Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Metabolomics in chemical ecology
Constanze
Kuhlisch
and
Georg
Pohnert
*
Friedrich Schiller University, Institute of Inorganic and Analytical Chemistry, Lessingstr. 8, D-07743 Jena, Germany. E-mail: Georg.Pohnert@uni-jena.de; Fax: +49 3641 948 172; Tel: +49 3641 948 170
First published on 30th April 2015
Abstract
Covering: up to December 2014
Chemical ecology elucidates the nature and role of natural products as mediators of organismal interactions. The emerging techniques that can be summarized under the concept of metabolomics provide new opportunities to study such environmentally relevant signaling molecules. Especially comparative tools in metabolomics enable the identification of compounds that are regulated during interaction situations and that might play a role as e.g. pheromones, allelochemicals or in induced and activated defenses. This approach helps overcoming limitations of traditional bioassay-guided structure elucidation approaches. But the power of metabolomics is not limited to the comparison of metabolic profiles of interacting partners. Especially the link to other –omics techniques helps to unravel not only the compounds in question but the entire biosynthetic and genetic re-wiring, required for an ecological response. This review comprehensively highlights successful applications of metabolomics in chemical ecology and discusses existing limitations of these novel techniques. It focuses on recent developments in comparative metabolomics and discusses the use of metabolomics in the systems biology of organismal interactions. It also outlines the potential of large metabolomics initiatives for model organisms in the field of chemical ecology.
1 Introduction
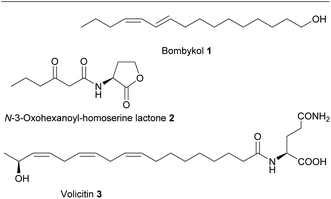
The ecological understanding of natural products that mediate interactions within and between organisms can be considered as the core of the discipline chemical ecology.1 Knowledge of the nature of chemicals that mediate such interactions is central in this research area. The identification of active metabolites opens up possibilities from fundamental work in bioassays over general investigations of the physiology and ecology of an organism to the manipulation of entire ecosystems. Signaling molecules that mediate interactions between organisms have motivated many activities in basic research but also the commercial interest e.g. for pest control is a driving force in the search of such bioactive metabolites.2 It is thus not surprising that the focus of numerous groups working in chemical ecology is the structure elucidation of these active principles. Since the early days of this discipline the central tool for such endeavors has been bioassay-guided structure elucidation. The work flow has first been successfully established for the identification of the insect pheromone bombykol (1) by Butenandt and colleagues in 1959.3 It is based on the separation of an active crude extract by e.g. chromatography, selective extraction or crystallization and testing of the resulting fractions in bioassays. Active fractions are further purified and again tests of activity guide the further selection until a pure active compound can be submitted to structure elucidation.4 This time-consuming and often tedious process can involve multiple rounds of separation and testing until the identification of a signal molecule is achieved. Besides the high workload the approach bears also the risk that activity of labile substances is lost during handling. Compounds that are only active in more complex formulations or specific mixtures, are also not recognized due to the involved purification steps and require combinatorial tests for activity verification.5 Despite these drawbacks bioassay-guided structure elucidation was the main available technique for signal identification in chemical ecology over decades. Indeed, besides bombykol most hallmark molecules in chemical ecology have been identified by this approach (Scheme 1).6,7With the advent of –omics techniques additional tools became available for the search for active principles. These include, among many others, the use of microarray methods, targeted knockout experiments based on the identification of candidate genes, and proteomics.8–10 But especially metabolomics techniques provide a novel approach to the identification of molecules of relevance in chemical interactions. In fact, long before the discipline of metabolomics emerged, comparison of metabolic profiles that were created from extracts of organisms in different physiological states or ecological stress situations were used to spot candidate metabolites that are upregulated in an interaction context. This is e.g. demonstrated by the identification of volatiles in response to insect herbivory.11 In a procedure that would today be categorized as metabolic profiling, volatiles in the headspace of plants were collected in controls and plants suffering herbivory. Comparison of gas chromatography/mass spectrometry (GC-MS) profiles led to the identification of several herbivory-induced compounds that could later be assigned to ecological functions in an indirect chemical defense. The collection and comprehensive analysis of plant volatiles from control organisms and those under herbivore pressure revealed not only the up regulation of terpenoid biosynthesis but also motivated further bioassays that led to a wide-ranging understanding of induced plant defenses. The involvement of the plant hormone jasmonic acid and its amino acid conjugates in the induction of metabolic pathways towards plant defensive metabolites was also discovered using such profiling tools.12 It is striking to see that even in early publications figures are presented that would meet the reporting standards of modern metabolomics. Despite the fact that data mining tools were not as elaborate as today these studies enabled the investigation of regulatory patterns that allowed spotting candidate molecules or pathways relevant in species interactions.
