitis C inhibition in liver cells†
Abirami
Lakshminarayanan
ab,
B. Uma
Reddy
b,
Nallani
Raghav
c,
Vijay Kumar
Ravi
c,
Anuj
Kumar
b,
Prabal K.
Maiti
*c,
A. K.
Sood
*c,
N.
Jayaraman
*a and
Saumitra
Das
*b
aDepartment of Organic Chemistry, Indian Institute of Science, Bangalore 560012, India. E-mail: jayaraman@orgchem.iisc.ernet.in
bDepartment of Microbiology and Cell Biology, Indian Institute of Science, Bangalore 560012, India. E-mail: sdas@mcbl.iisc.ernet.in
cDepartment of Physics, Indian Institute of Science, Bangalore 560012, India. E-mail: asood@physics.iisc.ernet.in; maiti@physics.iisc.ernet.in
First published on 28th September 2015
Abstract
A RNAi based antiviral strategy holds the promise to impede hepatitis C viral (HCV) infection overcoming the problem of emergence of drug resistant variants, usually encountered in the interferon free direct-acting antiviral therapy. Targeted delivery of siRNA helps minimize adverse ‘off-target’ effects and maximize the efficacy of therapeutic response. Herein, we report the delivery of siRNA against the conserved 5′-untranslated region (UTR) of HCV RNA using a liver-targeted dendritic nano-vector functionalized with a galactopyranoside ligand (DG). Physico-chemical characterization revealed finer details of complexation of DG with siRNA, whereas molecular dynamic simulations demonstrated sugar moieties projecting “out” in the complex. Preferential delivery of siRNA to the liver was achieved through a highly specific ligand–receptor interaction between dendritic galactose and the asialoglycoprotein receptor. The siRNA-DG complex exhibited perinuclear localization in liver cells and co-localization with viral proteins. The histopathological studies showed the systemic tolerance and biocompatibility of DG. Further, whole body imaging and immunohistochemistry studies confirmed the preferential delivery of the nucleic acid to mice liver. Significant decrease in HCV RNA levels (up to 75%) was achieved in HCV subgenomic replicon and full length HCV-JFH1 infectious cell culture systems. The multidisciplinary approach provides the ‘proof of concept’ for restricted delivery of therapeutic siRNAs using a target oriented dendritic nano-vector.
Introduction
Hepatitis C virus (HCV) is an important human pathogen primarily infecting the liver cells. The cascade of events caused by the entry of HCV positive strand RNA to hepatocytes includes Internal Ribosome Entry Site (IRES) mediated translation to produce an assortment of viral proteins, of which few are known to be involved in the virion assembly along the lipid droplets of ER.1,2 Virus entry into the cells is aided further by envelope proteins E1 and E2 interacting with hepatocytes.1 Detailed investigations to understand important events of HCV life cycle have allowed exploring various strategies against this virus. Several conventional approaches are reported to combat HCV infection which include (i) blocking the virus entry,3,4 (ii) inhibiting the interaction of essential host factors, such as, La, miR 122 with HCV RNA by use of peptides,5,6 miravirsin;6,7 to inhibit 5′ IRES mediated translation, (iii) inhibiting replication using protease or polymerase inhibitors, such as boceprevir, telaprevir and sofosbuvir8 and small molecules like punicalin, punicalagin,9 (iv) inhibiting virion assembly and release.10,11 However, interferon based therapy combined with serine protease inhibitors is prominent in current treatment against HCV. Despite high potency of different antivirals, the emergence of drug resistant variants of HCV is a major concern.12 In order to overcome drug resistance, alternate approaches have emerged in the past few years to inhibit viral translation and replication processes, by the use of antisense oligonucleotides,13,14 DNAzymes,15 and siRNAs16,17 and targeting the conserved 5′-untranslated region (UTR) of HCV.The use of siRNA to inhibit HCV in JFH-1 infectious cell culture showed viral clearance rates up to 80%.16 Further, the delivery of short hairpin RNA (shRNA), using Sendai virosome, showed specific inhibition of HCV IRES mediated translation in vivo. The membrane fusion glycoprotein of the virosome, having exposed galactose residues specifically targets hepatocytes and mediates localized delivery of the shRNA.14 A nanosome–siRNA formulation was also demonstrated recently to inhibit the viral replication.17 These studies exemplify the importance of siRNA mediated inhibition to combat HCV infection and highlight the advantages of targeted delivery.
Target specific delivery of siRNA assumes greater importance, especially in the in vivo systems, in order to enhance inhibitory activities, minimize off-target effects and avoid adverse effects on healthy organs. Although the use of pseudoviruses, virus-like particles, adenoviruses and virosomes was studied as delivery systems for antiviral agents inherent immunogenicity and virulence of these vectors raise concerns in their therapeutic developments.19 Non-viral vectors have emerged as efficient alternatives to deliver nucleic acids both in vitro and in vivo.19 A recent entry to the repertoire of non-viral vectors is the dendritic macromolecule or dendrimers, characterized by the presence of a branches-upon-branches structural motif.20 We have demonstrated earlier the efficient nucleic acid delivery and non-toxic properties of novel synthetic dendritic vectors, namely, the poly(propyl ether imine) (PETIM) dendrimers.21–23 In an effort to deliver antiviral siRNA against HCV specifically to liver cells, we undertook a study of the liver target-specific PETIM dendrimer as a delivery system. The target-specific nature of the dendrimer delivery system took advantage of the ligand–receptor interaction between a sugar ligand, namely, the galactoside ligand and its cognate asialoglycoprotein receptor (ASGPR), which is expressed abundantly in the liver cells.24 In the present study, a third generation galactose-functionalized PETIM dendrimer (DG) was synthesized and studied for its ability to interact with siRNA by biophysical techniques and molecular dynamic (MD) simulations. Immunofluorescence based confocal microscopy showed that the delivered siRNA co-localized with the HCV viral proteins NS5B and NS3 which participate in HCV replication. The study demonstrates, for the first time, that siRNA delivered using DG is localized in the perinuclear region which is the site for HCV replication. The target-specific PETIM dendrimer is found to deliver a chosen anti-HCV siRNA into the liver cells preferentially and inhibit HCV proliferation.
Methods
Synthesis of galactose functionalized PETIM dendrimer
Benzobromogalactose25 (0.50 g, 0.76 mmol) in CH2Cl2 (30 mL) was added to a solution of dendritic alcohol21,26 (0.050 g, 9.6 μmol), Ag2CO3 (0.20 g, 0.72 mmol) and AgClO4 (0.050 g, 0.085 mmol), stirred for 30 h at room temperature, filtered, filtrate concentrated in vacuo and purified by column chromatography (Al2O3, CH2Cl2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 95
MeOH = 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5). The resulting product in tetrahydrofuran
5). The resulting product in tetrahydrofuran![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH (1
MeOH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (10 mL) was added with NaOMe in MeOH (1 M) (0.02 mL), stirred at room temperature for 16 h, neutralized with an Amberlite ion-exchange (H+) resin, filtered, concentrated in vacuo and the resulting solution subjected to dialysis (MW cut-off 3.5 kDa) to afford DG, as a foamy solid. Yield: 0.057 g (57%, after two steps); 1H NMR (D2O, 400 MHz) δ 1.87–1.92 (br, 132 H, CH2–CH2–CH2), 3.15 (br, 132 H, N–CH2–CH2 and CH2–CH2–N), 3.37 (t, 24 H, J = 8.4 Hz,), 3.47–3.62 (br, 228 H), 3.62–3.80 (br, 72 H), 4.26 (d, 24 H, J = 4 Hz); 13C NMR (D2O, 100 MHz) δ 23.3, 31.3, 36.9, 60.9, 68.6, 70.0, 70.7, 72.8, 75.1, 102.7.
