Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Nitrogen and phosphorus co-doped graphene quantum dots: synthesis from adenosine triphosphate, optical properties, and cellular imaging†
Arundithi
Ananthanarayanan
a,
Yue
Wang
b,
Parimal
Routh
a,
Mahasin Alam
Sk
a,
Aung
Than
a,
Ming
Lin
c,
Jie
Zhang
c,
Jie
Chen
a,
Handong
Sun
b and
Peng
Chen
*a
aBioengineering Program, School of Chemical and Biomedical Engineering, Nanyang Technological University, 70 Nanyang Drive, Singapore 637457. E-mail: ChenPeng@ntu.edu.sg
bDivision of Physics and Applied Physics, School of Physical and Mathematical Sciences, Nanyang Technological University, 21 Nanyang Link, Singapore 637371
cInstitute of Materials Research and Engineering, A*STAR (Agency for Science, Technology and Research), 3 Research Link, Singapore 117602
First published on 3rd April 2015
Abstract
Graphene quantum dots (GQDs) are emerging zero-dimensional materials promising a wide spectrum of applications, particularly, as superior fluorescent reporters for bio-imaging and optical sensing. Heteroatom doping can endow GQDs with new or improved photoluminescence properties. Here, we demonstrate a simple strategy for the synthesis of nitrogen and phosphorus co-doped GQDs from a single biomolecule precursor (adenosine triphosphate – ATP). Such ATP-GQDs exhibit high fluorescence quantum yield, strong two-photon upconversion, small molecular weight, high photostability, and good biocompatibility. Furthermore, transferrin conjugated ATP-GQDs have been used for imaging and real-time tracking of transferrin receptors in live cells.
Introduction
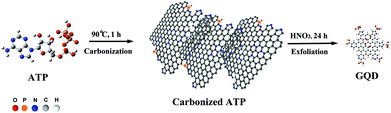
Graphene quantum dots (GQDs) are a new class of zero-dimensional nanomaterials, which have shown immense potential for bioimaging, optical sensing, energy conversion, and catalysis.1–4 Owing to their tunable photoluminescence (PL), photostability, molecular size, biocompatibility, and the ease of conjugation with biomolecules, GQDs are particularly attractive for bioimaging.5–15 It has been shown that heteroatom doping can endow GQDs with tailored or new PL as well as other properties.15–17 Heteroatom-doped GQDs can be synthesized by exfoliating pre-doped graphene or graphene oxide sheets,18–21 but such top-down methods are tedious because of the need for multiple steps and because they are usually of low doping efficiency. Alternatively, doped GQDs can be obtained by pyrolyzing small organic molecules, with heteroatoms naturally inherited from the precursor molecules.22–25Herein, we report a simple and cost-effective method for the synthesis of GQDs co-doped with nitrogen, and phosphorus by carbonization and subsequent chemical exfoliation of a single precursor molecule – adenosine triphosphate (ATP) (Scheme 1, also see the Experimental section). The dually doped GQDs show high quantum yield (∼27.5%) and strong two-photon excitation with a two-photon absorption cross-section (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 GM). Carbonaceous micro-sheets resulting from ATP carbonization exhibit good catalytic properties towards oxygen reduction, and heteroatom-doped GQDs conjugated with transferrin have been employed to image and track transferrin receptors in live cells.
000 GM). Carbonaceous micro-sheets resulting from ATP carbonization exhibit good catalytic properties towards oxygen reduction, and heteroatom-doped GQDs conjugated with transferrin have been employed to image and track transferrin receptors in live cells.