Since these early days of metabolic profiling the discipline of metabolomics has emerged and matured. Even if some approaches are still in their infancy, the multiple facets of metabolomics are now emerging as central tools in chemical ecology for screening the pool of natural products and for the elucidation of regulative principles and pathways. Per definition the metabolome represents all metabolites of a given species, but comprehensive monitoring of such a structurally diverse set of compounds is technically often not feasible.13 Thus several sub-disciplines have emerged that use the power of modern analytical instrumentation paired with elaborate statistical analysis suited mainly to recognize dynamic metabolic processes. Principal approaches and techniques in metabolomics have been reviewed extensively over the last years and will not be a subject of this contribution. We, however, want to refer to selected reviews for further reading concerning the general concepts in metabolomics,13–16 specialized techniques that play an important role in chemical ecology including mass spectrometry17,18 as well as nuclear magnetic resonance (NMR).19–21 A wide array of statistical methods is applied routinely within metabolomics and already reviewed under different aspects including data pre-processing, uni- and multivariate analyses and visualization aspects.22–24
Comparative metabolomics provides the best-suited tools to unravel chemically mediated species interactions.25 This approach will accordingly be the initial focus of this review. In contrast to previous studies where a targeted analysis of the induction of single compounds or specific groups of compounds was performed, comparative metabolomics allows to monitor global changes and to unravel multiple pathways that are affected by environmental stimuli using elaborate algorithms on complex data sets. Knowledge about the production of specific compounds or classes of metabolites during interaction situations might help to make educated guesses of their function. As a consequence this can motivate following bioassays to verify such hypotheses.26 In addition, this review will cover concepts where multiple –omics techniques are combined in a systems biology approach to obtain a comprehensive picture of an organisms' response in interaction situations. We will also introduce aspects how the comprehensive metabolomics initiatives for model species might be fruitfully applied in the chemical ecology. Taken together, we aim to classify and evaluate metabolomics as a potential future driver in the search for the nature, role and regulation of signaling molecules.
2 The comparative metabolomics approach
This rather descriptive approach is the fundamental metabolomics tool in chemical ecology. Comparison of metabolomes for the investigation of chemical interactions does not necessarily require a comprehensive monitoring of as many compounds as possible. If the nature of a chemical interaction is known, the work plan can be dramatically simplified by a pre-selection of specific groups of analytes. Thus, for example the metabolomics-enabled search for airborne pheromones of insects would surely not require extraction of the insects themselves for a metabolic profiling but rather be restricted to a comprehensive monitoring of emitted volatiles in the headspace. In contrast, elucidation of plant defense metabolites might exclusively require monitoring metabolites in the plant's tissue or on its surface. Numerous specialized techniques have been introduced focusing on specific groups of the exometabolome (exuded metabolites) e.g. volatiles, surface associated metabolites or dissolved metabolites from aquatic organisms. But also the endometabolome (internal metabolites) within specific cells, tissue or the whole organism can be addressed selectively. Another way to perform a pre-selection is the inclusion of functional considerations in the initial analytical approach. Thus, it has been successfully demonstrated that the search for compounds acting as metallophores, which mediate metal complexation and uptake in microorganisms and thereby shape organismal interactions, can be pre-selected by biasing analytical techniques.27,28 A pre-identification of metallophores in the exometabolome can be realized by recording liquid chromatography/mass spectrometry (LC-MS)-runs of the extracts in presence and absence of metals. The shift in masses resulting from the complexation of the metal can be used in a chemometric approach to pre-select only those metabolites relevant for metal interactions.27,28 Alternatively, isotope-assisted data mining approaches were recently introduced that spot specific isotopic patterns of metals in complexes.29 A pre-selection can also be made if specific pathways are suspected to be relevant in organismal interactions. This approach has e.g. been demonstrated for the elucidation of defense metabolites of the moss Dicranum scoparium. Here, initial evidence suggested that lipid-derived oxylipins mediate a wound activated chemical defense against herbivorous slugs.30 Administration of stable isotope labeled fatty acids allowed to identify upregulated metabolites using automated routines of data mining thereby pointing towards a novel set of previously unrecognized highly reactive metabolites.31 Genetic tools can make such pathway-associated approaches even more powerful. This was shown in the structure elucidation of myxoprincomide, a natural product from Myxococcus xanthus. The knockout of non-ribosomal peptide synthetase (NRPS) genes and subsequent comparative metabolic profiling led to the identification of this novel metabolite of which the function in the natural environment is, however, still undetermined.32 The emerging possibilities arising from the systematic pairing of metabolomics and genome mining opens up new avenues for the discovery of natural products.332.1 Comparative metabolomics in plant sciences