1) (10 mL) was added with NaOMe in MeOH (1 M) (0.02 mL), stirred at room temperature for 16 h, neutralized with an Amberlite ion-exchange (H+) resin, filtered, concentrated in vacuo and the resulting solution subjected to dialysis (MW cut-off 3.5 kDa) to afford DG, as a foamy solid. Yield: 0.057 g (57%, after two steps); 1H NMR (D2O, 400 MHz) δ 1.87–1.92 (br, 132 H, CH2–CH2–CH2), 3.15 (br, 132 H, N–CH2–CH2 and CH2–CH2–N), 3.37 (t, 24 H, J = 8.4 Hz,), 3.47–3.62 (br, 228 H), 3.62–3.80 (br, 72 H), 4.26 (d, 24 H, J = 4 Hz); 13C NMR (D2O, 100 MHz) δ 23.3, 31.3, 36.9, 60.9, 68.6, 70.0, 70.7, 72.8, 75.1, 102.7.
UV-visible spectroscopy
siRNA (1 μM) in 1X siRNA buffer (Dharmacon) was titrated with DG (25 mg mL−1) and the resultant solution equilibrated for 20 min before the UV-visible spectrum was recorded. Titrations were carried out up to a weight ratio (R) of 100 in a 10 mm stoppered quartz cuvette at a constant temperature of 25 °C. UV-Vis absorption spectra were recorded on a Perkin-Elmer Lambda35 spectrophotometer.Ethidium bromide displacement assay
A solution of siRNA (1 μM) in siRNA buffer was incubated for 10 min with EB at a molar ratio of 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 siRNA
1 siRNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EB. The fluorescence spectrum was recorded by exciting at 520 nm and recording the emission in the 535–800 nm region using a 10 mm quartz cuvette on a Varian Cary Eclipse Fluorimeter. The siRNA-EB solution was titrated with DG (25 mg mL−1) in siRNA buffer and the fluorescence spectrum recorded after 20 min equilibration.
EB. The fluorescence spectrum was recorded by exciting at 520 nm and recording the emission in the 535–800 nm region using a 10 mm quartz cuvette on a Varian Cary Eclipse Fluorimeter. The siRNA-EB solution was titrated with DG (25 mg mL−1) in siRNA buffer and the fluorescence spectrum recorded after 20 min equilibration.
Zeta potential measurements
Aqueous solutions of siRNA–dendrimer complexes at R were prepared by mixing siRNA (100 nM) with required amounts of DG (10 mg mL−1) in siRNA buffer and equilibrated at 25 °C for 45 min. Zeta potential and electrophoretic mobility measurements were carried out on a Brookhaven ZetaPals instrument at a controlled temperature of 25 ± 0.5 °C. Each data was taken as an average of three runs with each run comprising ten measurements. The zeta potentials of siRNA alone (100 nM) and dendrimer alone (193 μM) were also measured.Atomic force microscopy
DG (0.77 nM), siRNA (50 nM) and their complexes at R 1, 50 and 100 were prepared for AFM imaging by drop-casting 10 μL of each sample on mica, drying under vacuum and imaging using a VeecodiInnova AFM (Bruker, USA) under tapping mode in air. The resonance frequency of a tapping mode cantilever was set at 300 kHz and the scanning rate was fixed at 1 Hz. The z-height distributions were analyzed using Nanoscope Analysis software.Molecular dynamic simulations
The PETIM dendrimer was built using a dendrimer builder toolkit (DBT).27 The beta galactose structure was Gaussian optimized with the HF 6-31 g basis set and was added to the hydroxy terminated PETIM dendrimer to derive the galactose functionalized PETIM dendrimer. Using the antechamber module of Amber 12,28 the RESP charges are calculated and GAFF atom types are assigned for the dendrimer.29 ff99bsc030 with parmbsc031 correction was used to describe inter- and intra molecular interactions involving siRNA molecules. The solute (DG and siRNA) was solvated using the xleap module of Amber maintaining a buffer of 15 Å on all sides. The TIP3P water model was used for solvation.32 Of the non-protonated dendrimer and siRNA, only siRNA required counter-ions to charge neutralize the system. In non-protonated cases the system was charge neutralized with 44 Na+ ions. The system was then energy minimized for 1000 steps using the steepest descent method and further 2000 steps by the conjugate gradient method, while the solute was fixed using a harmonic constraint of 500 kcal mol−1 Å−2, to eliminate bad contacts with the solvent. Further 3000 steps of conjugate gradient minimization was carried out, reducing the harmonic constraint by 5 kcal mol−1 Å−2 at every 600 steps from 20 kcal mol−1 Å−2 to 0 kcal mol−1 Å−2. Subsequently, an equilibration run of 50 ps in the NPT ensemble was performed before going into a production run of 100 ns in the NPT ensemble.33–35 All the simulations were carried out using the PMEMD module of Amber12.36Cell culture
Cell monolayers were maintained in Dulbecco's modified Eagle's medium (DMEM) (Sigma Aldrich) supplemented with 10% heat-inactivated foetal bovine serum (FBS) (Invitrogen) and 1% penicillin–streptomycin at 37 °C in 5% CO2. Hygromycin B (25 mg/mL−1) was used as an additional supplement for stable maintenance of replicon 2a harbouring Huh 7 cell line.Transfection of siRNA–DG complexes in mammalian cells
1 × 105 cells were grown on sterilized 18 mm cover slips in a 12 well plate to 65% confluency. DG–siRNA complexes were prepared at various ratios using 50 nM siRNA and required amounts of dendrimers by incubating in Opti-MEM (Sigma Aldrich, India) for 45 min. The cells were incubated with the complexes in Opti-MEM for 6 h. After transfection, cells were replenished with complete growth medium. In the case of experiments where free sugars were used, a stock of 1 mM D-galactose or D-mannose was prepared by dissolving the appropriate amount of sugar in sterile double distilled water and the solution was filtered through 0.2 μm pores before use. Cells were pre-incubated with the free sugars in Opti-MEM for 20 min before the addition of the complexes for transfection.Confocal microscopy and immunofluorescence
Post transfection, cells were washed with PBS, fixed with 4% paraformaldehyde solution, permeated with 0.1% Triton X and the nuclei stained with DAPI. After blocking in 3% BSA (bovine serum albumin) solution for 40 min, cells were washed with PBS and mounted onto slides using glycerol. Fluorescent images were obtained by using excitation wavelengths of 350 nm, 488 nm and 657 nm (Zeiss Confocal microscope). Images were processed using ZEN10 software and quantified using Image J. Immunofluorescence was carried out by incubating transfected cells, with NS5B or NS3 primary antibody (Thermo Fisher, India) diluted to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 in 3% BSA for 2 h at 4 °C. Cells were then washed with PBS and incubated at room temperature with secondary antibodies at 1
200 in 3% BSA for 2 h at 4 °C. Cells were then washed with PBS and incubated at room temperature with secondary antibodies at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution (Alexa 488 tagged antirabbit antibody for NS5B and Alexa 657 tagged antimouse antibody for NS3) for 30 min.
