 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Combinatorial chemistry in nematodes: modular assembly of primary metabolism-derived building blocks
Stephan H.
von Reuss
*a and
Frank C.
Schroeder
*b
aMax Planck Institute for Chemical Ecology, Department of Bioorganic Chemistry, Jena, Germany. E-mail: svonreuss@ice.mpg.de
bBoyce Thompson Institute and Department of Chemistry and Chemical Biology, Cornell University, Ithaca, NY 14853, USA. E-mail: Schroeder@cornell.edu
First published on 10th June 2015
Abstract
Covering: up to 2015
The nematode Caenorhabditis elegans was the first animal to have its genome fully sequenced and has become an important model organism for biomedical research. However, like many other animal model systems, its metabolome remained largely uncharacterized, until recent investigations demonstrated the importance of small molecule-based signalling cascades for virtually every aspect of nematode biology. These studies have revealed that nematodes are amazingly skilled chemists: using simple building blocks from conserved primary metabolism and a strategy of modular assembly, C. elegans and other nematode species create complex molecular architectures to regulate their development and behaviour. These nematode-derived modular metabolites (NDMMs) are based on the dideoxysugars ascarylose or paratose, which serve as scaffolds for attachment of moieties from lipid, amino acid, carbohydrate, citrate, and nucleoside metabolism. Mutant screens and comparative metabolomics based on NMR spectroscopy and MS have so-far revealed several 100 different ascarylose (“ascarosides”) and a few paratose (“paratosides”) derivatives, many of which represent potent signalling molecules that can be active at femtomolar levels, regulating development, behaviour, body shape, and many other life history traits. NDMM biosynthesis appears to be carefully regulated as assembly of different modules proceeds with very high specificity. Preliminary biosynthetic studies have confirmed the primary metabolism origin of some NDMM building blocks, whereas the mechanisms that underlie their highly specific assembly are not understood. Considering their functions and biosynthetic origin, NDMMs represent a new class of natural products that cannot easily be classified as “primary” or “secondary”. We believe that the identification of new variants of primary metabolism-derived structures that serve important signalling functions in C. elegans and other nematodes provides a strong incentive for a comprehensive re-analysis of metabolism in higher animals, including humans.
1 Introduction: chemical communication in nematodes
The nematodes (roundworms) constitute one of the largest groups of animals, both with respect to the number of different species as well as individuals.1 Though rarely entering the spotlight, parasitic nematodes are responsible for significant agricultural losses2 and are the causative agents of widespread human disease in developing countries.3,4 In addition, the free-living bacteriovorous nematode Caenorhabditis elegans has become one of the most important model organisms for biomedical research. Not least as a result of the early (1998) sequencing of the C. elegans genome,5 our understanding of nematode biology and genetics has increased dramatically over the past 20 years. It has become apparent that much of C. elegans biology is governed by evolutionarily conserved signalling pathways, which enabled exploring ancient features of the regulatory circuitry that controls organismal development, reproduction, longevity, and metabolism.Surprisingly, nematode biochemistry and metabolism remained largely uncharacterized until recent investigations demonstrated the central importance of small molecule-based signalling cascades for nematode behaviour and development. Interorganismal chemical communication in nematodes was already suspected by Greet in 1964 who reported that males and females of the free living Panagrolaimus rigidus, when separated by a cellophane barrier, moved towards members of the opposite sex but not to members of the same sex, suggesting the release of gender-specific attractants, e.g. sex pheromones.6 Subsequent research demonstrated the presence of small molecule signals modulating gender-specific attraction in more than 40 representative nematodes including free living, entomopathogenic, plant-parasitic, and zoo-parasitic species.7–18 However, despite considerable efforts to isolate and identify nematode pheromones, molecular structures remained elusive for many decades.
Recent research focusing on the free-living Caenorhabditis elegans, the necromenic Pristionchus pacificus, and a few other species revealed that nematodes are amazingly skilled chemists: using simple building blocks from conserved primary metabolism and a strategy of modular assembly, they create complex molecular architectures, the nematode-derived modular metabolites (NDMMs) to regulate their development and behaviour. In this review, we will describe the discovery of this modular library, summarize current knowledge regarding the biosynthesis and perception of these components and outline possible implications for small-molecule signalling in higher organisms. Earlier reviews of this field focussed primarily on the biological activity of the initially discovered members of this family of signaling molecules.19,20
2 The discovery of ascaroside-based signalling in the model organism Caenorhabditis elegans
The bacteriovorous C. elegans, first described by Émile Maupas as Rhabditis elegans21 was introduced as a model organism for biological research by Sydney Brenner in 1963, given its ease in cultivation, short life cycle, and simple body plan.22C. elegans is an androdioecious nematode species, that is, it is composed of self-fertile hermaphrodites and males. Hermaphrodites constitute more than 99% of worms under laboratory conditions, and the ability to propagate worms from self-fertile hermaphrodites greatly simplifies genetics and mutant creation. Over the past 30 years, C. elegans research has contributed to many scientific milestones, including the dissection of programmed cell death,23 systematic genome cloning,24 first complete genome sequencing of a multicellular organism,5 discovery of RNA interference,25 and the use of green fluorescence protein (GFP),26 and C. elegans has emerged as an extremely successful model system. However, as is the case for many other animal model systems, its metabolome and biochemistry remained largely uncharacterized.The first hint that small molecule signalling may play an important role in C. elegans life history emerged from studies of a peculiar state of developmental diapause called dauer, from the German word for “enduring”. Under unfavourable conditions such as lack of food, high temperature, or high population density, C. elegans larvae abort reproductive development and instead enter a specialized, non-feeding and highly stress resistant larval stage that is optimized for dispersal and long-term survival (Fig. 1a). When environmental conditions improve, these dauer larvae resume normal development to reproductive adults.27 Formation of dauer larvae is regulated by conserved signalling pathways, including insulin and TGF-β signalling as well as steroid hormone biosynthesis converging on the nuclear hormone receptor DAF-12, a liver-X and vitamin D receptor homologue.28–31 Given that dauer formation results in greatly extended lifespan, the mechanisms that regulate entry and exit from the dauer stage have become an important focus of aging research.28
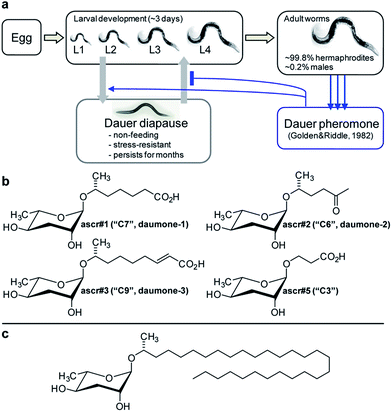 | ||
| Fig. 1 Initial identification of ascarosides in nematodes. (a) C. elegans life cycle. (b) Ascarosides with potent dauer inducing activity. (c) Lipid-like ascaroside from Ascaris spp. | ||
In 1982 Golden and Riddle reported that dauer formation is modulated by a worm-derived small molecule signal that is constitutively excreted by the worms and “may be a hydroxylated, short-chain fatty acid or a mixture of closely related compounds”.32,33 The nematode origin of this pheromone was proven unambiguously by showing that its production depends on a gene (of then unknown identity) named daf-22.34 However, the molecular structure of the dauer pheromone remained elusive for more than 20 years until several laboratories reported the identification of a family of dauer inducing glycosides derived from L-α-3,6-dideoxymannose, a sugar known as ascarylose, and a variety of hydroxylated short-chain fatty acids.35–38
Ascarylose-based glycosides were first described as an “unsaponifiable matter” that accounted for about 25% of the total lipid content of the human intestinal parasitic nematode Ascaris lumbricoides.39 At the time of this initial report (1912 !), the chemical structure of the A. lumbricoides-derived lipids could not be determined, and it was not until 1957 that the exact structures of the sugar and fatty acid parts of these ascarosides were determined (Fig. 1c).40 The A. lumbricoides-derived ascarosides feature very long aliphatic side chains (predominantly 29 and more carbons in length), are produced in copious amounts and are presumed to form part of a protective coating of Ascaris eggs.