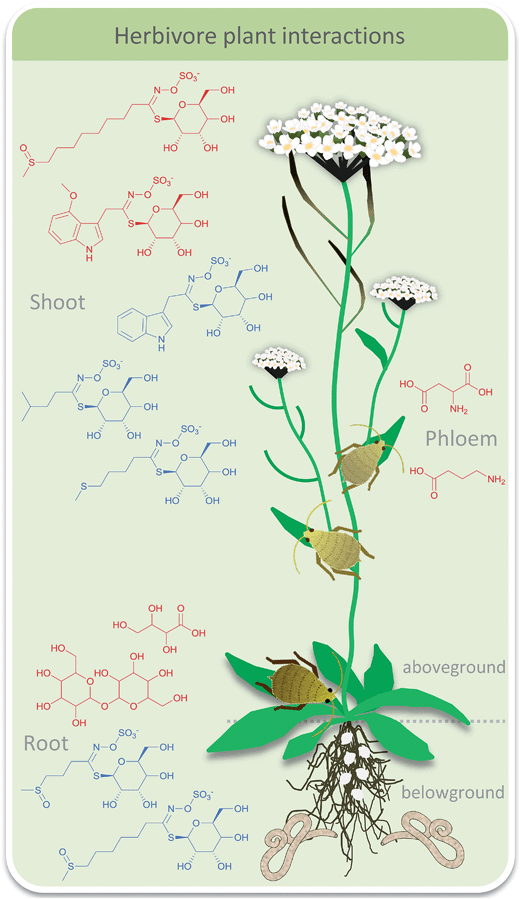
Plant interactions so far often have been studied by targeted analysis of a few model organisms. But especially within plant sciences comparative metabolomics has recently also been applied fruitfully. Metabolomics approaches using several platforms can be used on more diverse sets of plants to elucidate (common) interaction patterns. Plants can respond to a multitude of challenges by adapting their metabolic repertoire. These can include simple environmental stress situations, such as drought or light stress or infestation with pathogens and herbivory. But even herbivore interactions can be complex with different herbivores grazing on the same plant inducing different defense mechanisms at the same time. Thereby they also interact with each other in an indirect way via induced qualitative and quantitative changes of primary and secondary metabolites throughout the plant. All of these types of challenges can be addressed by comparative metabolomics. However the planning of experiments is crucial. For the understanding of simple responses the comparison of plant control and plants with grazer A might be sufficient. If a comprehensive picture of the defense potential of a plant shall be obtained additional investigations of the effects of a second grazer B as well as additional grazer A-grazer B-plant systems are required. On top, additional environmental stress factors can be considered to obtain a comprehensive picture.A well-studied system is the grazing of aphids and root feeding nematodes on Arabidopsis thaliana. Single effects of each grazer have already been studied. Targeted analysis of glucosinolates in response to feeding bioassays have been introduced thereby paving the way for more complex experimental set-ups.34 Such targeted profiling focuses on a small set of known candidate metabolites with similar chemical properties whereas untargeted approaches open up future perspectives in plant chemical ecology by uncovering previously unidentified lines of defense. Recently the mutual influence of above- and below-ground herbivores that is mediated by plant chemistry was addressed.34,35 Comparative metabolic profiling was used to monitor effects of grazing of the aphid Brevicoryne brassicae and the nematode Heterodera schachtii on A. thaliana (Fig. 1). Metabolic profiling of primary metabolites in the shoots by GC-MS and metabolic fingerprinting of shoot and root tissue by LC-MS were conducted and analyzed via multivariate statistics. Aphid grazing had a significant local effect in the shoot tissue. Only minor systemic effects were observed in the root tissue but changes of glucosinolates indicated an induced defense. As a consequence, aphid grazing led to reduced nematode infestation. In contrast, nematodes did not evoke any significant metabolite alterations locally or systemically thus having no influence on aphid population.34 The confounding effect of nitrate fertilization in this plant-mediated herbivore interaction was considered in a follow-up study.36 Again, only aphid grazing influenced nematodes but in a complex pattern. Under lower nitrate conditions aphid grazing increased nematode population whereas under higher nitrate fertilization nematodes were reduced as seen before. Metabolic profiling of root primary metabolites via GC-MS gave 88 compounds of which 54 could be identified. Multivariate statistics revealed that only marginal changes were observed under aphid grazing which might indicate a very sensitive reaction of nematodes. In a study where the specialist B. brassicae was replaced by the generalist Myzus persicae nematodes negatively influenced aphid abundance under lower nitrate availability.37 Targeted metabolic profiling of amino acids and glucosinolates was conducted via GC-MS followed by principal component analysis (PCA) and multivariate analysis of variance (MANOVA). In response to nematode feeding the amino acid composition in the phloem changed as well as the glucosinolate composition in the leaves. The latter was correlated with aphid abundance.
The aspect of diurnality of changes in above- and below-ground metabolite patterns in response to herbivory was addressed in the wild tobacco plant Nicotiana attenuata.38 Herbivory was simulated by wounding of the tobacco plants and administration of the oral secretion of the herbivorous caterpillar Manduca sexta. Metabolic fingerprinting showed that different compounds were oscillating in leaves compared to roots. In fact, only ten ions of 182 oscillating in leaves and 179 oscillating in roots (LC-MS) were rhythmic in both tissues. The response to simulated herbivory of seven selected metabolites was investigated in detail (unknown disaccharide, lyciumoside I, phenylalanine, tyrosine, coumaroyl tyramine, feruloyl putrescine, N-feruloyl tyramine, 12-oxo-phytodienoic acid, and jasmonic acid) and compared to transcript analyses, revealing a highly tissue-specific accumulation. It was concluded that the pronounced diurnal rhythm in the generalized and specialized metabolism that mediates the plant's responses to herbivory might play an important role in orchestrating its response to herbivory.