500 dilution (Alexa 488 tagged antirabbit antibody for NS5B and Alexa 657 tagged antimouse antibody for NS3) for 30 min.
Real time PCR to assess HCV RNA levels14
Huh 7 cells harbouring the HCV were transfected with siHCV–DG complexes as described previously. At the required time, cells were harvested in TRIzol reagent (Sigma Aldrich), and total cellular RNA was extracted and reverse transcribed with the HCV forward primer and GAPDH reverse primer. The resulting cDNA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 diluted) was subjected to quantitative PCR using a SYBR green real-time assay mixture (Thermo Scientific).
10 diluted) was subjected to quantitative PCR using a SYBR green real-time assay mixture (Thermo Scientific).
In vitro transcription of JFH1 RNA
The HCV-pJFH1 construct was linearized with the restriction enzyme XbaI. The linear DNA was used as a template for in vitro transcription to synthesize JFH1 RNA using a Ribomax Large scale RNA production T7 kit (Promega).JFH1 replication inhibition assay37
Huh 7.5 cells were transfected with 25 nM and 50 nM of siHCV using DG. Four hours later, cells were transfected with HCV-JFH1 RNA using lipofectamine 2000 (Invitrogen). After 48 h and 72 h, cells were harvested for total cellular RNA isolation. The relative levels of HCV RNA were quantified by real time PCR.Haemolysis assay38
500 μL blood was drawn by ocular bleeding into microfuge tubes containing 50 μL of 6% EDTA as an anticoagulant. The whole blood was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm to separate plasma and leukocytes. 1.78 × 105 cells were suspended in 1 mL of PBS and required amounts of dendrimer (1 mg, 5 mg and 10 mg) solutions were added to the RBCs. Triton X (1% v/v) was added in order to induce 100% haemolysis. The suspensions were incubated in a 37 °C water bath. After incubation time, the RBC suspensions were centrifuged at 10
000 rpm to separate plasma and leukocytes. 1.78 × 105 cells were suspended in 1 mL of PBS and required amounts of dendrimer (1 mg, 5 mg and 10 mg) solutions were added to the RBCs. Triton X (1% v/v) was added in order to induce 100% haemolysis. The suspensions were incubated in a 37 °C water bath. After incubation time, the RBC suspensions were centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min, the supernatant collected and the absorbance recorded at 540 nm. Percentage haemolysis was calculated as follows:
000 rpm for 5 min, the supernatant collected and the absorbance recorded at 540 nm. Percentage haemolysis was calculated as follows:| % Haemolysis = (Absorbance of sample − Absorbance of control)/(Absorbance of Trition X − Absorbance of control)×100 |
In vivo toxicity and histopathology
The mice were divided into six groups of three mice each. Group 1 served as the control; mice were administered with 100 μL of sterile distilled water. Mice in groups 2 to 6 were injected with 100 μL DG at 50, 100, 200, 350 and 500 mg per kg body weight (b.w.). All mice were observed for morphological and behavioural responses, food–water intake, body weight changes and mortality for 14 days. Subsequently, they were sacrificed, organs were excised and fixed in 10% NBF (neutral buffered formalin) for 72 h. The tissues were then dehydrated in graded alcohol series of ethanol, mounted in paraffin blocks, sectioned to 5 μm thickness, deparaffinized, rehydrated and stained with haematoxylin–eosin, and examined using a Zeiss microscope.In vivo bioimaging and immunohistochemistry
Mice were injected with DG-HCVLucDNA or DG–HCVLuc–siRNA complexes via tail vein injection. Luciferase expression in different organs of the mice was tracked by using a Xenogen In Vivo Imaging System (IVIS) by intraperitoneal administation of luciferin (150 mg per kg b.w.). The mice were sacrificed on the 5th day post treatment, and tissue samples were isolated and processed as described above. The presence of luciferase proteins in tissues was detected using primary antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution, goat polyclonal antibody G7451, Promega) and horseradish peroxidase tagged secondary antibody (1
100 dilution, goat polyclonal antibody G7451, Promega) and horseradish peroxidase tagged secondary antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 dilution anti-goat IgG). The sections were treated with 3,3′-diaminobenzidine stained with haematoxylin and imaged.
200 dilution anti-goat IgG). The sections were treated with 3,3′-diaminobenzidine stained with haematoxylin and imaged.
Ethical clearance
All animal experiments were performed in compliance with the relevant laws and institutional guidelines and approved by the ‘Institutional Animal Ethics Committee’ (IAEC) of Indian Institute of Science. Healthy Balb/c mice (4–7 weeks old) were selected from the inbred colony and maintained under a 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 light
8 light![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) dark cycle at a temperature of 20 ± 1 °C. The animals were fed with food pellets and water ad libitum.
dark cycle at a temperature of 20 ± 1 °C. The animals were fed with food pellets and water ad libitum.
Results
Functionalization of PETIM dendrimers with D-galactose
The PETIM dendrimer constitutes a tertiary amine as the branching site, and an ether as the linker separating the branching sites by n-propyl moieties.26 The third generation nitrogen-cored dendrimer presenting 24 hydroxyl group functionalities at its periphery (G3(OH)24) was used in the present study.23,26 Functionalization of the hydroxyl-group terminated PETIM dendrimer was performed through the glycosylation using benzobromogalactose in the presence of Ag2CO3/AgClO4 (ESI Scheme S1†) (Fig. 1). The galactopyranoside-derivatized DG is soluble in water. The purity of DG was ascertained by 1H and 13C NMR spectroscopy analysis (ESI Fig. S2 and S3†). A comparison of 1H NMR integrations of 4.26 ppm (H-1 of sugar) and 1.86 ppm (–CH2–CH2–CH2–) of the dendrimer structure showed that ∼21–23 sugar moieties functionalize a dendrimer molecule. A phenol-sulfuric acid assay further confirmed the extent of galactose functionalization (ESI Fig. S4†).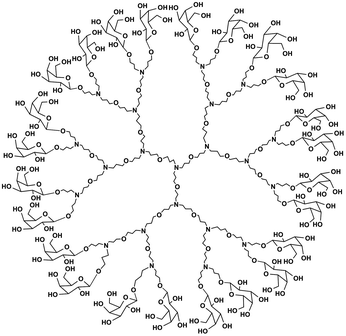 | ||
| Fig. 1 Molecular structure of DG. The molecular structure of galactopyranoside dendrimer (DG) with 24 peripheral galactose units. | ||
Efficient complexation of siRNA with DG
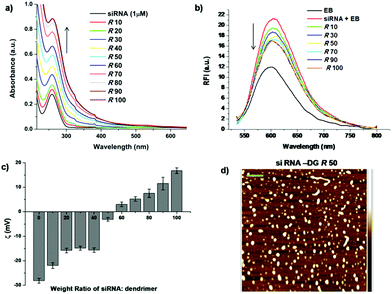
The ability of DG to interact with siRNA and the nature of this interaction were assessed by gel retardation assay, zeta potential measurements, UV-visible spectroscopy, ethidium bromide intercalation assay and visualization of the complex by atomic force microscopy (AFM). UV-Visible spectroscopy analysis provided an indication of siRNA-DG complexation, deduced from the changes in siRNA absorbance at 260 nm occurring due to the interaction of siHCV with DG (Fig. 2a). Displacement of intercalated ethidium bromide (EB) from siRNA upon titration with DG, monitored by the decreasing fluorescence intensity of EB, also indicated formation of a stable complex between siHCV and DG (Fig. 2b). Complex formation between siHCV and DG was also apparent from the gel retardation assay, wherein, a gradual retardation and disappearance of the siRNA band, visualized on a 2% formaldehyde-agarose gel indicated the formation of siRNA–dendrimer complex (ESI Fig. S5†).To study the charge neutralization upon complex formation, zeta potential measurements were performed. The zeta potential of siRNA, DG and their complexes at varying weight ratios (R) was determined (Fig. 2c). A charge of −28.15 ± 0.94 mV on siRNA and +15.13 ± 0.95 mV on DG was obtained. Increasing concentration of DG in the complex resulted in an increase in the zeta potential up to +16.8 ± 1.15 mV at R 100. The electroneutrality of the complex occurred at R 50, which is in agreement with the gel retardation assay. The shape and size distribution of siRNA-DG complexes were characterized by atomic force microscopy (AFM), wherein, siRNA appeared globular with a z-height of ∼1 nm, while DG was spherical with a height of 0.1–2 nm. DG–siRNA complexes at R 50 consisted of particles of various shapes and height profiles ranging from 1.5 to 3.25 nm (Fig. 2d). Complexes formed at R 100 showed a z-height distribution of 3–8 nm and very few particles showed height profiles up to 25 nm (ESI Fig. S6†).