In contrast, the dauer-inducing ascarosides identified from C. elegans incorporate short fatty acid-derived side chains of only 3–9 carbons (Fig. 1b) and are produced at low (nano- or micromolar) concentrations. Activity-guided fractionation of worm culture media with dauer-inducing activity resulted in the isolation of ascr#1 (daumone#1, C7),35† the methyl ketone ascr#2 (daumone#2, C6), ascr#3 (daumone#3, C9),36 and ascr#5 (C3),37 which were shown to induce dauer formation at near-physiological concentrations. However, even mixtures of synthetic samples of these compounds failed to fully reproduce the activity of unfractionated worm media, suggesting that there must be additional dauer-inducing components that had been missed by activity-guided fractionation. As described in the following section, the use of comparative metabolomics enabled the identification of important additional dauer pheromone components, including the so far most-potent dauer-inducing ascaroside, ascr#8 (Fig. 2).38 Ascr#8 represents the first example for an ascaroside that incorporates additional moieties beyond the ascarylose sugar and the fatty acid-derived side chain, and thus provided a first glimpse at the vast diversity of multi-modular ascarosides – the NDMMs – later identified from C. elegans, P. pacificus, and other nematodes.
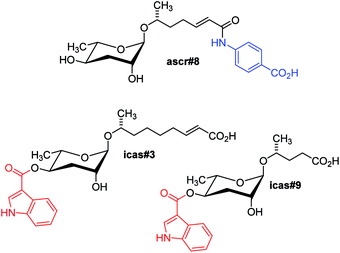 | ||
| Fig. 2 Modular ascarosides regulating development and mediating behavioural phenotypes in C. elegans, including building blocks derived from tryptophan (red) and folate (blue) metabolism. | ||
Soon after identification of the first dauer-inducing ascarosides, it became clear that ascarosides regulate many other aspects of nematode life history, including social behaviours and adult lifespan. For example, mixtures of ascr#2, its 2-glucoside ascr#4, ascr#3, and ascr#8 (Fig. 1b & 2) were found to act as potent male-specific attractants.38,41 These hermaphrodite-produced ascarosides strongly synergize in male attraction, that is, mixtures of these compounds were found to be much more attractive than when tested individually, even at elevated concentrations. Behavioural responses to these ascarosides were found to be highly sex-specific: whereas males are attracted by ascr#2 and ascr#3, these compounds act as deterrents for hermaphrodites. Generally, ascaroside concentrations required for behavioural responses (pico- to nanomolar) are much lower than those required for dauer induction.38
In addition, ascr#2 and ascr#3 (Fig. 1b) have been shown to regulate adult lifespan in a manner that is distinct from dauer induction. When exposed to nanomolar concentrations of these ascarosides after reaching adulthood, hermaphrodite worms live up to 25% longer than untreated controls.42 Other, yet unidentified ascarosides that are produced by male worms appear to shorten hermaphrodite lifespan.43
2.1 The role of comparative metabolomics for NDMM discovery
As described in the preceding paragraphs, the initial discovery of ascarosides as components of the C. elegans dauer and mating pheromone relied on activity-guided fractionation of worm culture media extracts.35–37,41 The ascarosides' synergistic activities, however, suggested that alternative methods for their detection may be required because compounds that act synergistically are likely to be missed by fractionation-based approaches since their activity will be greatly attenuated or abolished during purification. In fact, it was found that the initially identified ascarosides (ascr#1–5, Fig. 1b) did not fully reconstitute the activity of the natural pheromone blend. Therefore, identification of additional pheromone components was pursued using comparative metabolomics-based strategies that do not primarily rely on bioassays of fractionated material. Essential for the use of comparative metabolomics for pheromone identification was the earlier characterization of viable C. elegans mutants, e.g. daf-22, that do not produce dauer or sex pheromone.18,34 Comparative analysis of spectroscopic data derived from pheromone-containing wild-type exo-metabolome samples with data sets obtained from pheromone-deficient daf-22 exo-metabolome samples could thus reveal spectroscopic features that are daf-22 dependent and represent the sought-after additional pheromone components.For the purpose of comparing wild-type and daf-22 metabolome samples, Differential analysis of 2D NMR spectra (DANS), a method that relies on comparison of high-resolution dqfCOSY spectra, proved particularly useful.38 DANS revealed a series of daf-22-dependent NMR signals that enabled the identification of several additional ascarosides, including ascr#8 (Fig. 2) which incorporates a p-aminobenzoic acid (PABA) moiety likely derived from folate metabolism.38 Ascr#8 was then shown to be highly active in promoting male attraction in synergism with ascr#2 and ascr#3 (Fig. 1b), in addition to strong dauer-inducing activity. Synthetic blends of ascr#2, ascr#3, and ascr#8 at near-physiological concentrations were found to fully reconstitute the behavioural activity of unfractionated exo-metabolome, demonstrating the utility of DANS in connecting biological activity and molecular structures.38
In addition, DANS revealed a group of daf-22 dependent indole carboxy ascarosides (e.g. icas#3 and icas#9), which carry an indole-3-carboxylate moiety at the 4-position of the ascarylose and were subsequently shown to act as highly potent hermaphrodite aggregation pheromones (Fig. 2).44 The first indole ascaroside, icas#9, had previously been identified via activity-guided fractionation as a minor component of the dauer pheromone;45 however, extensive bioassays revealed that icas#9 and icas#3 act as potent attractants and aggregation pheromones at concentrations significantly lower than those required for dauer formation.44 Prior to the identification of the indole ascarosides, the existence of an aggregation pheromone in C. elegans had not been suspected, demonstrating how comparative metabolomic analyses and subsequent identification of novel biogenic small molecules can lead to the discovery of new and unexpected phenotypes.
2.2 Targeted metabolomics reveals structural and functional diversity
The finding that important behavioural phenotypes in C. elegans were elicited by picomolar concentrations of indole ascarosides, demonstrated the requirement to develop more sensitive methods for ascaroside screening. Considering the inherently low sensitivity of NMR spectroscopy in comparison to mass spectrometry (MS) a highly sensitive MS/MS technique for selective detection of known and yet unidentified ascarosides in highly complex metabolome extracts was developed, which depends on the production of a characteristic C3H5O2 fragment ion at m/z 73 upon collision induced dissociation (CID) of negatively charged ascaroside-based ions.46 Comparative MS/MS-based precursor ion screening of C. elegans wild type and mutant metabolomes enabled a functional characterization of several genes implicated in ascaroside biosynthesis (see following sections) and revealed a larger family of NDMMs incorporating building blocks derived from additional primary metabolic pathways, in addition to previously overlooked homologues and isomers.46 For example, targeted MS/MS analyses of the C. elegans metabolome revealed 4-(2-methylbutenoyl)- and 4-hydroxybenzoyl-substituted ascarosides, mbas#3 and hbas#3, respectively (Fig. 3). Whereas the 4-hydroxybenzoyl moiety is likely derived from tyrosine catabolism, the origin of the 2-methylbutenoic (tiglic) acid moiety in mbas#3 is less clear, as it could be derived from several different known pathways, for example isoleucine catabolism or mitochondrial short-chain fatty acid metabolism. hbas#3, which is produced at much lower concentrations (100–1000 fold less) than simple ascarosides such as ascr#3, was shown to be an extremely potent hermaphrodite attractant that is active even at low femtomolar concentrations.46 The biological functions of mbas#3 have not yet been investigated in detail.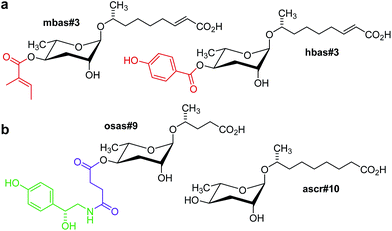 | ||
| Fig. 3 (a) mbas#3 and hbas#3, two modular ascarosides including tigloyl and p-hydroxybenzoyl modules putatively derived from amino acid or short-chain fatty acid metabolism (red) that were identified via targeted metabolomics in mixed stage C. elegans cultures.46 (b) ascr#10, a male-produced hermaphrodite attractant, and osas#9, a compound produced abundantly by starved L1 larvae,47 incorporating succinyl (magenta) and octopamine (green) moieties.49 | ||
Furthermore, targeted ascaroside profiling using precursor ion screening enabled the identification of life-stage and sex-specific ascarosides. It should be noted that most of the original work on small-molecule signalling in C. elegans was based on the analysis and biological testing of extracts derived from large liquid cultures containing worms of all ages and life stages, including eggs and larvae. However, given that ascarosides serve a great variety of biological functions, it seemed likely that different life stages would produce different ascaroside blends. Comparative MS/MS analysis of metabolome extracts of samples of a few hundred hand-picked C. elegans males and hermaphrodites facilitated the identification of ascr#10 (Fig. 3) as a highly potent, specifically male-produced hermaphrodite attractant.47 In addition, HPLC-MS analysis of different larval stages showed that production of one of the main dauer-inducing ascarosides, ascr#2, greatly increases before entering the dauer stage, whereas dauer larvae themselves produce little or no ascarosides.48 MS/MS analysis of different developmental stages also resulted in the identification of new types of modular ascaroside derivatives. For example, osas#9 and osas#10, two ascarosides incorporating octopamine-N-succinyl moieties, were found to be produced specifically by L1 larvae and function as a dispersal signal in the absence of food (Fig. 3).49 In nematodes, octopamine serves as a neurotransmitter in a manner similar to serotonin in higher animals, and thus osas#9 and osas#10 appear to connect neurotransmitter signalling with ascaroside-based interorganismal communication.