The role of the plant hormone jasmonic acid (JA) for herbivore defense was addressed in a Nicotiana attenuata mutant.39 If a jasmonate O-methyltransferase from A. thaliana is expressed in N. attenuata, it methylates JA, thereby acting as a sink for this plant hormone. This mutant was more vulnerable to grazing in the field. Targeted analyses of known defense compounds nicotine, diterpene glycosides and trypsin proteinase inhibitors but also untargeted metabolic profiling of volatile and non-volatile leaf metabolites was conducted. 42 of the most abundant and consistently detected volatile organic carbons (VOCs) in GC × GC/MS were compared between the mutant and the wild type after wounding. Early and late response VOCs could be separated clearly via PCA and partial least squares discriminant analysis (PLS-DA) due to increase of terpenes and hexenylesters and decrease of non-esterified green leaf volatiles during late response. In the late response mutants were separated from wild type due to reduced VOC emission (including cis-3-hexen-1-ol, cis-3-hexenyl-butyrate, -acetate, α-terpineol, trans-α-bergamotene and β-myrcene). Also the metabolic profiles (LC-MS) of secondary metabolites of mutant and wild type leaves were separated (PLS-DA) due to reduction of defense compounds such as nicotine, diterpene glycosides and phenylpropanoid–putrescine and –spermidine conjugates as well as some unknown metabolites in the mutant plants. This study convincingly demonstrated the multiple effects of jasmonic acid in a plant's defense and the power of metabolomics approaches to unravel such complex responses.
Gaquerel et al.40 followed the effect of silencing of a hydroxycinnamoyl transferase (HCT)-like gene in N. attenuata, which was found to be induced in a previous transcript screening after herbivory.41 Targeted analysis of well characterized herbivory-responsive caffeoyl- and feruloyl-based putrescine and spermidine phenolamides (PAs) did not show any pronounced silencing effects. However, an unknown PA-type did. This motivated an untargeted metabolomics approach using LC-MS. Up to 12% deregulation in insect-attacked leaves could be assigned to a diversion of coumaric acid units into the production of coumaroyl-containing PAs (Scheme 2). Samples could be separated according to genetic manipulation and herbivory treatment via PCA, and Venn diagrams showed increased metabolite numbers in silenced plants. All metabolites influenced by silencing were clustered hierarchically and annotation of the strongest regulated entries allowed the identification of several novel PA isomers and derivatives. The HTC-like gene thus encodes a coumaroylquinate-forming enzyme and mediates competition between the PA pathway and lignin production. This work is a nice example how metabolomics can be used to uncover a previously undescribed network of metabolites and help to unravel the interplay among herbivory and developmentally-controlled metabolic responses.
 | ||
| Scheme 2 Coumaroyl and caffeoyl spermidines and putrescines from A. thaliana. Specific positions of the residues were not assigned.40 | ||
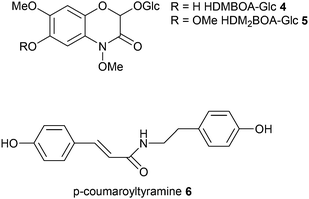
The herbivory effect of Spodoptera spp. on maize is well-investigated. Two recent studies now applied metabolomics to mine for hitherto poorly characterized responses.42,43 In those monocotyledonous plants 1,4-benzoxazin-3-ones (Bxs) are major defense compounds but the inducibility of specific metabolites of this class is rather poorly understood. Therefore Glauser et al.42 used unbiased LC-MS-profiling to screen plant material before and after herbivory. PCA clearly separated the treatments and by investigating the loadings 2-β-D-glucopyranosyloxy-4,7-dimethoxy-1,4-benzoxazin-3-one (HDMBOA-Glc 4) and 2-β-D-glucopyranosyloxy-4,7,8-trimethoxy-1,4-benzoxazin-3-one (HDM2BOA-Glc 5) appeared as strongest contributors to the separation in induced plants. Catabolism of these metabolites was addressed by monitoring also the digestive products in specialized and non-specialized herbivores. Since plants and herbivores differ in their Bxs-profiles digestive processes could be followed up based on PCA-evaluation of metabolic patterns. Consecutive analyses revealed the role of the highly dynamic activation of maize Bxs in chemical defense.
Local and systemic responses to leaf herbivory by S. littoralis were the topic of a recent survey of secondary metabolites from maize leaves, sap, roots and root exudates.43 A very comprehensive coverage using LC-MS in combination with unsupervised (PCA) and supervised (orthogonal (O)PLS-DA) data mining revealed more than 300 features in leaves and roots, 180 features in sap and 40 features in root exudates. Thirty-two differentially regulated compounds were identified from Spodoptera littoralis-infested maize seedlings and isolated for structure assignment by microflow NMR. Treatment-specific separation was only achieved for leaf samples and root exudates. Local leaf response consisted of an increase of lysophospholipids, azelaic acid, 1,3-benzoxazin-4-one derivatives, tryptophan and N-hydroxycinnamoyltyramines, whereas systemic response could be assigned to 1,3-benzoxazin-4-one derivatives increase in the sap and root exudates. Since N-hydroxycinnamoyltyramines were previously not identified in plant–herbivore interactions the authors tested the effect of the dominating p-coumaroyltyramine 6 on S. littoralis. This plant-derived natural product was metabolized by the larvae and increased larval growth, acting presumably as nitrogen source for the insect. The untargeted comparative metabolomics approach can thus lead to discovery of novel dynamically regulated metabolites and help to create novel hypotheses about herbivore–plant-interaction.