Results and discussion
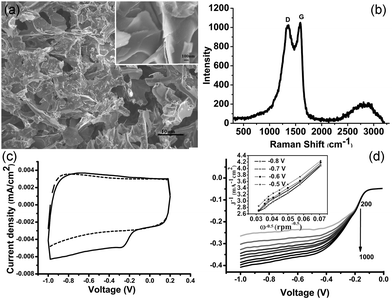
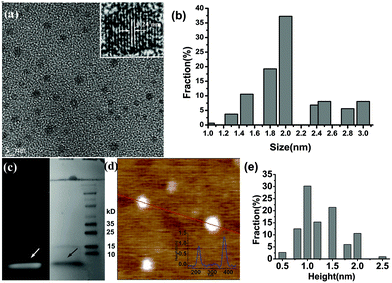
Adenosine triphosphate (ATP) is the universal energy currency in cells, containing cyclic carbon rings, nitrogen, phosphorus, and oxygen atoms. Carbonization of ATP molecules yields porous aggregates of carbonaceous micro-sheets (Fig. 1a), resulting from re-organization and fusion of the precursor molecules. The Raman spectrum of the carbonized ATP (cATP) shows prominent D and G bands (Fig. 1b), suggesting its graphitic nature with the presence of sp3 carbon due to covalent bonding with heteroatoms. Heteroatom doping is able to confer the inert graphene or graphitic materials with catalytic abilities.16,26 Indeed, cATP exhibits good catalytic properties towards oxygen reduction reaction (ORR) as evidenced by the prominent reduction peak at ∼−0.26 V in the presence of oxygen (Fig. 1c). Linear sweep voltammograms of cATP in oxygen-saturated 0.1 M KOH are obtained using a rotating disk electrode at various rotating speeds to determine the number of electrons transferred for the ORR process (Fig. 1d). Koutecky–Levich (K–L) plots at different potentials are shown in Fig. 1d inset. The slopes of the K–L curves suggest that the ORR process catalyzed by cATP is highly efficient through the 4-electron reduction pathway. Furthermore, the onset potential of the cATP electrode is around −0.14 V, which is desirably lower than the previously reported N-doped graphene.27–30 This experiment suggests that cATP is heteroatom-doped and has potential for fuel cell applications.Subsequently, cATP is exfoliated by refluxing in nitric acid. As revealed by transmission electron microscopy (TEM), carbon nanosheets ∼2.06 nm (±0.69, n = 161) in diameter have been produced (Fig. 2a and b). The high-resolution TEM image shows the lattice spacing of 0.24 nm corresponding to the d1120 plane of graphene (Fig. 2a inset). Gel electrophoresis also indicates that the obtained nanosheets have a narrow size-distribution and small molecular weight (≪10 kDa) (Fig. 2c). Atomic force microscopy (AFM) reveals that the average thickness of these carbon particles is ∼1.26 nm (±0.66, n = 231) corresponding to 1–3 layers of graphitic carbon (Fig. 2d and e). These observations demonstrate that cATP exfoliation yields a nearly homogeneous dispersion of GQDs.
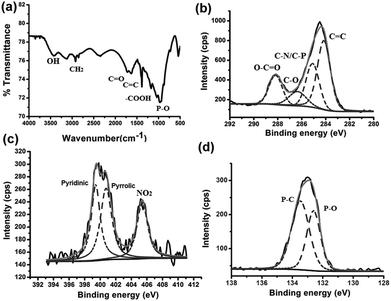
The Fourier transform infrared (FTIR) spectrum of ATP-GQD shows the characteristic peaks for C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (1722 cm−1), C
O (1722 cm−1), C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C (1629 cm−1), OH (3414 cm−1), –COOH (1388 cm−1), P–O (966 cm−1) and CH2 (2922 cm−1) (Fig. 3a). On further characterization by X-ray photoelectron spectroscopy (XPS), the high resolution C1s spectrum of GQDs can be well-fitted with the characteristic peaks for C
C (1629 cm−1), OH (3414 cm−1), –COOH (1388 cm−1), P–O (966 cm−1) and CH2 (2922 cm−1) (Fig. 3a). On further characterization by X-ray photoelectron spectroscopy (XPS), the high resolution C1s spectrum of GQDs can be well-fitted with the characteristic peaks for C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonds (284.2 eV),31 C–N/C–P
C bonds (284.2 eV),31 C–N/C–P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (285.1 eV),19,32 C–O (286.3 eV)33 and O–C
O (285.1 eV),19,32 C–O (286.3 eV)33 and O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (288.2 eV)19 (Fig. 3b). It correlates well with the FTIR results. The N1s spectrum consists of three peaks at 399.3, 400.5 and 405.1 eV, corresponding to the pyridinic,34 pyrrolic,34 and NO2 groups, respectively (Fig. 3c). The NO2 groups are likely introduced at the edges during the exfoliation with HNO3. Analysis of the P2p XPS spectrum gives two peaks at 132.6 eV and 133.5 eV (Fig. 3d), which can be assigned to P–O and P–C bonds.35,36 It is likely that these bonds belong to phosphonic acid groups (–PO3H2) at the edges because incorporation of P in the graphene lattice would introduce unstable structural distortion due to the longer length of the P–C bond than that of the C–C or C–N bond.37 As reported by XPS, the doping percentages of nitrogen and phosphorus are 6.2 (C/N ratio of 7.0) and 6.9 (C/P ratio of 6.3), respectively. Evidently, ATP-GQDs are doped with both N and P atoms. P-doped GQDs have yet to be synthesized, and this is the first demonstration of P-/N-co-doping. The N-doping level here is higher than the previously reported N-/B-co-doped GQDs.38 The interlayer spacing of ATP-GQDs analyzed by X-ray diffraction (XRD) is ∼0.36 nm (Fig. S1a†), which is slightly larger than that of graphite (0.34 nm) due to the presence of chemical groups. The Raman spectrum of ATP-GQD is similar to that of cATP (Fig. S1b†).