2.3 Modular assembly and structure–activity relationships
Extensive bioactivity screens using synthetic samples of the identified nematode signalling molecules have shown that even minor structural differences can result in starkly different activity profiles.50,51 For example, ascr#10, featuring a saturated 9-carbon side chain, strongly attracts hermaphrodites (Fig. 3), whereas ascr#3, which differs from ascr#10 only in that its side chain includes a double bond, acts as a deterrent (Fig. 1b).47 Similarly, modifications at the 4-position of the ascarylose scaffold often have a dramatic effect on biological activity. Whereas ascr#3 acts as a hermaphrodite repellent in higher concentrations, attachment of indole carboxyl or 4-hydroxybenzoyl moieties to its 4-position affords the potent aggregation pheromones icas#3 and hbas#3 (Fig. 3 and 4).44,46 The indole ascaroside icas#9, featuring a saturated 5-carbon side chain, additionally exhibits dauer inducing activity,45 whereas ascr#9, lacking the indole carboxy moiety, is not active in this assay.51 Extensive HPLC-MS analyses of worm exo-metabolome and worm body extract samples have demonstrated that attachment of the tryptophan-derived indole carboxy moiety is tightly controlled. For example, the potent attraction pheromones icas#3 and icas#9 are present in almost identical amounts (Fig. 2), whereas the unmodified ascr#3 is orders of magnitudes more abundant than the shorter chained ascr#9.46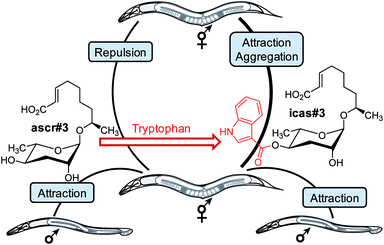 | ||
| Fig. 4 Attachment of a tryptophan-derived indole carboxy moiety to the deterrent ascr#3 results in the hermaphrodite attractant icas#3. In addition, ascr#3 is part of the male-attracting pheromone (see text).44 | ||
Attachment of additional building blocks at the carboxy terminus of the side chains can also modify biological function. As described above, ascr#8, which includes a 4-aminobenzoic acid moiety that most likely originates from bacterial folate metabolism38 is a potent male attractant (Fig. 2), whereas the corresponding unmodified ascaroside, ascr#7 (not shown), is inactive in this assay.38 Modification of the side chain carboxylic acid terminus may also be involved in the regulation of ascaroside secretion and storage. For example, a variety of β-glucosyl ester ascarosides (e.g. glas#3, Fig. 11) have been identified in C. elegans worm body extracts but could not be detected in exo-metabolomes samples.46 Overall, the observation that even seemingly minor structural changes in NDMMs can greatly affect biological activity strongly suggests that their biosynthesis must be precisely regulated.
3 Ascaroside signalling is highly conserved in nematodes
The production of ascarosides as well as their use as signalling molecules is highly conserved among nematodes, based on the recent analysis of a number of species representative of several groups of free living, entomopathogenic, and parasitic nematodes.52 Ascarosides featuring short to medium length side chains (C3–C11) are abundantly produced by the free-living C. elegans,46Panagrellus redivivus,53 and many other nematode species,52 whereas ascarosides incorporating longer side chains (C11–C20) have been identified in the entomopathogenic Heterorhabditis bacteriophora,52,54 the animal parasite Pelodera strongyloides,52 and several plant-parasitic nematode species.55 Ascarosides with very long-chain fatty acid residues (C21–C33) have been identified in the metabolomes of C. elegans peroxisomal β-oxidation mutants56 as well as in mammalian parasites of the genus Ascaris.40,57 Considering that many nematode species share identical habitats, overlap in the production of putative ascaroside-based signalling molecules raises interesting questions regarding the maintenance of species-specific communication. Preliminary comparative analysis of biological responses to ascarosides demonstrated that ascaroside functions are highly species-specific.52 In addition, more detailed analysis suggests that “simple ascarosides”, consisting of the sugar unit and the lipid side chain only, serve as templates for species-specific modifications and attachment of additional building blocks that are required for specific biological activities. For example, in the entomopathogenic nematode Heterorhabditis bacteriophora, the ethanolamide ascaroside easc#18 has been shown to serve as a potent dauer inducing signal, whereas the parent ascaroside, ascr#18 is much less active (Fig. 5).54 In another example, sex-specific side chain hydroxylation in the sour past nematode Panagrellus redivivus results in formation of the male-specific dihydroxy ascaroside dhas#18, which acts as a female attractant (Fig. 5), whereas male attraction is modulated by the widely distributed ascr#1 (Fig. 1b).53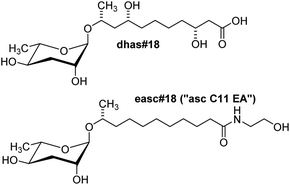 | ||
| Fig. 5 Structures of the side chain-hydroxylated dhas#18, a pheromone identified from the sour paste nematode P. redivivus,54 and easc#18, a component of the dauer pheromone of the entomopathogenic nematode H. bacteriophora.54 | ||
Given that the use of ascarosides as signalling molecules is highly conserved among nematodes, it seems possible that other organisms have evolved the capability to detect and respond to these nematode-specific molecular markers. Recent work showed that, in fact, ascarosides mediate cross-kingdom ecological interactions: in fungi that feed on nematodes, sensing of nematode-produced ascarosides was found to induce formation of specific mycelial structures that serve as “nematode traps”.58
3.1 Complex NDMM architectures from Pristionchus pacificus
The nematode Pristionchus pacificus occurs in association with scarab beetles and has been developed, alongside C. elegans, as a satellite model organism for the study of developmental and evolutionary biology.59P. pacificus was selected as a satellite model system to C. elegans because it is related, but also sufficiently distinct evolutionarily, to enable the comparative study of homologous biosynthetic, cellular, and developmental processes. In contrast to the free-living C. elegans, P. pacificus forms a close association with scarab beetles, in which dauer larvae persist on live beetles, without negative effect. Following death of the beetle, the larvae resume development and reproduce, feeding on bacteria that consume the beetle carcass. This “necromenic” association with beetles may represent a pre-adaptation to the evolution of parasitism.Previous work had suggested that, like in C. elegans, dauer formation in P. pacificus depends on small molecules. Furthermore, it had been shown that in P. pacificus a unique dimorphism of mouth development is likely controlled by inter-organismal small-molecule signalling. Under conditions of high population density, worms develop a wider mouth (“eurystomatous”) equipped with a tooth-like structure that enables a predatory lifestyle consuming other nematodes, whereas under low population density conditions worms develop a narrow mouth (“stenostomatous”) optimized for a bacterial diet.60,61 Since none of the ascarosides previously identified from C. elegans elicited dauer formation or mouth dimorphism in P. pacificus, it appeared that other, species-specific components must be responsible. Extensive analysis of active P. pacificus exo-metabolome fractions via NMR spectroscopy and MS/MS revealed a fascinatingly diverse array of multimodular NDMMs, incorporating building blocks from all major primary metabolic pathways including carbohydrate, lipid, amino acid, nucleoside, and neurotransmitter metabolism (Fig. 6).