The interaction between the fungal endophyte Fusarium verticillioides and the pathogen Ustilago maydis which are both growing on maize has been the subject of several studies.44,45 The endophyte thereby decreases the harmful effect of the pathogen.46 LC-MS metabolic profiling of isolated partners as well as of co-cultures of the fungi revealed that most secondary metabolites are constitutively produced by each species.44,45 Nevertheless the presence of F. verticillioides leads to reduction in U. maydis biomass. Comparative metabolomics was also employed to investigate leaf herbivory of Manduca sexta and Helicoverpa zea on tomato (Solanum lycopersicum) by GC-MS.47,48 Metabolites were identified based on library searches resulting in 56 and 60 identified primary metabolites respectively and further analyzed via PCA and Venn diagrams. Primary metabolites throughout the plant (apex, leaves, stem, and roots) showed a species- and tissue-specific plant response. Induced metabolic changes were strong in the apex and root tissues as well as in damaged leaves confirming a whole-plant response to damage.
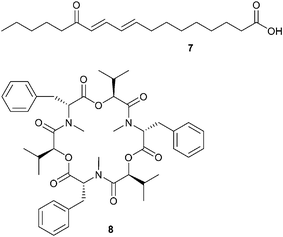
The metabolome of the endophytic fungus Paraconiothyrium variabile is of interest if the indirect interaction with its host plant Cephalotaxus harringtonia is concerned. P. variabile inhibits the phytopathogen Fusarium oxysporum thus providing protection for the host.49 During an in vitro competition situation of both fungi LC-MS profiling and analysis with the powerful software XCMS50 revealed, besides a comprehensive set of inactive constitutively produced metabolites, a series of induced compounds. These included 13-oxo-9,11-octadecadienoic acid 7, 13-hydroperoxy-9,11-octadecadienoic acid, and several unknowns (Scheme 3). Interestingly, nanogram amounts of 13-oxo-9,11-octadecadienoic acid reduced the production of the mycotoxin beauvericin 8 in the phytopathogen F. oxysporum thereby promoting the idea of an induced interference mechanism.
 | ||
| Scheme 3 13-Oxo-9,11-octadecadienoic acid 7 is upregulated in Paraconiothyrium variabile and suppresses formation of the mycotoxin beauvericin 8 in Fusarium oxysporum. | ||
Allelopathic interactions were investigated in a large study that addressed the effects of 16 Mediterranean plants on ovate goatgrass Aegilops geniculata using NMR-based comparative metabolic profiling.51 Extracts of the 16 donor plants were characterized in terms of chemical composition. The effects of plant extracts on A. geniculata revealed a biological activity for most studied species probably due to phenolic compounds such as flavonoids and hydroxycinnamate derivatives. In accordance to allelopathic activity, PCA analysis of NMR data of A. geniculate extracts clearly separated active treatment samples from controls and inactive treatment samples. This separation could be assigned to an increase in amino acids and organic acids and a decrease in oblongaroside, betaine, and cis-aconitic acid indicative for oxidative stress responses of the receiver plant. In accordance, hierarchical cluster analysis indicated three metabolite response groups: no, slight and strong metabolic changes due to treatment. This study is a nice example how such systemic approaches allow unravelling synergistic effects of allelopathic interactions that would have been difficult to spot in traditional bioassay-guided approaches.
Schweiger et al. analyzed the mutualistic interaction of arbuscular mycorrhiza (AM) with plant roots.52 This ancient fungi–plant-symbiosis is widespread occurring in >80% of land plants. Common vs. species-specific responses in leaves of five model and non-model plant species (Plantago lanceolata, P. major, Veronica chamaedrys, Medicago trunculatula, Poa annua) to the generalist AM fungi Rhizophagus irregularis were investigated. A comparative metabolomics approach was selected involving the targeted metabolic profiling of carbohydrates, organic acids and sugar alcohols via GC-MS, of amino acids via HPLC-fluorescence detection, and the untargeted fingerprinting via LC-MS of methanolic leaf extracts. Between 18% and 45% of the polar metabolome was shared among species and designated as core metabolome with the rest being highly species/taxon-specific metabolites. Those species-specific differences would superimpose treatment effects and therefore these were analyzed separately for each plant. Besides a few common features in the dicotyledonous plants, responses were very species-specific even though the core metabolome would have allowed a common response pattern. The low conservation of responses indicates a long time of specific plant–fungus coevolution that leads to specific manifestations in the expressed metabolic response.