O (288.2 eV)19 (Fig. 3b). It correlates well with the FTIR results. The N1s spectrum consists of three peaks at 399.3, 400.5 and 405.1 eV, corresponding to the pyridinic,34 pyrrolic,34 and NO2 groups, respectively (Fig. 3c). The NO2 groups are likely introduced at the edges during the exfoliation with HNO3. Analysis of the P2p XPS spectrum gives two peaks at 132.6 eV and 133.5 eV (Fig. 3d), which can be assigned to P–O and P–C bonds.35,36 It is likely that these bonds belong to phosphonic acid groups (–PO3H2) at the edges because incorporation of P in the graphene lattice would introduce unstable structural distortion due to the longer length of the P–C bond than that of the C–C or C–N bond.37 As reported by XPS, the doping percentages of nitrogen and phosphorus are 6.2 (C/N ratio of 7.0) and 6.9 (C/P ratio of 6.3), respectively. Evidently, ATP-GQDs are doped with both N and P atoms. P-doped GQDs have yet to be synthesized, and this is the first demonstration of P-/N-co-doping. The N-doping level here is higher than the previously reported N-/B-co-doped GQDs.38 The interlayer spacing of ATP-GQDs analyzed by X-ray diffraction (XRD) is ∼0.36 nm (Fig. S1a†), which is slightly larger than that of graphite (0.34 nm) due to the presence of chemical groups. The Raman spectrum of ATP-GQD is similar to that of cATP (Fig. S1b†).
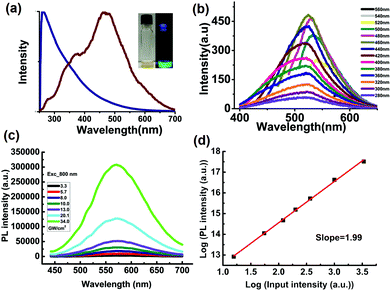
ATP-GQDs appear brown under bright light and show green fluorescence on illumination with a 365 nm UV lamp (Fig. 4a, inset). From the UV-Vis spectrum, it can be seen that, when adsorption extends to the visible light region, ATP-GQDs can more efficiently absorb short-wavelengths due to π–π* transitions of the aromatic sp2 domains in the graphene lattice31 (Fig. 4a). The photoluminescence (PL) excitation spectrum shows a prominent peak at 460 nm and a shoulder peak at 360 nm which is due to the σ–π and π–π* transitions originated from the carbene-like triplet state of the zig-zag edges of GQDs (Fig. 4a).39
In contrast to most reported GQDs,40–42 the PL of ATP-GQDs only exhibits a slight excitation dependent emission with the maximum emission at 520 nm when excited at 460 nm (Fig. 4b and S2†). It suggests the good uniformity of size and chemical composition of our ATP-GQDs.17 The quantum yield (QY) is calculated to be ∼27.5% using rhodamine 6G (QY 96%) as the reference, which is much higher than the non-doped GQDs.40 The improved QY could be attributable to the interplay of the different dopants in the graphene lattice. It has been proposed that the coexistence of n- and p-dopants can increase QY due to formation of p–n type photochemical diodes.43 In ATP-GQD, pyridinic and pyrrolic N atoms are strongly electron withdrawing conferring p-type doping characteristics.44 The electron-withdrawing –PO3H2 groups impose p-doping effects.45 In addition, the presence of oxygenated groups at the edges also gives rise to either weak n- (e.g. –OH) or p- (e.g. –COOH) doping effects.43
It is known that oxygenated groups on GQDs compromise QY because of non-radiative recombination at these sites.46 Li et al. have demonstrated that greenish-yellow luminescent GQDs prepared via a microwave-assisted method become blue with a much improved QY (from 11.7 to 22.9%).47 Consistently, we have shown that chemical reduction of ATP-GQDs using NaBH4 can largely increase QY to 53%, and at the same time, blue shift the emission peak to ∼475 nm (Fig. S3a†). But the emission spectrum is broadened, presumably due to the increased chemical heterogeneity after reduction. From the FTIR spectrum (Fig. S3b†), it can be observed that the –COOH, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, and P–O groups are eliminated by reduction while –OH groups are still present because of reduction of –COOH to –CH2OH. Clearly, the overall oxygen content is greatly decreased after chemical reduction. On the other hand, removing oxygenated groups makes GQDs more prone to aggregation and more difficult to be conjugated with biomolecules. Therefore, in the later bioimaging experiments, we have used the as-prepared ATP-GQDs without reduction.
O, and P–O groups are eliminated by reduction while –OH groups are still present because of reduction of –COOH to –CH2OH. Clearly, the overall oxygen content is greatly decreased after chemical reduction. On the other hand, removing oxygenated groups makes GQDs more prone to aggregation and more difficult to be conjugated with biomolecules. Therefore, in the later bioimaging experiments, we have used the as-prepared ATP-GQDs without reduction.