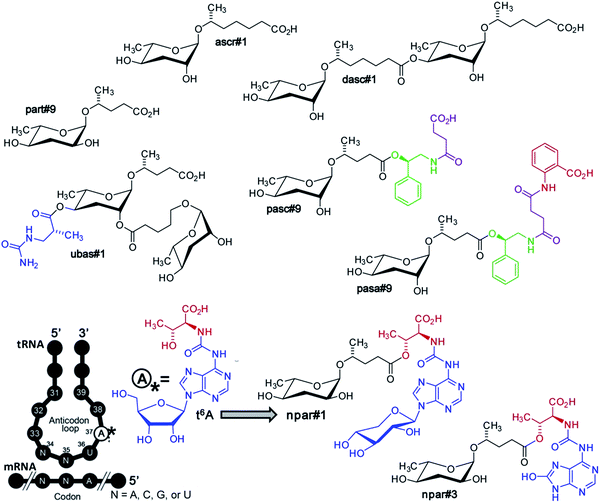 | ||
| Fig. 6 Ascaroside and paratoside-derived metabolites in P. pacificus. Major components of the P. pacificus exo-metabolome derived from assembly of building blocks from carbohydrate, lipid, amino acid (red), and nucleoside (blue) metabolism, as well as TCA cycle-derived succinate (magenta). Also shown is the highly conserved tRNA nucleoside, N6-threonylcarbamoyladenosine (t6A), a putative precursor of paratosides npar#1–3.62 | ||
Many of the building blocks in the P. pacificus NDMMs featured unexpected structural modifications. For example, whereas all known C. elegans NDMMs include L-ascarylose as the central scaffold, the P. pacificus metabolites are based on two different di-deoxysugars, L-ascarylose and the related L-paratose, a new sugar, the D-enantiomer of which having previously been reported from bacteria.63 Additional structural and functional diversity of the P. pacificus NDMMs derives from dimerization of ascarosides. For example, dasc#1, formally derived from dimerization of ascr#1, strongly induces the eurystomatous mouth form, whereas the monomeric ascr#1 is inactive (Fig. 7).62,64 Another dimeric ascaroside, ubas#1, which is formally derived from dimerization of two different ascarosides, the (ω-1)-side chain-functionalized ascr#9 and (ω)-functionalized oscr#9, contributes to dauer formation in some P. pacificus strains (Fig. 6). The structures of ubas#1 further includes an unusual ureido isobutyrate moiety that likely originates from metabolism of the nucleobase thymine. Nucleoside metabolism further contributes to dauer signalling in P. pacificus via a family of paratose-based NDMMs, npar#1–3, which incorporate a L-threonylcarbamoyl adenosine moiety that most likely originates from canonical threonylcarbamoyl adenosine (t6A), a highly conserved nucleoside found adjacent to the anticodon triplet of a subset of tRNAs (Fig. 6).62 Most surprisingly, however, a xylopyranoside residue was observed in npar#1 instead of the canonical ribofuranoside variant of t6A found in tRNA.62 Physiological concentrations of npar#1 strongly induced dauer formation in P. pacificus, whereas the corresponding unmodified paratoside part#9 was much less active62 Two additional derivatives have been described. npar#2 represents the N-deglycosylated analogue of npar#1, whereas in npar#3 the adenosine moiety of N-deglycosylated npar#1 is replaced by the corresponding 8-hydroxy derivative.65 Hydroxylated adenosine derivatives have been shown to be produced as a result of oxidative damage to nucleic acids,66 suggesting that npar#3 functions as an indicator of oxidative stress.
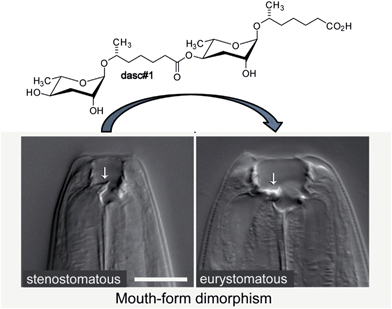 | ||
| Fig. 7 The dimeric ascaroside dasc#1 promotes development of the wide (eurystomatous) mouth form that enables a predatory lifestyle consuming other nematodes. | ||
P. pacificus also produces a family of NDMMs that, similar to osas#9 in C. elegans, incorporate putatively neurotransmitter-derived moieties, e.g. pasc#9 (Fig. 6). Analogous to the incorporation of N-succinyl-octopamine in osas#9 (Fig. 3), pasc#9, the most abundant NDMM in all P. pacificus strains analyzed so far, incorporates a N-succinyl-phenylethanolamine moiety, suggesting that N-succinylation plays a role in nematode neurotransmitter metabolism.49,62 In contrast to osas#9 in C. elegans (Fig. 3), the succinamide in pasc#9 is linked to the carboxy terminus of the side chain instead of the ascarylose.
Recently, a derivative of pasc#9 has been described in which an additional anthranilic acid likely derived from tryptophan is attached to the succinate unit. This compound, named pasa#9, represents the first NDMM composed of 5 different modules (Fig. 6).64 The structure of pasa#9 demonstrates that additional modules must not necessarily be incorporated via direct attachment to the ascaroside scaffold, but may also get linked to other modules, suggesting that NDMMs may be even more structurally diverse than previously suspected. Penta-modular pasa#9 and pasc#9 are accompanied by smaller amounts of the cyclic succinimide pasy#9 (not shown), which may represent a by-product of pasc#9 biosynthesis or could be derived from decomposition of pasa#9.62,64
Many of the new chemical structures proposed on the basis of spectroscopic analysis of the P. pacificus metabolome were structurally much more complex than the NDMMs previously identified from C. elegans and other nematodes (Fig. 6). As a result, confirmation of the proposed structures via total synthesis emerged as a major bottle neck in the characterization of P. pacificus NDMMs. For example, the synthesis of npar#1 was based on a sequence with 13 linear steps. Therefore, proposed structures were prioritized for synthesis based on their relative abundances and results from activity-guided fractionation; however, structures of a large number of additional NDMMs remain to be confirmed via synthesis which will then also facilitate their biological evaluation. As in the case of C. elegans, the identification of additional NDMMs in P. pacificus may lead to the discovery of new small-molecule mediated phenotypes.
3.2 Structure–activity relationships and natural variation in P. pacificus
NDMM biosynthesis in P. pacificus appears to be tightly regulated. ascr#1, including a seven-carbon side chain, forms the 4′-homodimer dasc#1, whereas an ascaroside featuring an (ω-1)-functionalized five-carbon side chain forms the scaffold for a heterodimer with an ascaroside featuring an (ω)-functionalized five-carbon side chain in ubas#1 (Fig. 6).62 Notably, homodimers of ascarosides with (ω)- or (ω-1)-functionalized five-carbon side chains are absent. Similarly, there are no ascaroside-based analogues of the paratose-based nucleobase-derived npar#1–3 (Fig. 6), and no paratose-based analogues of the ascarylose-based N-succinyl-phenylethanolamine-containing pasc#9, pasy#9, and pasa#9 have been found.62Correspondingly, the biological activity of the identified NDMMs is strongly structure-dependent.62 A screen of six different natural P. pacificus strains has further shown that bioactivity of different NDMMs varies greatly between different natural isolates.64 These observations indicate that NDMM receptors evolve rapidly, as also suggested by the finding that laboratory strains of C. elegans are less sensitive to dauer pheromone than the parent wildtype strain due to rapid accumulation of mutations in receptor-coding genes.67 Comparison of the metabolomes of the six different natural P. pacificus strains further revealed marked differences in the relative abundances of NDMM in different strains.64 For example, the ascaroside pasc#9 varied more than six-fold between strains. Several NDMMs, for example ubas#1 and ubas#2, were completely absent in two of the six analysed strains. Furthermore, it was found that some P. pacificus strains respond strongly to NDMMs they do not produce themselves, which suggested that NDMMs may play a role in communication and/or competition between different strains.64,68 Similarly, analysis of several C. elegans wild type isolates indicated that the production52 and dauer inducing activity69 of ascr#2 and ascr#3 is strongly strain-specific, suggesting the possibility of “dishonest” signalling between competing C. elegans strains.69
4 Ascaroside biosynthesis and primary metabolism
The NDMMs identified from C. elegans, P. pacificus, and other nematode species incorporate building blocks originating from diverse primary metabolic pathways and hence may encode information about the metabolic state of the producing organism. In addition to carbohydrate and fatty acid metabolism, which together give rise to the ascaroside core structures, integration of amino acid, neurotransmitter, and nucleoside metabolism as well as the citric acid cycle affords a diversity of complex architectures. Their de novo biosynthesis appears to proceed without participation of microbial metabolism, since all NDMMs identified from C. elegans and P. pacificus are also produced in axenic (bacteria-free) cultures,46,62 as long as all necessary building blocks – including those the nematodes do not produce themselves, such as bacteria-derived PABA – are provided in the media. Moreover, the highly specific assembly of primary metabolic building blocks, for example the combination of octopamine-derived moieties only with ascarosides of specific side chain lengths, suggests that NDMM biosynthesis is carefully controlled.L-Ascarylose (or L-paratose) are glycosidically connected to fatty acid-derived side chains of varying length and functionality that have been shown to originate from peroxisomal β-oxidation,45,46,56 producing homologous series of the glycolipid core structures. In some nematode species, the glycolipid profiles are dominated by one or two chain lengths, whereas other species have broader distributions.52 Although the origin of these fatty acid chains is not well understood, the general predominance of odd-numbered side chains indicates that these are derived from long-chain odd-numbered fatty acids via multiple steps of β-oxidation.