Metabolomics also allows the classification of responses of organisms by collecting “metabolic fingerprints” of a multitude of samples in defined states. Novel samples can then be compared to this data set allowing a categorization without any further ecological or physiological evaluation. This approach is especially used in plant sciences where stress responses are often analyzed by suitable interpretation of such metabolic patterns. Categorization of responses might also allow the identification of metabolites relevant in a specific interaction context thereby making this approach very valuable for the search of active metabolites in chemical ecology.
2.2 Comparative metabolomics in marine science
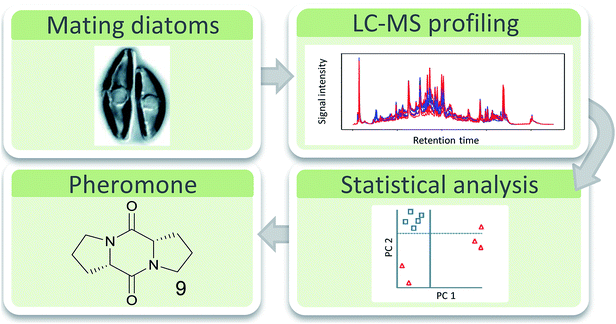
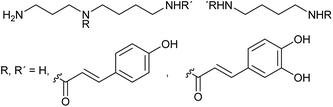
Especially in the chemical ecology of marine algae comparative metabolomics has recently gained importance e.g. to unravel anti-herbivory defense strategies,53 allelopathic interactions,54,55 bacteria–algae-symbioses,56 or to discover algal pheromones.57 However, compared to plants, defense strategies against herbivory are only poorly investigated in marine algae.58,59 Other disciplines in marine chemical ecology currently start to use this emerging tool box as well.60 Generally, the rather complex matrix of the seawater that carries signaling molecules represents a certain challenge if waterborne signals are concerned. In addition, there is no logical pre-selection for enrichment and extraction techniques of specific compound classes as it is the case for volatiles in insect signaling. It has been shown that the physical nature of metabolites that carry information in the seawater can range from highly polar zwitterionic metabolites61 to entirely hydrophobic hydrocarbons.62 The molecular weight is also not limiting the information content, since gases63 and small molecules64 as well as high molecular weight lipids65 and proteins66 can be active in interaction situations. Extraction techniques thus have to cover a broad spectrum of metabolites if those relevant in interactions shall be universally covered.Solid phase extraction and LC-MS analysis were suitable tools for the elucidation of the first diatom sex pheromone guided by comparative metabolomics.67 The pennate diatom Seminavis robusta reproduces mainly asexually by cell division. Due to the rigid biomineralized cell wall of the diatoms this leads to a gradual cell size reduction in the population. If the cell size drops below a specific value, sexual reproduction of the alga is enabled. In this process pheromones are involved that activate the mating behavior and attract the partner cells of opposite mating types.68 Comparative metabolic profiling of culture medium from cells that are not sexually active and those that call their mating partner revealed several upregulated metabolites as candidates for attraction pheromones.67 The highest upregulated metabolite (>100 times) was isolated by fractionation and further bioassays confirmed its activity as mate attractant. Structure elucidation via MS(/MS), synthesis and co-injection proved di-L-prolyl diketopiperazine 9 as pheromone that was as active in bioassays as the natural pheromone (Fig. 2).
To unravel its chemical defense, the metabolome of the red alga Gracilaria vermiculophylla in response to herbivory by the generalist isopod Idotea baltica was studied using a combination of GC-MS and LC-MS metabolomics and bioassays.53 Mechanical wounding significantly changed metabolic profiles as revealed by Permanova analysis, with some compounds being increased more than 100-fold. Metabolites that were upregulated in wounded tissue were predominantly eicosanoids (Scheme 4).
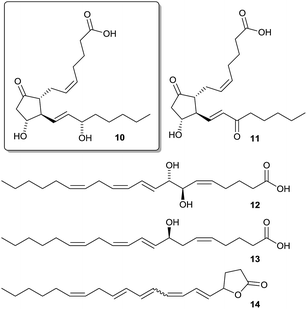 | ||
| Scheme 4 Wound-activated produced eicosanoids (PGA210, PGE211, 7,8-di-HETE 12, 8-HETE 13, and a previously unknown lactone 14) with the feeding deterrent prostaglandin A2 10 framed. | ||
A detailed investigation of the data set by canonical analysis of principal coordinates (CAP) showed 11 upregulated metabolites in LC-MS and 8 upregulated metabolites in GC-MS data including prostaglandins, hydroxylated fatty acids, and arachidonic acid derived lactones. Isotopic labelling confirmed that most upregulated metabolites were derivatives of arachidonic acid. In subsequent bioassays five of the upregulated eicosanoids were tested for feeding deterrence. Only prostaglandin A2 10 was deterrent while all other tested compounds rather attracted Idotea. This wound-activated defense differed from the induced defense which was systemically manifested in tissue neighboring feeding activity as revealed by CAP analysis in comparison to controls. While the activated response led to a change in eicosanoids, induced response affected mainly primary metabolites indicating a general stress effect, which calls for further investigations.