Structural analysis of siRNA–DG complexation by MD simulation
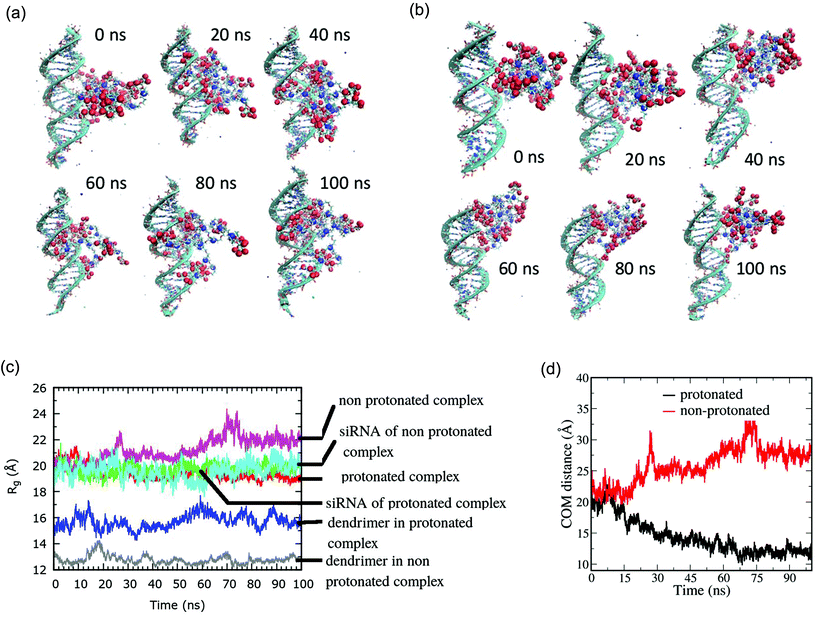
In order to understand the complexation and to visualize the dendriplexes, a fully atomistic MD simulation of the siRNA–DG complexation was performed. Simulation studies revealed that in case the dendrimer is not protonated, there is no complexation with siRNA. At physiological pH, tertiary amines at the outer shells (penultimate shell) are protonated, as pKa values are known to be in the range of 8–9. A lower pH is required generally for the tertiary amines at the interior and core for their protonation. In the MD studies, tertiary amines at the outer shells were protonated, as would be expected in experimental studies conducted at pH 7.4.39,40 When dendrimer amines are protonated, formation of a very tight complex was observed, demonstrating the role of electrostatic interactions in the complexation process. Fig. 3a and b shows molecular level pictures of the siRNA–DG complex as a function of simulation time from 0 to 100 ns. Simulations also revealed that the dendrimer preferentially binds to the major groove during complexation with siRNA. Instantaneous snapshots of the complex (ESI Fig. S7†) shown at few ns intervals reveal the major groove binding nature of the dendrimer.A graphical representation of the sizes of the complexes, as well as sizes of the siRNA and DG in the complex as the complexation proceeds is shown in Fig. 3c. The radius of gyration (Rg) of the siRNA–DG complex was found to be ∼2 nm and those of siRNA alone and dendrimer alone were 1.8 nm and 1.3 nm, respectively. The Rg of siRNA remained almost constant in all the conditions studied. In order to verify the complex formation between the protonated dendrimer and siRNA, the radial distribution of the dendrimer amines as well as sugar groups and phosphates of the siRNA was studied (ESI Fig. S8†). Further, a plot of the distance between the centre of mass (COM) of a dendrimer and siRNA as a function of simulation time showed that in the case of non-protonated dendrimer, the dendrimer moves away from the siRNA over simulation time. In contrast the protonated dendrimer binds tightly on the surface of siRNA, as seen by the very low COM distance between the two (Fig. 3d).
Preferential perinuclear delivery of siRNA into liver cells using DG
Initial transfection studies were carried out by monitoring the internalization of a green fluorescent tagged siRNA. Electroneutral complexes formed at R 50 (siRNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DG = 1
DG = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
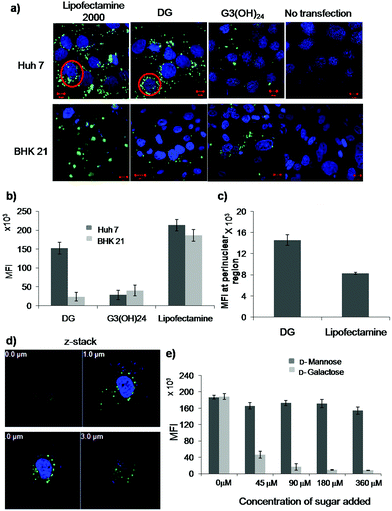
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50) showed preferential delivery of siRNA to liver derived Huh 7 cells over kidney derived BHK 21 cells (Fig. 4a). The siRNA delivery efficiency was compared to other delivery agents, namely, lipofectamine 2000 and non-galactose functionalized dendrimer G3(OH)24. A higher transfection efficiency obtained using DG when compared to G3(OH)24 highlights the role of the ASGPR–galactose interaction in siRNA-DG internalization (Fig. 4b).
50) showed preferential delivery of siRNA to liver derived Huh 7 cells over kidney derived BHK 21 cells (Fig. 4a). The siRNA delivery efficiency was compared to other delivery agents, namely, lipofectamine 2000 and non-galactose functionalized dendrimer G3(OH)24. A higher transfection efficiency obtained using DG when compared to G3(OH)24 highlights the role of the ASGPR–galactose interaction in siRNA-DG internalization (Fig. 4b).