In most ascarosides/paratosides, the fatty acid residue is attached to the dideoxysugar at the (ω-1)-position and the corresponding chiral centre always carries the (R)-configuration. Less common are (ω)-linked side chains, as in ascr#5, a major component of the C. elegans dauer pheromone, which carries an (ω)-linked 3-hydroxypropanoic acid residue (Fig. 1b),37 and several other compounds based on longer chained (ω)-linked side chains identified from both C. elegans and P. pacificus.46,62 As described in the following section, targeted metabolomic analysis of peroxisomal β-oxidation mutants revealed complete series of longer chain homologues with (ω-1)- and (ω)-hydroxylated side chains ranging from 3 to 21 carbons which suggested that (ω-1)- and (ω)-hydroxylation occurs upstream of peroxisomal β-oxidation.46
4.1 The role of peroxisomal β-oxidation
The participation of primary metabolic enzymes has been demonstrated only for a few of the building blocks in NDMMs: the origin of the octopamine moiety in osas#9 and osas#10 from canonical tyrosine metabolism (Fig. 10)49 and, importantly, side chain biosynthesis via peroxisomal β-oxidation (Fig. 8). Shortly after describing the dauer pheromone, Golden and Riddle in 1985 reported a gene named daf-22 that was required for dauer pheromone production.34 Following the identification of ascarosides as major dauer pheromone components, it was observed that daf-22 mutant worms do not produce short-chain ascarosides,41 and subsequent work revealed that daf-22 encodes a homologue of peroxisomal 3-ketoacyl-CoA thiolases that is expressed in the hypodermis and intestine.45 Mammalian homologues of daf-22 have been shown to catalyze the last step of peroxisomal fatty acid β-oxidation, a four-step iterative process that is evolutionarily highly conserved (Fig. 8).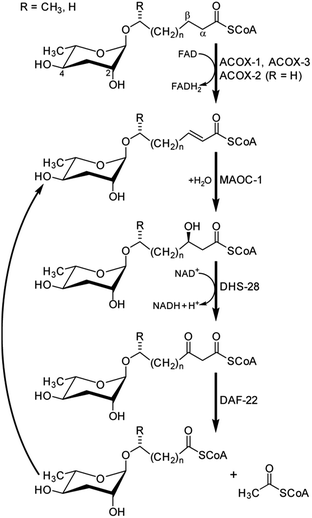 | ||
| Fig. 8 Side chain shortening of long-chain ascarosides via iterative four-step peroxisomal β-oxidation in C. elegans.46,71 | ||
Subsequent studies showed that C. elegans genes coding for homologues of enzymes catalyzing the remaining three enzymatic steps in peroxisomal β-oxidation also participate in ascaroside biosynthesis. This includes three acyl-CoA oxidases (acox-1,46,70acox-2 and acox-3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 71), the enoyl-CoA hydratase maoc-1,46 and the 3-hydroxyacyl-CoA dehydrogenase dhs-28.45,46 The role of acox-1, maoc-1, dhs-28, and daf-22 in chain shortening of ascarosides via peroxisomal β-oxidation was investigated using the ascaroside-targeted MS/MS screen described above.46 Mutation of acox-1, maoc-1, or dhs-28 was shown to result in accumulation of shunt metabolites with structural features supportive of the enzymatic functions of these genes, which originally had been predicted solely based on homology. For example, production of short-chain ascarosides (<9 carbon side chain) and ascarosides with α,β-unsaturated side chains, which are highly abundant in C. elegans wild type, is attenuated in acox-1 mutant metabolomes. Instead, acox-1 mutant worms accumulate large amounts of saturated longer-chain ascarosides (especially C9, C11, and C13), indicating that acox-1 serves as acyl-CoA oxidase introducing a double bond in the first step of peroxisomal β-oxidation.46 Additional work revealed that homodimers of the acyl-CoA oxidase acox-1 specifically act on the CoA ester of ascarosides with nine carbon side chains, whereas heterodimers of acox-3 and acox-1 act on ascaroside CoA esters of shorter carbon chains. Furthermore, homodimers of acox-2 appear to act specifically on ascaroside CoA esters of (ω)-oxygenated side chains and thus may control the biosynthesis of the dauer pheromone component ascr#5.71
71), the enoyl-CoA hydratase maoc-1,46 and the 3-hydroxyacyl-CoA dehydrogenase dhs-28.45,46 The role of acox-1, maoc-1, dhs-28, and daf-22 in chain shortening of ascarosides via peroxisomal β-oxidation was investigated using the ascaroside-targeted MS/MS screen described above.46 Mutation of acox-1, maoc-1, or dhs-28 was shown to result in accumulation of shunt metabolites with structural features supportive of the enzymatic functions of these genes, which originally had been predicted solely based on homology. For example, production of short-chain ascarosides (<9 carbon side chain) and ascarosides with α,β-unsaturated side chains, which are highly abundant in C. elegans wild type, is attenuated in acox-1 mutant metabolomes. Instead, acox-1 mutant worms accumulate large amounts of saturated longer-chain ascarosides (especially C9, C11, and C13), indicating that acox-1 serves as acyl-CoA oxidase introducing a double bond in the first step of peroxisomal β-oxidation.46 Additional work revealed that homodimers of the acyl-CoA oxidase acox-1 specifically act on the CoA ester of ascarosides with nine carbon side chains, whereas heterodimers of acox-3 and acox-1 act on ascaroside CoA esters of shorter carbon chains. Furthermore, homodimers of acox-2 appear to act specifically on ascaroside CoA esters of (ω)-oxygenated side chains and thus may control the biosynthesis of the dauer pheromone component ascr#5.71
In contrast to acox-1, maoc-1, and dhs-28, targeted metabolomic analyses via HPLC-MS/MS did not reveal any shunt metabolites whose structures could serve to corroborate the predicted function of daf-22 as a 3-ketoacyl-CoA thiolase.46 This was not unexpected, given that β-ketoacids such as they would be expected to form from accumulating substrates of DAF-22 (Fig. 8) easily decarboxylate or may undergo other chemical transformations. To identify such daf-22-dependent shunt metabolites, the daf-22 metabolome was re-analyzed using 2D NMR-based comparative metabolomics with a focus on spectroscopic signatures present in the mutant metabolome, but absent in wildtype. For this purpose, multivariate differential analysis by 2D NMR spectroscopy (mvaDANS) was developed, which integrates automatic crosspeak identification and binning with statistical analysis via principal component analysis. mvaDANS then revealed a series of very long-chain ascaroside ethanolamides and other long-chain ascarosides as shunt metabolites in daf-22 mutants (Fig. 9).56 Subsequent re-analysis of the metabolomes of other peroxisomal β-oxidation mutants revealed similar shunt metabolites. Formation of these long-chain ethanolamides in response to defects in daf-22 and other components of peroxisomal β-oxidation was associated with a severe depletion of endocannabinoid pools, demonstrating an unexpected interaction between peroxisomal lipid β-oxidation and the biosynthesis of endocannabinoids (Fig. 9), which are major regulators of lifespan in C. elegans.56
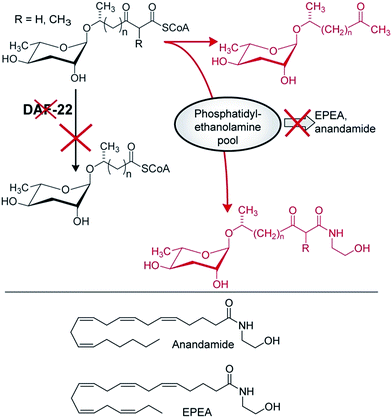 | ||
| Fig. 9 Interaction of endocannabinoid biosynthesis with peroxisomal β-oxidation. daf-22 mutation abolishes processing of long-chain ascaroside CoA esters, whose conversion into ascaroside ethanolamides is associated with reduced EPEA and anandamide production, likely due to depletion of phosphatidylethanolamine pools. Shown in red are shunt metabolites identified in daf-22 mutant endo-metabolomes via mvaDANS.56 | ||
4.2 Biogenesis of multimodular ascarosides
Additional modifications of the ascaroside core structure with other primary metabolism-derived moieties have a dramatic effect on the biological activities and appear to be tightly controlled, since, as discussed above, attachment of additional moieties is highly side chain length-specific and most multimodular ascarosides are produced primarily by specific life stages. Their structures suggest that the indolyl-3-carboxylate, 4-hydroxybenzoate, and tiglate moieties observed in C. elegans (Fig. 2 & 3) are of amino acid origin, which, however, has only been demonstrated for indole ascarosides, which incorporate isotope labelled tryptophan.44 No biosynthetic intermediates have been identified and the enzyme(s) responsible for linking tryptophan metabolism and indole ascaroside biosynthesis have not been identified. Addition of synthetic short-chain ascarosides to daf-22 mutant cultures (which do not produce short-chain ascarosides) revealed side chain length-specific conversion to the corresponding indole carboxy derivatives, suggesting that attachment of amino acid-derived indol-3-carboxylate moieties occurs downstream of peroxisomal β-oxidation.46 Additional evidence for the incorporation of amino acid-derived building blocks originates from the analysis of tyrosine decarboxylase (tdc-1) and tyramine β-hydroxylase (tbh-1) mutant metabolomes, which revealed that the production of octopamine-N-succinamide ascarosides (osas) by starved C. elegans L1 larvae depends on the production of the neurotransmitter octopamine from L-tyrosine via tdc-1 and tbh-1 (Fig. 10).49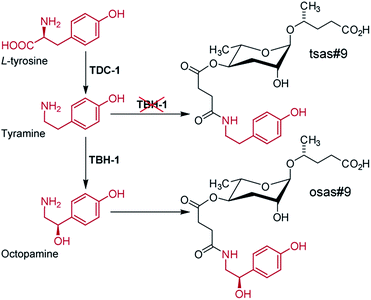 | ||