Based on the chemical/element composition analyses and observed quantum dot size, we propose a hypothetical model for ATP-GQD (Fig. S4†) and theoretically investigate its PL properties using density-functional theory (DFT) and time-dependent DFT (TDDFT) calculations. The hypothetical ATP-GQD (∼2 nm) contains 3 pyridinic N atoms, 3 pyrrolic N atoms, and 3 –PO3H2 groups. As the synthesized ATP-GQDs have rich oxygen content and correlate with XPS characterization, 1 epoxy group is added onto the graphene lattice, 1 –NO2 and 2 –COOH are attached to pyridine rings. The TDDFT calculations suggest that ATP-GQD exhibits the maximum emission at 556 nm which is close to the experimentally measured value (520 nm) (Fig. S4a†). Furthermore, our calculations suggest that chemical reduction (removal of oxygenated groups) can cause either a red- or blue-shift (Fig. S4b–f†), probably accounting for the observed broadening of the emission spectrum. In addition, it is likely that the reduction pathway from Fig. S4a–f† dominates, giving an emission peak of 496.6 nm which is close to the experimental value of 475 nm. The molecular weight of the hypothetical ATP-GQD is ∼1.4 kDa, which is much smaller than green fluorescence protein (GFP, ∼27 kDa).
A major drawback of the currently used organic fluorophores (e.g., GFP and FITC) is their poor photostability, which makes long-term real-time imaging difficult. As shown in Fig. S5,† under the same confocal illumination, FITC photobleaches quickly whereas the PL of ATP-GQD remains stable. Because ATP molecules abundantly exist in cells, it is not surprising that ATP-GQDs show good biocompatibility at concentrations much higher than needed for bioimaging experiments (Fig. S6†).
Furthermore, we examined the two-photon absorption properties of ATP-GQDs as two-photon fluorescence microscopy has advantages over normal one-photon fluorescence microscopy in terms of a larger penetration depth amenable for tissue imaging, a higher spatial resolution and a lower background signal due to the highly non-linear and localized adsorption process.48 Interestingly, ATP-GQD also exhibits good two-photon excitation properties. Under the excitation at 800 nm, which is far away from the one-photon absorption regime of GQD, strong up-converted PL is observed with the emission maximum at ∼560 nm (Fig. 4c). The quadratic dependence of the PL signal on the excitation intensity confirms the non-linear two-photon absorption and emission process (Fig. 4d).49 The Z-scan technique is employed to determine the two-photon absorption cross-section (σ) of our ATP-GQDs (Fig. S7†). The nonlinear absorption signal of the ATP-GQDs can be well-fitted by using the Z-scan theory,49,50 giving a σ value as high as ∼20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 GM. This value is two orders of magnitude higher than organic fluorphores51 and comparable to much larger-sized CdSe quantum dots.52
000 GM. This value is two orders of magnitude higher than organic fluorphores51 and comparable to much larger-sized CdSe quantum dots.52
Un-doped GQDs exhibit poor two-photon excitation properties due to the low absorption cross-section.53 It has been observed that the two-photon absorption cross-section of quadrupolar molecules increases due to the co-existence of electron withdrawing and donating groups which facilitates intramolecular charge transfer.54 A theoretical study also suggests that non-linear emission from GQDs could arise when electron-withdrawing NO2 and electron-donating NH2 groups at the edges form a donor-GQD-acceptor system.55 Liu et al. have demonstrated that dimethyl amine functionalized GQD shows strong two-photon absorption and attributed this to the electron-donating nature of the dimethyl amine groups.13 Hence conceivably, the co-existence of electron withdrawing and donating sites (which can form p–n type photochemical diodes) in heteroatom-doped ATP-GQD leads to its good two-photon upconversion properties.
ATP-GQD demonstrates excellent photostability under continuous one-photon or two-photon laser excitation (Fig. S8†). The lifetime measurements of ATP-GQDs under both one- (400 nm) and two-photon excitation show nearly identical PL decay dynamics (Fig. S9†), suggesting the common radiative recombination channels.50 The PL decay curves can be well-fitted by a double-exponential function: I(t) = A1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) exp
exp![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) (−t/τ1) + A2
(−t/τ1) + A2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) exp
exp![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) (−t/τ2), where τ1 and τ2 are the time constants of the two radiative decay channels; A1 and A2 are the corresponding amplitudes. From the best fit of the data, τ1 (A1) and τ2 (A2) are derived to be 320 ps (0.44) and 1.62 ns (0.56), respectively. The nanosecond-scale lifetime suggests that ATP-GQDs are suitable fluorescent probes for bio-imaging.9
(−t/τ2), where τ1 and τ2 are the time constants of the two radiative decay channels; A1 and A2 are the corresponding amplitudes. From the best fit of the data, τ1 (A1) and τ2 (A2) are derived to be 320 ps (0.44) and 1.62 ns (0.56), respectively. The nanosecond-scale lifetime suggests that ATP-GQDs are suitable fluorescent probes for bio-imaging.9
ATP-GQD is desirable as a fluorescent tag for bioimaging because of its high QY, small molecular weight, excellent photostability and good biocompatibility. A number of studies have provided proof-of-concept demonstrations to show the potential of GQDs for bioimaging, through simple non-specific cell imaging of passively taken GQDs. However, for useful biological studies, GQDs should be bio-functionalized for specific and real-time molecular imaging in live cells. Here, we conjugated ATP-GQD with a transferrin molecule for imaging and real-time tracking of transferrin receptors in human cervical cancer (HeLa) cells. Many cancerous cells overexpress transferrin receptors, which are responsible for internalization and recycling of iron-bound transferrin molecules. The gel electrophoresis experiment indicates that the molecular weight of transferrin-conjugated GQD (Tr-GQD) is similar to that of a bare transferrin molecule (80 kDa) implying one-to-one pairing between ATP-GQD and transferrin (Fig. S10†). The successful conjugation of transferrin is evidenced by staining of Tr-GQD by transferrin-binding Coomassie blue (Fig. S10†).