Allelopathy is an important mediator of competition in plankton69 and has been investigated focusing on the nature of allelopathic metabolites and the physiological response of the receiver.54,55 In a comparative study of the interaction of the diatoms Skeletonema costatum and Thalassiosira weissflogii in co-culture it was observed that S. costatum promoted the growth of T. weissflogii. Comparative metabolic profiling via LC-MS revealed significant metabolic changes in the intra- and extracellular metabolome of both algae compared to monocultures. In the medium eight S. costatum specific metabolites and one T. weissflogii specific metabolite were found; their identification still remains an open task. Intracellular metabolomics in co-culture showed both up- and down-regulated metabolites in S. costatum whereas in T. weissflogii only downregulation was observed compared to the respective controls. This study demonstrates that even without physical contact non-toxic diatom cells can influence interaction partners by modulation of their endo- and exometabolome. In this context a novel approach to characterize exudates from aquatic autotrophs by stable-isotope metabolic footprinting might prove helpful for a thorough characterization of algal exudates from the complex seawater background.70 Using a similar approach, the positive effect of the bacterium Dinoroseobacter shibae on the alga Thalassiosira pseudonana was investigated.56D. shibae slightly enhanced the growth of T. pseudonana in co-culture. Metabolic profiling of intracellular metabolites via GC-MS revealed more than 500 algal metabolites which could be separated according to treatment via principal coordinate analysis (PCO) and CAP. In accordance to the observed enhanced growth, algal primary metabolites were upregulated in presence of bacteria, among them amino acids such as serine, proline, phenylalanine and a glutamic acid derivative, as well as picolinic acid, some fatty acids and sugars. This study demonstrates a re-wiring of primary metabolism in the presence of bacteria, thereby underlying the complexity of plankton interactions.
2.3 Comparative metabolomics in nematode research
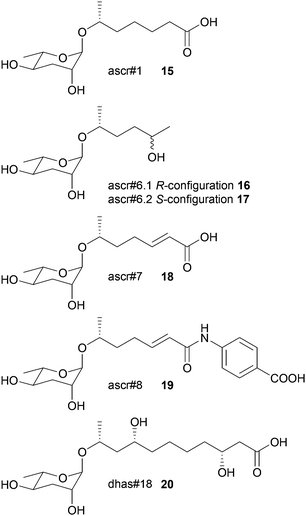
In nematode pheromone research comparative metabolic profiling has been applied mainly via MS/MS and NMR.71,72 This field has recently been comprehensively reviewed elsewhere and thus this chapter will focus only on the general principles of metabolomics in the elucidation of signal molecules in this context.73 The model organism Caenorhabditis elegans produces a class of glycolipids, the ascarosides, regulating diverse functions including development and behaviour.74,75 Ascarosides (e.g.15–20) are built of the dideoxysugar ascarylose and fatty acid-derived side chains of varying length. Since bioassay-guided fractionation often was unsuccessful due to synergistic functions of the metabolites that are lost during purification, comparative metabolomics proved to be the method of choice. The differential analysis by 2D-NMR spectroscopy (DANS) simplifies detection of ascarosides that are preferentially produced during interaction and regulation events.76 For this purpose DQF-COSY NMR spectra of the wild type were overlaid with spectra of a signaling-deficient mutant, daf-22. Since the biosynthesis of certain mating and dauer pheromone components is abolished in daf-22 worms a subtraction of the wild type and the mutant metabolome allowed identification of novel potentially active components. Major remaining signals represented mainly short-chained ascaroside structures including the four novel ascarosides ascr#6.1 16, ascr#6.2 17, ascr#7 18, and ascr#8 19.Additional HSQC- or HMBC-NMR, as well as HPLC-MS investigations and total synthesis confirmed the structures, and their function was verified in bioassays where ascr#8 proved to be a strong male-specific attractant and inducer of larval arrest.76 Using LC-MS-based targeted metabolomics it could be shown that C. elegans males produce ascaroside signatures that differ from those of hermaphrodites. Even minor structural modifications profoundly affect the ascarosides' signaling properties.77 These approaches proved to be fruitful in the elucidation of other functions and structures of ascarosides.72,78,79 The sex-specific attraction of the nematode Panagrellus redivivus was initially investigated by bioassay-guided fractionation followed by LC-MS, 1D- and 2D-NMR, and total synthesis.72 Males are attracted by ascr#1 (only produced by females) and females by dhas#18 (only produced by males). Targeted profiling of the exometabolomes of a mixed culture by LC-MS2 revealed four more ascarosides (ascr#10, ascr#3, bhas#18, and bhas#10); however, a biological function could not be identified. Further comparative profiling of males, females and mixed cultures revealed highly sex-specific ascaroside production. Males only exhibited bhas#18 and dhas#18 whereas females exhibited at least 6 different ascarosides. The study suggests that ascarosides can have different functions in different nematode species and can even serve as allelochemicals.