| Fig. 10 Model for the biosynthesis of the avoidance signal osas#9 in C. elegans. tbh-1 mutant worms produce large quantities of the shunt metabolite tsas#9 due to accumulation of the octopamine precursor tyramine.49 | ||
4.3 Ascaroside biosynthesis is strongly regulated
Ascaroside biosynthesis in C. elegans strongly depends on environmental factors such as temperature,70 population density,47 and food availability,46,49 as well as developmental stage48,49 and gender,47 but the underlying mechanisms are poorly understood. Increased production of ascarosides at elevated temperatures corresponds to increased expression of acyl-CoA oxidases acox-1,70acox-2, and acox-3.71 Furthermore, food availability has been shown to modulate the composition of ascaroside bouquets via differential expression of acox genes. Under non dauer inducing conditions (low population density and high food availability) increased expression of acox-2 results in increased production of (ω)-oxygenated short chained ascr#5.71 Males of C. elegans show reduced expression of acox-1 and consequently release larger amounts of the corresponding substrate, the saturated ascr#10 that acts as a highly potent hermaphrodite attractant.47 Similarly, in the sour paste nematode Panagrellus redivivus the female-attracting pheromone dhas#18 is exclusively produced by males (Fig. 5), whereas the shorter chain male-attracting ascr#1 is only produced by females (Fig. 1b), suggesting that gender-specific production of sex pheromones originates from gender-specific fine tuning of primary metabolic pathways such as peroxisomal β-oxidation.52Finally, comparative analysis of exo- and endo-metabolome (worm body extracts) samples revealed that C. elegans and P. pacificus exhibit significant control over ascaroside release.46,62,64 For example, ascaroside glucosyl esters (e.g. glas#3, Fig. 11) have been found only in the C. elegans endo-metabolome (worm body extract), but not the exo-metabolome, suggesting that such glucosides may be involved in ascaroside storage or transport.46
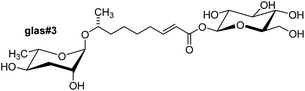 | ||
| Fig. 11 glas#3, an ascaroside glucosyl ester identified in the C. elegans endo-metabolome.46 | ||
5 Ascaroside perception and downstream signalling
The C. elegans genome encodes for a large diversity of GTP-binding protein (G protein)-coupled receptors (GPCRs), most of which are expressed in chemosensory neurons.72,73 Ascaroside induced developmental phenotypes such as dauer formation depend on at least five different types of chemosensory neurons (ADF, ASG, ASI, ASJ, ASK, and the male-specific CEM neurons),72 whereas behavioural phenotypes depend on ASK, ASI, and ADL neurons.41,44,74–76 Mutants for G protein alpha subunits gpa-2 and gpa-3 are dauer defective; further suggesting that ascaroside perception is mediated by GPCRs.77However, so far only a relatively small number of G-protein coupled receptors involved in ascaroside perception have been identified,67,78,79 whereas the diversity of identified ascaroside functions suggests that many more are yet to be discovered. SRBC-64 and SRBC-66 are two heterotrimeric GPCRs expressed in the ASK chemosensory neurons that mediate dauer formation in response to ascr#1, ascr#2, and ascr#3 (Fig. 1b).79 Two members of the serpentine chemoreceptor class g family, srg-36 and srg-37, discovered through genetic analysis of pheromone resistant C. elegans strains obtained from high density cultivation for several years, encode redundant GPCRs for the dauer inducing activity of ascr#5, demonstrating that adaptation to specific environments can entail rapid remodelling of the chemoreceptor repertoire. Deletion of a srg gene paralogous to srg-36 and srg-37 has also been linked to resistance to dauer formation in a laboratory Caenorhabditis briggsae strain, indicating that parallel genetic changes can affect life-history traits across species.67 Finally, heterodimers of DAF-37 and DAF-38, two GPCRs identified using DAF-8 immunoprecipitation, cooperatively mediate ascaroside perception. daf-37 mutants are defective in ascr#2-mediated dauer-induction but respond normally to other ascarosides, whereas daf-38 mutants are partially defective to several different ascarosides. Furthermore, cell-specific overexpression revealed that ascr#2-mediated dauer formation depends on DAF-37 expression in ASI neurons, whereas DAF-37 expression in ASK neurons regulates behaviour.78 Using a photoaffinity-labeled ascr#2 probe and amplified luminescence assays (AlphaScreen), it was further demonstrated that ascr#2 binds directly to DAF-37.78
Perception of natural dauer pheromone samples, i.e. ascaroside-containing exo-metabolome, has been shown to act upstream of several conserved signalling pathways, including the insulin, TGF-β, serotonin, and guanylyl cyclase pathways.27,31 However, it is not known if and to what extent downstream pathways of individual dauer-inducing ascarosides differ. Nonetheless, it has been shown that different phenotypes induced by the same ascaroside can depend on different downstream signalling components. For example, ascarosides ascr#2 and ascr#3 extend lifespan in adult worms via sir-2.1, a homologue of the mammalian histone-deacetylase SIRT-1,42 and the same two compounds also induce dauer via the insulin signalling pathway.36 However, sir-2.1 is not required for dauer induction by ascr#2 and ascr#3,27 and neither is insulin signalling required for ascr#2 and ascr#3-mediated longevity.42
6 Conclusion
Nematode-derived ascarosides and paratosides represent a “new” class of natural products that cannot easily be classified as “primary” or “secondary” metabolites. Much of their chemical appeal and originality derives from the fact that they incorporate well-known primary metabolites into new structures, sometimes with unexpected chemical modifications. Since the building blocks of NDMMs are directly from conserved primary metabolism, it seems possible that similar types of modular small molecule signals are produced by other metazoans. These possibilities provide strong incentives for a comprehensive re-analysis of vertebrate metabolomes, taking advantage of the approaches developed and validated using nematode model systems.Further elucidation of the biosynthesis of ascaroside-based NDMMs will reveal how input from conserved primary metabolism is transduced to create signals that regulate development, behaviour, and many other aspects of the life history of nematode model organisms. Moreover, since ascaroside signalling is highly conserved among nematodes, a detailed understanding of their biosynthesis and regulation may enable new approaches for the treatment of human nematode infections or the control of parasitic nematodes in agriculture. Lastly, it should be noted that ascaroside-based NDMMs are likely just one of several families of small molecule signals. Mass spectrometric analyses of the C. elegans metabolome have revealed evidence for several 1000 yet unidentified compounds,46 and nematode genomes feature many genes that, based on homology, may encode small-molecule biosynthetic pathways, including one gene with clear homology to microbial hybrid PKS/NRPSs (polyketide synthase/non-ribosomal peptide synthetases),80 and whose functions remain to be elucidated.