Importantly, the siRNA–DG complex was observed to be delivered preferentially to the perinuclear region in liver cells. The fluorescence intensity around the perinuclear region of the cell was considerably higher for the delivery mediated by the DG–siRNA complex, in comparison with the lipofectamine–siRNA complex (Fig. 4c). Further, siRNA internalization occurring in the perinuclear region was confirmed through z-stack imaging. The higher fluorescence intensity at the middle frame of the cell clearly indicated the internalization of siRNA occurring in the perinuclear region (Fig. 4d). The preferential perinuclear localization can be due to ASGPR mediated endocytosis, since ASGPR enters the cell during endocytosis and particularly delivers the bound ligand in the area surrounding the nucleus.41 In order to verify this hypothesis, the Huh 7 cells were pre-incubated with free D-galactose (a specific ligand for ASGPR) or D-mannose (a non-specific control) at different concentrations (45 μM, 90 μM, 180 μM and 360 μM) prior to the addition of siRNA–DG complexes. At 8 h post addition of the complexes, the cells were fixed with paraformaldehyde and imaged by CLSM. Internalization of siRNA–DG complexes was greatly reduced in cells pre-incubated with D-galactose due to saturation of the receptor sites with the ligand which prevented the binding of DG. Cells pre-incubated with D-mannose did not show appreciable difference in siRNA internalization when compared to control cells (Fig. 4e). Experiments were also carried out in BHK 21 cells, and no effect on siRNA internalization was observed (ESI Fig. S9†).
Localization of DG–siRNA complex at the site of replication and inhibition of viral RNA synthesis
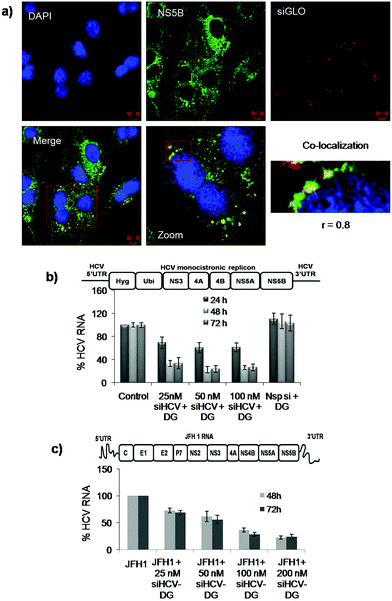
Following successful transfection mediated by DG in cultured liver cells, the study was extended to a functional assay, to demonstrate the applicability of preferential liver delivery in a disease model involving hepatitis C virus (HCV). HCV replicates along the lipid rafts in the perinuclear region of liver cells. The viral proteins NS5B and NS3 regulate the viral replication.1 In order to identify the localization site of the delivered siRNA, Huh 7 cells harbouring a HCV monocistronic replicon were transfected with siRNA and an immunofluorescence assay was performed. We also probed NS5B and NS3 proteins so as to investigate co-localization of siRNA with these proteins. Confocal microscopy images showed that labelled siRNA co-localized with both the viral proteins (Fig. 5a, ESI Fig. S10†), indicating that the siRNA delivered using DG was probably localized at the site of HCV RNA replication. Following this observation, the effect of DG-mediated delivery of a pre-characterized therapeutic siRNA known to inhibit HCV RNA translation and replication18 was studied. Anti HCV siRNA–DG complexes were prepared at R 50 using different concentrations of siRNA, ranging from 25 to 100 nM and transfected into Huh 7 cells harbouring the HCV replicon. About 77% reduction in HCV RNA levels was noted up to 72 h, indicating the successful delivery of siRNA using DG (Fig. 4b). siHCV-G3(OH)24 and siNsp–DG complexes were used as the non-targeting and non-specific siRNA (siNsp) controls, respectively. While transfection with the siHCV-G3(OH)24 complex showed only about 25% inhibition of HCV RNA, the non-specific siRNA did not show any effect on HCV RNA levels. Transfection with DG alone did not show any reduction in the levels of HCV RNA indicating that the dendrimer alone does not interfere with the virus replication (ESI Fig. S11†).The effectiveness of the siHCV–DG system was further validated in an infectious JFH1 cell culture system.37 Huh 7.5 cells were transfected with 25, 50, 100 and 200 nM siHCV using DG. Four hours later, the cells were transfected with JFH1 RNA and the relative RNA levels were quantified at 48 h and 72 h post transfection of siHCV. A dose dependent reduction in HCV RNA levels (up to 75%) was observed (Fig. 5c) which further confirmed the successful delivery of siRNA to inhibit HCV replication in the infectious cell culture system.
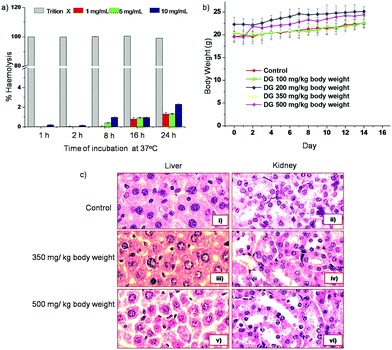
Low toxicity and high tolerance of DG in animal cells
The toxicity of the siHCV–DG complex was evaluated in order to extend the studies to in vivo systems. The cytotoxicity profile of DG was assayed by the MTT assay in four different cell types, namely, Huh 7 (liver cells), BHK 21 (kidney cells) A549 (lung carcinoma cells) and HeLa (cervical cancer cells). The studies showed that DG was completely non-toxic up to a concentration of 225 μg mL−1 (ESI Fig. S12†). In order to ensure that DG is not haemolytic, the release of haemoglobin upon incubation of RBCs with DG at different concentrations was monitored up to 24 h. Triton X, which is known to induce 100% haemolysis, was used as a positive control. Cells not treated with DG or Triton X were also assayed as controls for comparison.38 The results indicate that DG induces only a marginal haemolysis of 2.4% at a concentration of 10 mg mL−1 after 24 h of incubation (Fig. 6a).Toxicity studies in Balb/c mice were carried out by intravenous tail vein injection of DG at doses of 100–500 mg per kg b.w., and monitoring their body weight, behavioural responses, food–water intake and mortality for 14 days. No statistically significant differences in the body weight were detected between mice injected with DG up to a dose of 350 mg per kg b.w. compared to control mice (with no dendrimer treatment). Additionally, no morbidity, weight loss or changes in the gross organ weights, and organ morphology (liver and kidney) were detected. Fifty percent death was observed in the case of mice treated with a dose of 500 mg per kg b.w. of DG (Fig. 6b). On the 14th day post injection, the mice were sacrificed and dissected. Organs (liver and kidneys) were isolated and processed for their histopathological examination. The haematoxylin and eosin-staining based histopathological studies showed no alterations in the histoarchitecture of liver as compared to control groups up to a dose of 350 mg per kg b.w. The tissues of mice treated with a dose of 500 mg kg−1 showed larger inter cellular spaces between the hepatocytes (Fig. 6c). These results clearly indicate that intravenous administration of DG is non-toxic up to a dose of 350 mg per kg b.w.