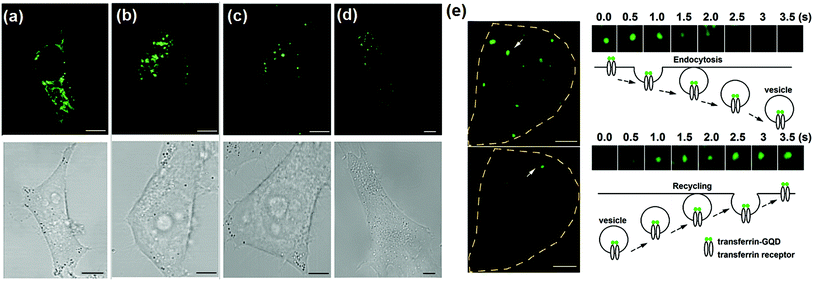
Using confocal microscopy, it is found that ATP-GQDs can be readily and non-specifically taken up by HeLa cells (Fig. 5a). The internalized ATP-GQDs scatter in the cytoplasm. In comparison, the uptaken Tr-GQDs are segregated near the nucleus because of the specific binding of Tr-GQDs with transferrin receptors on the cell membrane triggers endocytosis and compartmentalization in the recycling endosomes (a network of tubular structures adjacent to the nucleus) as shown in Fig. 5b.56 When excess free transferrin molecules are introduced together with Tr-GQDs, fewer fluorescent puncta could be observed within the cells due to competitive inhibition, confirming the specific binding between Tr-GQDs and transferrin receptors (Fig. 5c). Furthermore, Tr-GQD staining is much weaker in human fibroblasts (ATCC) (Fig. 5d) because these non-cancerous cells have less transferrin receptors expressed. Using total internal reflection fluorescence microscopy (TIRFM) which evanescently illuminates only the thin plasmalemmal region (<200 nm thick), it is further observed that Tr-GQD/transferrin complexes on the cell membrane are quickly endocytosed and many endocytic events are observed within 3 min of imaging (Fig. 5e). Individual endocytic (Fig. 5e, right top) and exocytic events (Fig. 5e, right bottom) of the vesicles containing internalized complexes can also be resolved.
Experimental
Synthesis of ATP-GQDs
ATP powder (1 g, Acros Organics) was carbonized by heating in a quartz tube furnace at 900 °C for 1 h under an argon atmosphere. This was followed by refluxing in HNO3 (6 M, Honeywell Chemicals) for 24 h. The solution was then cooled and centrifuged at 7500g for 20 min. The supernatant was collected and the excess HNO3 was removed by evaporation at 200 °C. The resulting powder was subsequently dissolved in DI water and dialyzed using a cellulose ester dialysis membrane (500–1000 Da MWCO, Spectra/Pro Biotec) for 2 days to remove ions and other impurities.Oxygen reduction reaction (ORR)
The carbonized ATP samples were washed with DI water 6 times to remove ions or other impurities and dried at 50 °C in a hot air oven. The carbonized ATP dispersion (2 mg ml−1) was drop-cast onto a polished glassy carbon electrode. The ORR measurements were conducted with a CHI 660D electrochemical workstation (Chenhua), using a nitrogen or oxygen saturated KOH (0.1 M) solution as the electrolyte.Characterization
Samples were characterized by field-emission scanning electron microscopy (JSM6700-FESEM, JEOL), Raman spectroscopy (WITeck CRM200), atomic force microscopy (MFP-3D AFM, Asylum Research), high-resolution transmission-electron microscopy (JEOL-2010), Fourier transform infrared spectroscopy (Perkin-Elmer Spectrum GX FTIR system), X-ray photoelectron spectroscopy (Theta Probe), X-ray diffraction analysis (Bruker D8 Advanced Diffraction meter using Cu Kα radiation), UV-vis absorption spectroscopy (UV-2450 spectrophotometer, Shimadzu), photoluminescence spectroscopy (LS-55 fluorescence spectrometer, Perkin-Elmer) and polyacrylamide gel electrophoresis and imaging (Mini-Protean Tetra Cell, Bio-Rad and ProXPRESS 2D, Perkin-Elmer). The two-photon excited PL properties at 800 nm excitation were investigated using a femtosecond amplified-pulsed laser with a repetition rate of 1000 Hz and a pulse-width of 100 fs.50 The lifetime measurements were carried out for the one- and two-photon excitation using an Optronics streak camera system.57Cell imaging