7 Notes and references
- M. Blaxter and G. Koutsovoulos, Parasitology, 2014, 1 Search PubMed.
- N. A. Mitkowski and G. S. Abawi, Nematology, 2003, 5, 77 CrossRef PubMed.
- P. J. Hotez, P. J. Brindley, J. M. Bethony, C. H. King, E. J. Pearce and J. Jacobson, J. Clin. Invest., 2008, 118, 1311 CrossRef CAS PubMed.
- D. W. Crompton and M. C. Nesheim, Annu. Rev. Nutr., 2002, 22, 35 CrossRef CAS PubMed.
- The C. elegans Sequencing Consortium, Science, 1998, 282, 2012 Search PubMed.
- D. N. Greet, Nature, 1964, 204, 96 CrossRef PubMed.
- L. W. Bone and H. H. Shorey, J. Chem. Ecol., 1978, 4, 595 CrossRef CAS.
- C. D. Green, Helminthological Abstracts Series B: Plant nematology, 1980, 49, 81 Search PubMed.
- R. N. Huettel, J. Nematol., 1986, 18, 3 CAS.
- B. M. MacKinnon, Parasitol. Today, 1987, 3, 156 CrossRef CAS.
- H. Jaffe, R. N. Huettel, A. B. Demilo, D. K. Hayes and R. V. Rebois, J. Chem. Ecol., 1989, 15, 2031 CrossRef CAS PubMed.
- J. Aumann, E. Dietsche, S. Rutencrantz and H. Ladehoff, Int. J. Parasitol., 1998, 28, 1691 CrossRef CAS.
- J. Aumann and M. Hashem, Fundam. Appl. Nematol., 1993, 16, 43 Search PubMed.
- J. Aumann, H. Ladehoff and S. Rutencrantz, Fundam. Appl. Nematol., 1998, 21, 119 CAS.
- J. M. Neves, N. Simoes and M. Mota, Nematologica, 1998, 44, 95 CrossRef PubMed.
- M. G. Jonz, E. Riga, A. J. Mercier and J. W. Potter, Nematology, 2001, 3, 55 CrossRef CAS PubMed.
- E. E. Lewis, B. Barbarosa and R. Gaugler, J. Nematol., 2002, 34, 328 Search PubMed.
- J. M. Simon and P. W. Sternberg, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 1598 CrossRef CAS PubMed.
- A. H. Ludewig and F. C. Schroeder, Ascaroside signaling in C. elegans, in WormBook, ed. The C. elegans Research Community, 2013, DOI:10.1895/wormbook.1.155.1.
- F. C. Schroeder, ACS Chem. Biol., 2006, 1, 198 CrossRef CAS PubMed.
- É. Maupas, Arch. Zool. Exp. Gen., 1900, 8, 463 Search PubMed.
- S. Brenner, Genetics, 1974, 77, 71 CAS.
- R. E. Ellis, J. Y. Yuan and H. R. Horvitz, Annu. Rev. Cell Biol., 1991, 7, 663 CrossRef CAS PubMed.
- A. Coulson, J. Sulston, S. Brenner and J. Karn, Proc. Natl. Acad. Sci. U. S. A., 1986, 83, 7821 CrossRef CAS.
- A. Fire, S. Q. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver and C. C. Mello, Nature, 1998, 391, 806 CrossRef CAS PubMed.
- M. Chalfie, Y. Tu, G. Euskirchen, W. W. Ward and D. C. Prasher, Science, 1994, 263, 802 CAS.
- P. J. Hu, Dauer, in WormBook, ed. The C. elegans Research Community, 2007, DOI:10.1895/wormbook.1.144.1.
- N. Fielenbach and A. Antebi, Genes Dev., 2008, 22, 2149 CrossRef CAS PubMed.
- D. L. Motola, C. L. Cummins, V. Rottiers, K. K. Sharma, T. Li, Y. Li, K. Suino-Powell, H. E. Xu, R. J. Auchus, A. Antebi and D. J. Mangelsdorf, Cell, 2006, 124, 1209 CrossRef CAS PubMed.
- P. Mahanti, N. Bose, A. Bethke, J. C. Judkins, J. Wollam, K. J. Dumas, A. M. Zimmerman, S. L. Campbell, P. J. Hu, A. Antebi and F. C. Schroeder, Cell Metab., 2014, 19, 73 CrossRef CAS PubMed.
- S. S. Lee and F. C. Schroeder, PLoS Biol., 2012, 10, e1001307 CAS.
- J. W. Golden and D. L. Riddle, Science, 1982, 218, 578 CAS.
- J. W. Golden and D. L. Riddle, J. Nematol., 1982, 14, 443 Search PubMed.
- J. W. Golden and D. L. Riddle, Mol. Gen. Genet., 1985, 198, 534 CrossRef CAS.
- P. Y. Jeong, M. Jung, Y. H. Yim, H. Kim, M. Park, E. Hong, W. Lee, Y. H. Kim, K. Kim and Y. K. Paik, Nature, 2005, 433, 541 CrossRef CAS PubMed.
- R. A. Butcher, M. Fujita, F. C. Schroeder and J. Clardy, Nat. Chem. Biol., 2007, 3, 420 CrossRef CAS PubMed.
- R. A. Butcher, J. R. Ragains, E. Kim and J. Clardy, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 14288 CrossRef CAS PubMed.
- C. Pungaliya, J. Srinivasan, B. W. Fox, R. U. Malik, A. H. Ludewig, P. W. Sternberg and F. C. Schroeder, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 7708 CrossRef CAS PubMed.
- F. Flury, Arch. Exp. Pathol. Pharmakol., 1912, 67, 275 CAS.
- C. Fouquey, J. Polonsky and E. Lederer, Bull. Soc. Chim. Biol., 1957, 39, 101 Search PubMed.
- J. Srinivasan, F. Kaplan, R. Ajredini, C. Zachariah, H. T. Alborn, P. E. A. Teal, R. U. Malik, A. S. Edison, P. W. Sternberg and F. C. Schroeder, Nature, 2008, 454, 1115 CrossRef CAS PubMed.
- A. H. Ludewig, Y. Izrayelit, D. Park, R. U. Malik, A. Zimmermann, P. Mahanti, B. W. Fox, A. Bethke, F. Doering, D. L. Riddle and F. C. Schroeder, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 5522 CrossRef CAS PubMed.