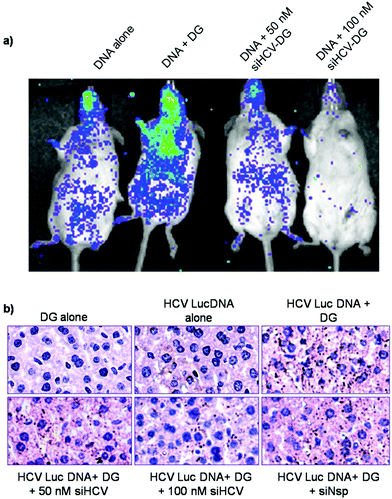
Preferential delivery of DG–siRNA complex into mouse liver
Upon determining the biocompatible nature of DG, the biodistribution of DNA–DG complexes (5′ HCVIRESLuc) was studied by in vivo bioluminescence imaging (BLI). Balb/c mice (5–6 weeks old) were injected intravenously with HCVLucDNA–DG complexes in the presence or absence of siHCV at R 50. After 48 h, the mice were injected with the luciferase substrate luciferin and whole body imaging was carried out to track the luciferase expression in the mice. BLI showed that the DNA was delivered to the mice liver in greater amounts compared to other organs indicating preferential liver targeting (Fig. 7a). The results of the bio-imaging were further confirmed by immunohistochemistry (IHC) studies which showed higher levels of luciferase localized in the liver and brain tissues when compared to other organs (ESI Fig. S13†). In the presence of siHCV, luciferase expression was greatly reduced compared to the control. Liver tissues of mice injected with siHCV–HCVLuc–DG complexes showed a dose dependent decrease in luciferase expression compared to those injected with only HCVLuc–DG complexes (Fig. 7a). The results of BLI were confirmed by IHC wherein, luciferase antibody was used to probe the luciferase production in the mice liver tissues. The formation of a black–brown precipitate in tissues of mice treated with HCVLuc–DG complexes indicated successful delivery of DNA to the liver (Fig. 7b). Tissues of mice treated with siHCV–HCVLuc–DG complexes showed a net decrease in luciferase production observed by the lower amounts of brown precipitates formed. These results were consistent with that observed in the bioluminescence imaging assay. Further, the use of a non-specific siRNA in the DG–HCVLuc complex did not affect the luciferase production reiterating the specificity of siHCV to the HCV 5′ UTR. Mice injected with either DNA alone or DG alone were used as controls to rule out background signals. Taken together, the results clearly establish the potential use of DG for delivery of therapeutic siRNA to inhibit HCV infection.Discussion
Our study provides a ‘proof-of-concept’ approach for the use of siRNA technology for combating HCV infection based on a newly developed dendritic galactoside nanovector. Functionalization of the dendrimer with galactose units imparted the ability of target-oriented delivery which is one of the key attributes to the successful action of the desired antiviral. The formation of a stable complex with intact siRNA by the delivery vector is essential to ensure crossing of cellular barriers and successful delivery of siRNA at the target site to downregulate gene expression.19 Gel retardation assay, zeta potential measurements, ethidium bromide displacement assay and UV-Vis spectroscopy confirmed the formation of stable siRNA–DG complexes. Atomic force microscopy imaging showed that majority of DG particles had height profiles below 1 nm, similar to those observed in the case of PAMAM42 and PETIM dendrimers.21 The shape of siRNA and its non-aggregating behaviour were consistent with the reports of siRNA AFM imaging.43,44 The morphologies of DG–siRNA complexes at different weight ratios were visualized as dome shaped, spherical and globular structures. The particle size distribution at R 50 was also monodispersed, possibly as a result of net neutral charge of the complex, as determined by the zeta potential measurements.MD simulation showed that most of the galactose residues are projected away from the siRNA whereas dendrimer nitrogens (highlighted as blue spheres in Fig. 3a) are involved in electrostatic complex formation with siRNA. The galactose residues are free to interact with the asialoglycoprotein receptor (ASGPR) when these complexes are used for the transfection. The radius of gyration (Rg) values obtained from simulation studies are in reasonable agreement with those obtained from AFM imaging. For example, the z-height profile of the complex varied between 1.5 and 3.25 nm which is in the same range as the Rg value of the complex. The dendrimer structure was seen to undergo a change becoming more compact and orienting itself such that it accommodates itself into the major groove. The constant value of Rg for siRNA indicates that siRNA maintains its structural integrity in the complex. The analysis of the radial distribution function (RDF) of the dendrimer amines as well as, galactose oxygens with respect to phosphate groups of siRNA showed complex formation between the protonated dendrimer and siRNA. This was further confirmed by the analysis of the change in the distance between the COM of dendrimer and siRNA with respect to simulation time.
Initial nucleic acid delivery experiments involved the transfection of fluorescent-tagged siRNA using DG at three ratios, R 25, R 50 and R 100. Comparison of these three ratios revealed that the delivery at R 50 was more efficient. The use of low N/P ratios to mediate effective siRNA transfection was demonstrated recently45 and our observation was on similar lines to that study. Due to the electroneutrality of the siRNA–DG complex at R 50, the entry inside the cell would occur preferentially by receptor-mediated endocytosis, involving the ASGPR–galactoside interaction. A comparison of transfection with a native hydroxyl functionalized dendrimer and lipofectamine, showed that DG preferentially localized in the perinuclear region of liver cells with much higher fluorescence intensities. The z-stack imaging helped rule out surface adherence and ascertain ASGPR mediated endocytosis in liver cells. ASGPR belongs to the class of receptors that enter the cells during endocytosis and deliver the bound ligand in the area surrounding the nucleus. Saunier and co-workers had earlier shown co-localization of GFP-labelled ASGPR and red fluorescence labelled galactoprotein which binds to ASGPR in the perinuclear region.41 These studies showed that ASGPR internalized along with its bound ligand in the perinuclear regions of the cell. The present study demonstrates that siRNA, delivered specifically to the liver cells by receptor mediated endocytosis, is localized in the perinuclear area.
The role of ASGPR in internalization was further validated by the competition assay involving the pre-incubation of liver cells with D-galactose or D-mannose. Ligand–receptor interactions are highly specific due to the ability of receptor to recognize subtle conformational differences of the ligand which alters the ligand-binding affinity.46 ASGPR specifically binds to D-galactose over D-mannose by conformational specific hydrogen bonding interactions. Hence cells pre-incubated with D-galactose showed decreased siRNA uptake compared to incubation with D-mannose.
Perinuclear localization of siRNA gains importance to inhibit HCV RNA replication, since it is known that the replication occurs along the lipid droplets of the endoplasmic reticulum in the perinuclear region of hepatocytes.1,2 The viral proteins NS5B, the RNA dependent RNA polymerase (RdRp) and NS3 which exhibit RNA helicase activity are known to regulate the viral replication.1 Both proteins are a part of the HCV replication complex. We demonstrated here that siRNA tagged with a fluorophore co-localizes with NS5B and NS3 proteins of the HCV virus replication complex. The proximity of siRNA localization at the site of viral RNA replication ensures the effective action of siRNA, which was proven by delivering siHCV using DG in replicon harbouring cells and probing the viral RNA levels. The siRNA against HCV inhibits the replication of viral mRNA through targeted binding to the 5′-UTR of HCV RNA. The use of 5′ shDNA (short hairpin DNA), a precursor for the in situ generation of the siRNA to inhibit HCV IRES mediated translation and hence HCV RNA replication inhibition, was demonstrated earlier.14 In the present study, inhibition of HCV RNA up to ∼77% was obtained by the use of siRNA–DG delivery systems. The preferential perinuclear delivery of siRNA in the vicinity of HCV virus replication site might contribute to the decrease in HCV RNA levels.