Human cervical cancer cells (HeLa, ATCC) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Gibco). For the imaging experiments, HeLa cells were seeded on a Lab-Tek II chambered cover glass and grown overnight. Prior to imaging, the cells were incubated with ATP-GQDs for 2 h or Tr-GQDs for 30 min at 37 °C. The cells were rinsed thrice and incubated in a bath solution (150 mM NaCl, 5 mM KCl, 1.1 mM MgCl2, 2.6 mM CaCl2, 10 mM HEPES, and 10 mM glucose, pH 7.4) during imaging. Confocal microscope (Zeiss LSM 510) with a 63× oil objective and a 488 nm laser was used to obtain the confocal images. Time-lapse digital images were recorded using a Zeiss Axiovert-200M inverted total-internal-reflection fluorescence microscope system (Carl Zeiss, Germany) equipped with a 100× objective lens (numerical aperture, NA = 1.45) and a charge coupled device (CCD) camera with a pixel size of 0.248 μm. Using MetaMorph 6.3 (Universal Imaging Corp., Downingtown, PA), time-lapse images of 361 frames (0.5 s per frame) were acquired at 37 °C, and analyzed using ImageJ (National Institute of Health, USA).Conclusions
In summary, we have devised a simple strategy to synthesize N and P co-doped GQDs using a biomolecule (ATP) as the precursor. Such novel QDs exhibit high brightness (QY ∼27.5% and ∼53.0% after chemical reduction), low molecular weight (∼1.4 kDa), good two-photon excitation properties (absorption cross-section as high as 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 GM), excellent one-photon and two-photon stability, good biocompatibility, high water solubility, and the ease to conjugation with biomolecules. We further demonstrate the potential of ATP-GQD for real-time molecular tracking in live cells whereby uncovering the dynamic cellular events, in this case distribution, trafficking, and recycling of transferrin receptors.
000 GM), excellent one-photon and two-photon stability, good biocompatibility, high water solubility, and the ease to conjugation with biomolecules. We further demonstrate the potential of ATP-GQD for real-time molecular tracking in live cells whereby uncovering the dynamic cellular events, in this case distribution, trafficking, and recycling of transferrin receptors.
Acknowledgements
We thank the Ministry of Education of Singapore under an AcRF Tier 2 grant (MOE2014-T2-2-003, ARC31/14), Singapore National Research Foundation under its CBRG grant (NMRC/CBRG/0070/2014) administrated by the Singapore Ministry of Health's National Medical Research Council, and Agency for Science, Technology and Research under its SERC grant (SERC#1021700137) for their support.Notes and references
- L. Li, G. Wu, G. Yang, J. Peng, J. Zhao and J. J. Zhu, Nanoscale, 2013, 5, 4015 RSC.
- J. Shen, Y. Zhu, X. Yang and C. Li, Chem. Commun., 2012, 48, 3686 RSC.
- H. Sun, L. Wu, W. Wei and X. Qu, Mater. Today, 2013, 16, 433 CrossRef PubMed.
- Z. P. Zhang, J. Zhang, N. Chen and L. T. Qu, Energy Environ. Sci., 2012, 5, 8869 Search PubMed.
- M. M. Xie, Y. J. Su, X. N. Lu, Y. Z. Zhang, Z. Yang and Y. F. Zhang, Mater. Lett., 2013, 93, 161 CrossRef PubMed.
- L. Lin and S. Zhang, Chem. Commun., 2012, 48, 10177 RSC.
- D. B. Shinde and V. K. Pillai, Chem. – Eur. J., 2012, 18, 12522 CrossRef PubMed.
- X. Wu, F. Tian, W. X. Wang, J. Chen, M. Wu and J. X. Zhao, J. Mater. Chem. C, 2013, 1, 4676 RSC.
- J. Peng, W. Gao, B. K. Gupta, Z. Liu, R. Romero-Aburto, L. Ge, L. Song, L. B. Alemany, X. Zhan, G. Gao, S. A. Vithayathil, B. A. Kaipparettu, A. A. Marti, T. Hayashi, J. J. Zhu and P. M. Ajayan, Nano Lett., 2012, 12, 844 CrossRef CAS PubMed.
- Y. Q. Dong, C. Q. Chen, X. T. Zheng, L. L. Gao, Z. M. Cui, H. B. Yang, C. X. Guo, Y. W. Chi and C. M. Li, J. Mater. Chem., 2012, 22, 8764 RSC.