- T. J. Maures, L. N. Booth, B. A. Benayoun, Y. Izrayelit, F. C. Schroeder and A. Brunet, Science, 2014, 343, 541 CrossRef CAS PubMed.
- J. Srinivasan, S. H. von Reuss, N. Bose, A. Zaslaver, P. Mahanti, M. C. Ho, O. G. O'Doherty, A. S. Edison, P. W. Sternberg and F. C. Schroeder, PLoS Biol., 2012, 10, e1001237 CAS.
- R. A. Butcher, J. R. Ragains, W. Li, G. Ruvkun, J. Clardy and H. Y. Mak, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 1875 CrossRef CAS PubMed.
- S. H. von Reuss, N. Bose, J. Srinivasan, J. J. Yim, J. C. Judkins, P. W. Sternberg and F. C. Schroeder, J. Am. Chem. Soc., 2012, 134, 1817 CrossRef CAS PubMed.
- Y. Izrayelit, J. Srinivasan, S. L. Campbell, Y. Jo, S. H. von Reuss, M. C. Genoff, P. W. Sternberg and F. C. Schroeder, ACS Chem. Biol., 2012, 7, 1321 CrossRef CAS PubMed.
- F. Kaplan, J. Srinivasan, P. Mahanti, R. Ajredini, O. Durak, R. Nimalendran, P. W. Sternberg, P. E. Teal, F. C. Schroeder, A. S. Edison and H. T. Alborn, PLoS One, 2011, 6, e17804 CAS.
- A. B. Artyukhin, J. J. Yim, J. Srinivasan, Y. Izrayelit, N. Bose, S. H. von Reuss, Y. Jo, J. M. Jordan, L. R. Baugh, M. Cheong, P. W. Sternberg, L. Avery and F. C. Schroeder, J. Biol. Chem., 2013, 288, 18778 CrossRef CAS PubMed.
- F. C. Schroeder, Chem. Biol., 2015, 22, 7 CrossRef CAS PubMed.
- K. A. Hollister, E. S. Conner, X. Zhang, M. Spell, G. M. Bernard, P. Patel, A. C. de Carvalho, R. A. Butcher and J. R. Ragains, Bioorg. Med. Chem., 2013, 21, 5754 CrossRef CAS PubMed.
- A. Choe, S. H. von Reuss, D. Kogan, R. B. Gasser, E. G. Platzer, F. C. Schroeder and P. W. Sternberg, Curr. Biol., 2012, 22, 772 CrossRef CAS PubMed.
- A. Choe, T. Chuman, S. H. von Reuss, A. T. Dossey, J. J. Yim, R. Ajredini, A. A. Kolawa, F. Kaplan, H. T. Alborn, P. E. Teal, F. C. Schroeder, P. W. Sternberg and A. S. Edison, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 20949 CrossRef CAS PubMed.
- J. H. Noguez, E. S. Conner, Y. Zhou, T. A. Ciche, J. R. Ragains and R. A. Butcher, ACS Chem. Biol., 2012, 7, 961 CrossRef CAS PubMed.
- P. Manosalva, M. Manohar, S. H. von Reuss, S. Chen, A. Koch, F. Kaplan, A. Choe, R. J. Micikas, X. Wang, K.-H. Kogel, P. W. Sternberg, V. M. Williamson, F. C. Schroeder and D. F. Klessig, Nature Commun., 2015 Search PubMed , accepted for publication .
- Y. Izrayelit, S. L. Robinette, N. Bose, S. H. von Reuss and F. C. Schroeder, ACS Chem. Biol., 2013, 8, 314 CrossRef CAS PubMed.
- J. P. Bartley, E. A. Bennett and P. A. Darben, J. Nat. Prod., 1996, 59, 921 CrossRef CAS PubMed.
- Y. P. Hsueh, P. Mahanti, F. C. Schroeder and P. W. Sternberg, Curr. Biol., 2013, 23, 83 CrossRef CAS PubMed.
- C. Dieterich, S. W. Clifton, L. N. Schuster, A. Chinwalla, K. Delehaunty, I. Dinkelacker, L. Fulton, R. Fulton, J. Godfrey, P. Minx, M. Mitreva, W. Roeseler, H. Tian, H. Witte, S. P. Yang, R. K. Wilson and R. J. Sommer, Nat. Genet., 2008, 40, 1193 CrossRef CAS PubMed.
- A. Ogawa, G. Bento, G. Bartelmes, C. Dieterich and R. J. Sommer, Development, 2011, 138, 1281 CrossRef CAS PubMed.
- J. Sommer Ralf and A. Ogawa, Curr. Biol., 2011, 21, R758 CrossRef CAS PubMed.
- N. Bose, A. Ogawa, S. H. von Reuss, J. J. Yim, E. J. Ragsdale, R. J. Sommer and F. C. Schroeder, Angew. Chem., Int. Ed., 2012, 51, 12438 CrossRef CAS PubMed.
- D. A. L. Davies, A. M. Staub, I. Fromme, O. Luderitz and O. Westphal, Nature, 1958, 181, 822 CrossRef CAS PubMed.
- N. Bose, J. M. Meyer, J. J. Yim, M. G. Mayer, G. V. Markov, A. Ogawa, F. C. Schroeder and R. J. Sommer, Curr. Biol., 2014, 24, 1536 CrossRef CAS PubMed.
- J. J. Yim, N. Bose, J. M. Meyer, R. J. Sommer and F. C. Schroeder, Org. Lett., 2015, 17, 1648 CrossRef CAS PubMed.
- S. Kanvah, J. Joseph, G. B. Schuster, R. N. Barnett, C. L. Cleveland and U. Landman, Acc. Chem. Res., 2009, 43, 280 CrossRef PubMed.
- P. T. McGrath, Y. Xu, M. Ailion, J. L. Garrison, R. A. Butcher and C. I. Bargmann, Nature, 2011, 477, 321 CrossRef CAS PubMed.
- M. G. Mayer and R. J. Sommer, Proc. R. Soc. B, 2011, 278, 2784 CrossRef PubMed.
- S. A. Diaz and M. Viney, Ecol. Evol., 2014, 4, 2058 Search PubMed.
- H. J. Joo, K. Y. Kim, Y. H. Yim, Y. X. Jin, H. Kim, M. Y. Kim and Y. K. Paik, J. Biol. Chem., 2010, 285, 29319 CrossRef CAS PubMed.
- X. Zhang, L. Feng, S. Chinta, P. Singh, Y. Wang, J. K. Nunnery and R. A. Butcher, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 3955 CrossRef CAS PubMed.
- C. I. Bargmann, Chemosensation in C. elegans, in Wormbook, ed. The C. elegans Research Community, 2006, DOI:10.1895/wormbook.1.123.1.
- C. I. Bargmann, Nature, 2006, 444, 295 CrossRef CAS PubMed.
- H. Jang, K. Kim, S. J. Neal, E. Macosko, D. Kim, R. A. Butcher, D. M. Zeiger, C. I. Bargmann and P. Sengupta, Neuron, 2012, 75, 585 CrossRef CAS PubMed.
- E. Z. Macosko, N. Pokala, E. H. Feinberg, S. H. Chalasani, R. A. Butcher, J. Clardy and C. I. Bargmann, Nature, 2009, 458, 1171 CrossRef CAS PubMed.
- J. Q. White and E. M. Jorgensen, Neuron, 2012, 75, 593 CrossRef CAS PubMed.
- R. R. Zwaal, J. E. Mendel, P. W. Sternberg and R. H. Plasterk, Genetics, 1997, 145, 715 CAS.
- D. Park, I. O'Doherty, R. K. Somvanshi, A. Bethke, F. C. Schroeder, U. Kumar and D. L. Riddle, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 9917 CrossRef CAS PubMed.
- K. Kim, K. Sato, M. Shibuya, D. Zeiger, R. Butcher, J. Ragains, J. Clardy, K. Touhara and P. Sengupta, Science, 2009, 326, 994 CrossRef CAS PubMed.
- R. V. O'Brien, R. W. Davis, C. Khosla and M. E. Hillenmeyer, J. Antibiot., 2014, 67, 89 CrossRef PubMed.
Footnote |
| † For ascaroside nomenclature see www.smid-db.org |
| This journal is © The Royal Society of Chemistry 2015 |