Dendrimer based delivery systems are injected intravenously for in vivo studies, encountering blood in the first instance.38 In order to verify that intravenously injected compounds do not cause a haemolysis by rupturing the RBC membranes, a haemolytic assay was performed. The haemolysis induced by DG was observed to be negligible when compared to the dendritic vectors like PAMAM dendrimers, (60% haemolysis), PPI dendrimer (80% haemolysis) and polyethylenimine (PEI) based polymers (17–30% haemolysis).41,47–49 These results showed that DG is safe to be injected intravenously. Further experiments in female Balb/c mice showed acute tolerance dose for DG up to 350 mg per kg b.w. A comparison with reported studies on other dendritic systems47 shows DG to be much less toxic, since even a dose of 350 mg per kg b.w. is very well tolerated by mice, which, to our knowledge, is perhaps the most non-toxic synthetic delivery vector in vivo reported so far.
In vivo whole body imaging showed a preferential biodistribution of the complexes to the liver. This observation was confirmed further by immunohistochemistry (IHC) of the isolated mice organs. Localization of large quantities of luciferase enzymes in the liver when compared to other organs indicated the preferential targeting of DG to the mouse liver. Previous studies using radiolabeled glycolipids and PAMAM dendrimers showed that liver uptake of delivery vehicle 15 min to 1 h post i.v. injection was 60–90%. Minor or negligible uptake by other organs was also reported.49 The present study establishes the targeted delivery and successful expression of a reporter gene using a non-radiolabeled dendrimer, even after 84 h post injection. Reports of siRNA being delivered to tumour cells in vivo using engineered PAMAM or PPI dendrimers showed only a modest success.50,51 Interestingly, luminescence signals were observed in the brain tissues which were further validated by IHC experiments. Crossing of the blood–brain barrier by hydroxyl-terminated PAMAM dendrimers was demonstrated recently.52 Experiments are currently under progress to decipher the mechanism and specificity for delivery into neuronal cells. The use of siRNA in the complexes resulted in a net decrease in the luciferase production probed by BLI, as well as, by IHC assays. The siRNA binds to the 5′-UTR and prevents the HCV IRES mediated translation of the subsequent luciferase gene, thereby reducing the luminescence signal, especially in the liver. This resulted in the successful delivery of siHCV preferentially to the liver cells by using DG.
Conclusion
RNAi technology and dendrimer based delivery are being used as popular strategies to combat diseases, such as, cancer and HIV.53–55 Significant progress has been made to implement this technology to overcome HCV infection.54,56 Our results establish that siRNA–DG is a biocompatible system for preferential liver targeted delivery. The ability of DG to deliver siRNA in the perinuclear region near the HCV replication complex and observation of a significant decrease in JFH1 RNA levels authenticates the siRNA–DG complex as an efficient delivery system. Our study shows that DG as a targeted delivery vector can be used for the effective delivery of antivirals against HCV. The novel siRNA–DG nano-system appears to be a promising option for HCV related therapeutics.Acknowledgements
Authors would like to thank Ralf Bartenschlager, Charles M. Rice and Takaji Wakita for sharing plasmid constructs and cell lines. We thank the Central Animal Facility, Confocal imaging facility, Department of Microbiology and Cell Biology and Centre for Nanoscience and Engineering (CeNSE), IISc. Current work was supported by grants from the Department of Biotechnology, India to SD, NJ, AKS, PKM; and from the Science and Engineering Research Board, DST, India to BUR and NJ. AL and AK thank Council of Scientific and Industrial Research (CSIR), India for a research fellowship. SD also acknowledges a J. C Bose fellowship from DST, India.References
- D. Moradpur, F. Penin and C. M. Rice, Nat. Rev. Microbiol., 2007, 5, 453 CrossRef PubMed.
- Y. Ma, M. Anantpadma, J. M. Timpe, S. Shanmugam, S. M. Singh, S. M. Lemon and M. Yi, J. Virol., 2011, 85, 86 CrossRef CAS PubMed.
- S. Das, R. K. Shetty, A. Kumar, R. N. Shridharan, R. Tatineni, G. Chi, A. Mukherjee, S. Das, S. M. Subbarao and A. A. Karande, PLoS One, 2013, 8, e53619 CAS.
- C. Ji, Y. Liu, C. Pamulapati, S. Bohini, G. Fertig, M. Schraeml, W. Rubas, M. Brandt, S. Ries, H. Ma and K. Klumpp, Hepatology, 2015, 61, 1136 CrossRef CAS PubMed.
- A. K. Manna, A. Kumar, U. Ray, S. Das, G. Basu and S. Roy, Antiviral Res., 2013, 97, 223 CrossRef CAS PubMed.
- T. J. Liang and M. G. Ghany, N. Engl. J. Med., 2013, 368, 1907 CrossRef CAS PubMed.
- R. E. Lanford, E. S. Hildebrandt-Eriksen, A. Petri, R. Persson, M. Lindow, M. E. Munk, S. Kauppinen and H. Ørum, Science, 2010, 327, 198 CrossRef CAS PubMed.
- P. J. Pockros and J. S. Au, Clin. Pharmacol. Ther., 2014, 95, 78 Search PubMed.
- B. U. Reddy, R. Mullick, A. Kumar, G. Sudha, N. Srinivasan and S. Das, Sci. Rep., 2014, 24, 5411 Search PubMed.
- T. L. Foster, G. S. Thompson, A. P. Kalverda, J. Kankanala, M. Bentham, L. F. Wetherill, J. Thompson, A. M. Barker, D. Clarke, M. Noerenberg, A. R. Pearson, D. J. Rowlands, S. W. Homans, M. Harris, R. Foster and S. Griffin, Hepatology, 2014, 59, 408 CrossRef CAS PubMed.
- Y. Gao, X. Yu, B. Xue, F. Zhou, X. Wang, D. Yang, N. Liu, L. Xu, X. Fang and H. Zhu, PLoS One, 2014, 28, e90333 Search PubMed.
- E. Poveda, D. L. Wyles, A. Mena, J. D. Pedreira, A. Castro-Iglesias and E. Cachay, Antiviral Res., 2014, 108, 181 CrossRef CAS PubMed.
- M. M. Verstegen, Q. Pan and L. J. van der Laan, Adv. Exp. Med. Biol., 2015, 848, 1 CrossRef.
- N. Subramanian, P. Mani, S. Roy, S. V. Gnanasundram, D. P. Sarkar and S. Das, J. Gen. Virol., 2009, 90, 1812 CrossRef CAS PubMed.
- S. Roy, N. Gupta, N. Subramanian, T. Mondal, A. C. Banerjea and S. Das, J. Gen. Virol., 2008, 89, 1579 CrossRef CAS PubMed.
- C. Chevalier, A. Saunier, Y. Benureau, D. Fléchet, D. Delgrange, F. Colbère-Garapin, C. Wychowski and A. Martin, Mol. Ther., 2013, 2, 1452 Search PubMed.
- P. K. Chandra, A. K. Kundu, S. Hazari, S. Chandra, L. Bao, T. Ooms, G. F. Morris, T. Wu, T. K. Mandal and S. Dash, Mol. Ther., 2012, 20, 1724 CrossRef CAS PubMed.
- R. Kanasty, J. R. Dorkin, A. Vegas and D. Anderson, Nat. Mater., 2013, 12, 967 CrossRef CAS PubMed.
- J.-P. Behr, Acc. Chem. Res., 2012, 45, 980 CrossRef CAS PubMed.
- X. Liu, P. Rocchic and L. Peng, New J. Chem., 2012, 36, 256 RSC.