- W. J. Xie, Y. Y. Fu, H. Ma, M. Zhang and L. Z. Fan, Acta Chim. Sin., 2012, 70, 2169 CrossRef.
- L. Zhang, Y. Xing, N. He, Y. Zhang, Z. Lu, J. Zhang and Z. Zhang, J. Nanosci. Nanotechnol., 2012, 12, 2924 CrossRef PubMed.
- Q. Liu, B. Guo, Z. Rao, B. Zhang and J. R. Gong, Nano Lett., 2013, 13, 2436 CrossRef PubMed.
- X. T. Zheng, A. Than, A. Ananthanaraya, D.-H. Kim and P. Chen, ACS Nano, 2013, 7, 6278 CrossRef PubMed.
- X. T. Zheng, A. Ananthanarayanan, K. Q. Luo and P. Chen, Small, 2015, 11, 1620 CrossRef PubMed.
- X. Wang, G. Sun, P. Routh, D.-H. Kim, W. Huang and P. Chen, Chem. Soc. Rev., 2014, 43, 7067 RSC.
- M. A. Sk, A. Ananthanarayanan, L. Huang, K. H. Lim and P. Chen, J. Mater. Chem. C, 2014, 2, 6954 RSC.
- Y. Li, Y. Zhao, H. H. Cheng, Y. Hu, G. Q. Shi, L. M. Dai and L. T. Qu, J. Am. Chem. Soc., 2012, 134, 15 CrossRef PubMed.
- M. Li, W. B. Wu, W. C. Ren, H. M. Cheng, N. J. Tang, W. Zhong and Y. W. Du, Appl. Phys. Lett., 2012, 101 Search PubMed.
- T. Palaniselvam, M. O. Valappil, R. Illathvalappil and S. Kurungot, Energy Environ. Sci., 2014, 7, 1059 Search PubMed.
- C. F. Hu, Y. L. Liu, Y. H. Yang, J. H. Cui, Z. R. Huang, Y. L. Wang, L. F. Yang, H. B. Wang, Y. Xiao and J. H. Rong, J. Mater. Chem. B, 2013, 1, 39 RSC.
- J. Ju and W. Chen, Biosens. Bioelectron., 2014, 58, 219 CrossRef PubMed.
- D. Qu, M. Zheng, P. Du, Y. Zhou, L. Zhang, D. Li, H. Tan, Z. Zhao, Z. Xie and Z. Sun, Nanoscale, 2013, 5, 12272 RSC.
- D. Qu, M. Zheng, L. Zhang, H. Zhao, Z. Xie, X. Jing, R. E. Haddad, H. Fan and Z. Sun, Sci. Rep., 2014, 4 Search PubMed.
- L. Lin, M. Rong, S. Lu, X. Song, Y. Zhong, J. Yan, Y. Wang and X. Chen, Nanoscale, 2015, 7, 1872 RSC.
- H. Wang, T. Maiyalagan and X. Wang, ACS Catal., 2012, 2, 781 CrossRef.
- I.-Y. Jeon, H.-J. Choi, S.-M. Jung, J.-M. Seo, M.-J. Kim, L. Dai and J.-B. Baek, J. Am. Chem. Soc., 2012, 135, 1386 CrossRef PubMed.
- L. T. Qu, Y. Liu, J. B. Baek and L. M. Dai, ACS Nano, 2010, 4, 1321 CrossRef PubMed.
- Z. J. Lu, S. J. Bao, Y. T. Gou, C. J. Cai, C. C. Ji, M. W. Xu, J. Song and R. Y. Wang, RSC Adv., 2013, 3, 3990 RSC.
- Z. H. Sheng, L. Shao, J. J. Chen, W. J. Bao, F. B. Wang and X. H. Xia, ACS Nano, 2011, 5, 4350 CrossRef CAS PubMed.
- Y. Li, Y. Zhao, H. Cheng, Y. Hu, G. Shi, L. Dai and L. Qu, J. Am. Chem. Soc., 2012, 134, 15 CrossRef CAS PubMed.
- S. H. Liao, P. L. Liu, M. C. Hsiao, C. C. Teng, C. A. Wang, M. D. Ger and C. L. Chiang, Ind. Eng. Chem. Res., 2012, 51, 4573 CrossRef.
- H. Jabeen, V. Chandra, S. Jung, J. W. Lee, K. S. Kim and S. Bin Kim, Nanoscale, 2011, 3, 3583 RSC.
- Z. Luo, S. Lim, Z. Tian, J. Shang, L. Lai, B. MacDonald, C. Fu, Z. Shen, T. Yu and J. Lin, J. Mater. Chem., 2011, 21, 8038–8044 RSC.
- J. N. Hart, P. W. May, N. L. Allan, K. R. Hallam, F. Claeyssens, G. M. Fuge, M. Ruda and P. J. Heard, J. Solid State Chem., 2013, 198, 466 CrossRef PubMed.
- K. S. Prasad, R. Pallela, D. M. Kim and Y. B. Shim, Part. Part. Syst. Char., 2013, 30, 557 CrossRef CAS PubMed.
- H.-m. Wang, H.-x. Wang, Y. Chen, Y.-j. Liu, J.-x. Zhao, Q.-h. Cai and X.-z. Wang, Appl. Surf. Sci., 2013, 273, 302 CrossRef PubMed.
- S. Dey, A. Govindaraj, K. Biswas and C. N. R. Rao, Chem. Phys. Lett., 2014, 595, 203 CrossRef PubMed.
- Y. Li, Y. Hu, Y. Zhao, G. Shi, L. Deng, Y. Hou and L. Qu, Adv. Mater., 2011, 23, 776 CrossRef PubMed.
- Y. Dong, C. Chen, X. Zheng, L. Gao, Z. Cui, H. Yang, C. Guo, Y. Chi and C. M. Li, J. Mater. Chem., 2012, 22, 8764 RSC.
- X. Wu, F. Tian, W. Wang, J. Chen, M. Wu and J. X. Zhao, J. Mater. Chem. C, 2013, 1, 4676 RSC.
- A. Ananthanarayanan, X. Wang, P. Routh, B. Sana, S. Lim, D.-H. Kim, K.-H. Lim, J. Li and P. Chen, Adv. Funct. Mater., 2014, 24, 3021 CrossRef PubMed.
- T.-F. Yeh, C.-Y. Teng, S.-J. Chen and H. Teng, Adv. Mater., 2014, 26, 3297 CrossRef PubMed.
- T. Schiros, D. Nordlund, L. Palova, D. Prezzi, L. Y. Zhao, K. S. Kim, U. Wurstbauer, C. Gutierrez, D. Delongchamp, C. Jaye, D. Fischer, H. Ogasawara, L. G. M. Pettersson, D. R. Reichman, P. Kim, M. S. Hybertsen and A. N. Pasupathy, Nano Lett., 2012, 12, 4025 CrossRef PubMed.
- I. Gillaizeau-Gauthier, F. Odobel, M. Alebbi, R. Argazzi, E. Costa, C. A. Bignozzi, P. Qu and G. J. Meyer, Inorg. Chem., 2001, 40, 6073 CrossRef.
- S. Zhu, J. Zhang, S. Tang, C. Qiao, L. Wang, H. Wang, X. Liu, B. Li, Y. Li, W. Yu, X. Wang, H. Sun and B. Yang, Adv. Funct. Mater., 2012, 22, 4732 CrossRef PubMed.
- L.-L. Li, J. Ji, R. Fei, C.-Z. Wang, Q. Lu, J.-R. Zhang, L.-P. Jiang and J.-J. Zhu, Adv. Funct. Mater., 2012, 22, 2971 CrossRef PubMed.
- G. S. He, L.-S. Tan, Q. Zheng and P. N. Prasad, Chem. Rev., 2008, 108, 1245 CrossRef CAS PubMed.
- Y. Wang, X. Yang, T. C. He, Y. Gao, H. V. Demir, X. W. Sun and H. D. Sun, Appl. Phys. Lett., 2013, 102, 021917 CrossRef PubMed.
- Y. Wang, V. D. Ta, Y. Gao, T. C. He, R. Chen, E. Mutlugun, H. V. Demir and H. D. Sun, Adv. Mater., 2014, 26, 2954 CrossRef PubMed.
- W. R. Zipfel, R. M. Williams and W. W. Webb, Nat. Biotechnol., 2003, 21, 1369 CrossRef PubMed.
- S.-C. Pu, M.-J. Yang, C.-C. Hsu, C.-W. Lai, C.-C. Hsieh, S. H. Lin, Y.-M. Cheng and P.-T. Chou, Small, 2006, 2, 1308 CrossRef PubMed.
- A. Zhu, C. Ding and Y. Tian, Sci. Rep., 2013, 3, 2933 Search PubMed.
- W.-H. Lee, M. Cho, S.-J. Jeon and B. R. Cho, J. Phys. Chem. A, 2000, 104, 11033 CrossRef.
- Z.-J. Zhou, Z.-B. Liu, Z.-R. Li, X.-R. Huang and C.-C. Sun, J. Phys. Chem. C, 2011, 115, 16282 CAS.
- A. Nagabhushana, M. L. Chalasani, N. Jain, V. Radha, N. Rangaraj, D. Balasubramanian and G. Swarup, BMC Cell Biol., 2010, 11, 4 CrossRef PubMed.
- Y. Wang, K. S. Leck, V. D. Ta, R. Chen, V. Nalla, Y. Gao, T. He, H. V. Demir and H. Sun, Adv. Mater., 2015, 27, 169 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Supplementary figures related to characterization, computational studies and protein conjugation. See DOI: 10.1039/c5nr01519g |
| This journal is © The Royal Society of Chemistry 2015 |