- A. Lakshminarayanan, V. K. Ravi, R. Tatineni, Y. B. R. D. Rajesh, V. Maingi, K. S. Vasu, N. Madhusudhan, P. K. Maiti, A. K. Sood, S. Das and N. Jayaraman, Bioconjugate Chem., 2013, 24, 1612 CrossRef CAS PubMed.
- U. P. Thankappan, S. N. Madhusudhana, A. Desai, G. Jayamurugan, Y. B. R. D. Rajesh and N. Jayaraman, Bioconjugate Chem., 2011, 22, 115 CrossRef CAS PubMed.
- T. R. Krishna, M. Belwal, U. S. Tatu and N. Jayaraman, Tetrahedron, 2005, 61, 4281 CrossRef CAS PubMed.
- L. A. J. M. Sliedregt, P. C. N. Rensen, E. T. Rump, P. J. van Santbrink, M. K. Bijsterbosch, A. R. P. M. Valentijn, G. A. van der Marel, J. H. van Boom, T. J. C. van Berkel and E. A. L. Biessen, J. Med. Chem., 1999, 42, 609 CrossRef CAS PubMed.
- R. K. Ness, H. G. Fletcher Jr. and C. S. Hudson, J. Am. Chem. Soc., 1950, 72, 2200 CrossRef CAS.
- T. R. Krishna and N. Jayaraman, J. Org. Chem., 2003, 68, 9694 CrossRef CAS PubMed.
- V. Maingi, V. Jain, P. V. Bharatam and P. K. Maiti, J. Comput. Chem., 2012, 33, 1997–2011 CrossRef CAS PubMed.
- D. A. Case, T. A. Darden, T. E. Cheatham III, C. L. Simmerling, J. Wang, R. E. Duke, R. Luo, R. C. Walker, W. Zhang, K. M. Merz, B. Roberts, S. Hayik, A. Roitberg, G. Seabra, J. Swails, A. W. Götz, I. Kolossváry, K. F. Wong, F. Paesani, J. Vanicek, R. M. Wolf, J. Liu, X. Wu, S. R. Brozell, T. Steinbrecher, H. Gohlke, Q. Cai, X. Ye, J. Wang, M.-J. Hsieh, G. Cui, D. R. Roe, D. H. Mathews, M. G. Seetin, R. Salomon-Ferrer, C. Sagui, V. Babin, T. Luchko, S. Gusarov, A. Kovalenko and P. A. Kollman, AMBER 12, University of California, San Francisco, 2012 Search PubMed.
- J. M. Wang, R. M. Wolf, J. W. Caldwell, P. A. Kollman and D. A. Case, J. Comput. Chem., 2004, 25, 1157–1174 CrossRef CAS PubMed.
- J. Wang, P. Cieplak and P. A. Kollman, J. Comput. Chem., 2000, 21, 1049–1074 CrossRef CAS.
- A. Perez, I. Marchan, D. Svozil, J. Sponer, T. E. Cheatham III, C. A. Laughton and M. Orozco, Biophys. J., 2007, 92, 3817–3829 CrossRef CAS PubMed.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, J. Chem. Phys., 1983, 79, 926–935 CrossRef CAS PubMed.
- B. Nandy and P. K. Maiti, J. Phys. Chem. B, 2011, 115, 217 CrossRef CAS PubMed.
- V. Vasumati and P. K. Maiti, Macromolecules, 2010, 43, 8264 CrossRef.
- P. K. Maiti and B. Bagchi, Nano Lett., 2006, 6, 2478 CrossRef CAS PubMed.
- R. Salomon-Ferrer, A. W. Goetz, D. Poole, S. L. Grand and R. C. Walker, J. Chem. Theory Comput., 2013, 9, 3878 CrossRef CAS.
- T. Kato, T. Date, A. Murayama, K. Morikawa, D. Akazawa and T. Wakita, Nat. Protoc., 2006, 1, 2334 CrossRef CAS PubMed.
- B. Klajnert, S. Pikala and M. Bryszewska, Proc. R. Soc. London, Ser. A, 2010, 466, 1527 CrossRef CAS.
- R. C. van Duijvenbode, M. Borkovec and G. J. M. Koper, Polymer, 1998, 39, 2657–2664 CrossRef CAS.
- D. Cakara, J. Kleimann and M. Borkovec, Macromolecules, 2003, 36, 4201–4207 CrossRef CAS.
- B. Saunier, M. Triyatni, L. Ulianich, P. Maruvada, P. Yenand and L. D. Kohn, J. Virol., 2003, 77, 546 CrossRef CAS.
- J. Li, L. T. Piehler, D. Qin, J. R. Baker and D. A. Tomalia, Langmuir, 2000, 16, 5613 CrossRef CAS.
- H. G. Abdelhady, Y.-L. Lin, H. Sun and M. E. H. El-Sayed, PLoS One, 2013, 8, e61710 CAS.
- P. Ofek, W. Fischer, M. Calderón, R. Haag and R. Satchi-Fainaro, FASEB J., 2010, 24, 3122 CrossRef CAS PubMed.
- M. Zheng, G. M. Pavan, M. Neeb, A. K. Schaper, A. Danai, G. Klebe, O. M. Merkel and T. Kissel, ACS Nano, 2012, 6, 9447 CrossRef CAS PubMed.
- N. Jayaraman, Chem. Soc. Rev., 2009, 38, 3463 RSC.
- K. L. Allion, Y. Xie, N. El-Gendy, C. J. Berkland and M. L. Forrest, Adv. Drug Delivery Rev., 2009, 61, 457 CrossRef PubMed.
- V. Russ, M. Gunther, A. Halama, M. Ogris and E. Wnage, J. Controlled Release, 2008, 132, 131 CrossRef CAS PubMed.
- N. Malik, R. Wiwattanapatapee, R. Klopsch, K. Lorenz, H. Frey, J. W. Weener, E. W. Meijer, W. Paulus and R. Duncan, J. Controlled Release, 2000, 65, 133 CrossRef CAS.
- O. Taratula, O. B. Garbuzenko, P. Kirkpatrick, I. Pandya, R. Savla, V. P. Pozharov, H. He and T. Minko, J. Controlled Release, 2009, 140, 284 CrossRef CAS PubMed.
- X. Liu, C. Liu, J. Zhou, C. Chen, F. Qu, J. J. Rossi, P. Rocchi and L. Peng, Nanoscale, 2015, 7, 3867 RSC.
- M. K. Mishra, C. A. Beaty, W. G. Lesniak, S. P. Kambhapati, F. Zhang, M. A. Wilson, M. E. Blue, J. C. Troncoso, S. Kannan, M. V. Johnston, W. A. Baumgartner and R. M. Kannan, ACS Nano, 2014, 8, 2134 CrossRef CAS PubMed.
- D. H. Kim and J. J. Rossi, Nat. Rev. Genet., 2007, 8, 173 CrossRef CAS PubMed.
- J. Haasnoot, E. M. Westerhout and B. Berkhout, Nat. Biotechnol., 2007, 25, 1435 CrossRef CAS PubMed.
- B. Nandy, D. H. Bindu, N. M. Dixit and P. K. Maiti, J. Chem. Phys., 2013, 139, 024905 CrossRef PubMed.
- D. Grimm and M. A. Kay, Gene Ther., 2006, 13, 563 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Spectral data and experimental details. See DOI: 10.1039/c5nr02898a |
| This journal is © The Royal Society of Chemistry 2015 |