The biosynthesis of ascarosides was elucidated by comparative LC-MS2 profiling of wild type and several peroxisomal beta-oxidation mutants. Based on a series of elaborate experiments a working model of ascaroside biogenesis was proposed that involves an origin from very long side chain precursors that are transformed via peroxisomal beta-oxidation.71 A picture arises of ascarosides as a modular library derived from carbohydrate metabolism, peroxisomal beta-oxidation and amino acid catabolism. Interested readers are referred to the contribution “combinatorial chemistry in nematodes: modular assembly of primary metabolism-derived building blocks” by Stephan H. von Reuss and Frank C. Schroeder within this issue of NPR.
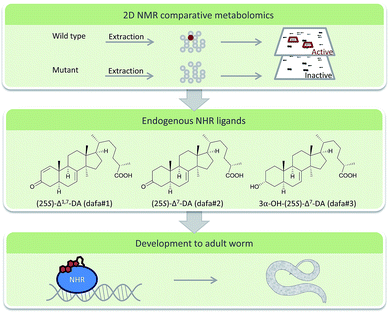
DANS-based comparative metabolomics of daf-12 and daf-9 C. elegans mutants revealed steroids with unexpected structural features that were further characterized after HPLC purification.80 The compounds that are ligands of nuclear hormone receptors (NHR) of C. elegans extend our knowledge of transcriptional regulators in metazoans significantly (Fig. 3). More elaborate algorithms for comparative metabolic profiling via NMR have been already applied for comparison of the nematodes Pristionchus pacificus and Panagrellus redivivus and await application in the chemical ecology of this system.81
2.4 Comparative metabolomics unravels pheromones in social insects
Social insects like bees, ants, wasps or bumblebees use chemical signals for diverse chemical interactions such as nest-,82 sex-,83,84 or caste-recognition,85–87 or as appeasement signal in mutualistic nest sharing.88 Mainly cuticular hydrocarbons (CHCs) serve for chemical communication within and between species. These rather simple natural products are produced in complex mixtures that can hardly be resolved using traditional bioassay-guided fractionation. However, comparative metabolomics proves to be an ideal tool to spot relevant CHCs that are present in specific species or members of one species or that are regulated in interaction situations.Reproduction in eusocial insects and thus development of reproductive queens and non-reproductive workers is regulated by pheromones which are primarily non-volatile, saturated hydrocarbons.85 Comparative profiling of queen and worker cuticular extracts were investigated in the wasp Vespula vulgaris,89 the bumblebee Bombus terrestris,90 and the ant Cataglyphis iberica. Candidate pheromones were selected based on the assumption that they are more abundant in queens compared to workers.85 The identified structures were tested in ovary regression and development bioassays and it was found that active compounds were all long-chain linear and methyl-branched saturated hydrocarbons (n-C27, n-C29, 3-MeC29 in the wasp and ant species, and n-C25 in the bumblebee).85
Primitively eusocial insects – such as permanently social polygynous epiponin wasps – can show caste flexibility until the adult stage. Queen selection either happens under physical attack86 or peacefully.91 Regarding the first case, after queen removal Polybia micans workers show aggression towards young, potential queen candidates. Prospective queens with larger ovaries also have higher amounts of queen caste-specific hydrocarbons thus probably increasing worker aggression. After a few days some are accepted and start as new queens. Comparative metabolic profiling of CHCs via GC-MS revealed a general dominance of linear alkanes but also showed differences between caste, age and reproductive status. C25 and 3-MeC25 could be identified as queen caste-specific. Further bioassays would be needed to confirm the potential roles. A similar but peaceful social behavior was found within the closely related Synoeca surinama, however dominant CHCs differ considerably.91 Altogether 22 CHCs (alkenes or linear alkanes) were identified. Stepwise discriminate function analysis separated queens from >4 days old workers. C25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was increased in all queens in all colonies, whereas workers had longer hydrocarbons. Thus it seems that after queen removal, juvenile hormone modifies the CHC blend of young workers with an increase in C25
1 was increased in all queens in all colonies, whereas workers had longer hydrocarbons. Thus it seems that after queen removal, juvenile hormone modifies the CHC blend of young workers with an increase in C25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 matching that of reproductive queens.
1 matching that of reproductive queens.
Worker reproduction in ants is reduced through policing behavior, such as physical aggression or egg eating. Based on the observation that CHCs regulate such aggression Smith et al. investigated the ant Aphaenogaster cockerelli where worker-produced eggs are not policed.92A. cockerelli has large colonies with only one queen and workers with active ovaries producing trophic eggs. CHCs were collected via solid phase microextraction from ants and via hexane extraction from eggs, measured by GC-MS and analyzed via nonmetric multidimensional scaling (nMDS). Interestingly, this species was found to have no distinct signaling pattern for worker-produced eggs which was confirmed by policing assays. In contrast, cuticular profiles of queens, workers and reproductive workers showed quantitative and qualitative differences which allow a separation via nMDS. These patterns were further investigated and confirmed via bioassays: reproductive cheaters could be mimicked by applying synthetic pentacosane, a compound typical of fertile individuals on nonreproductive workers. Pentacosane treatment induced nestmate aggression in colonies where queens were present.93
Within the trap-jaw ant Odontomachus brunneus large population differences within CHC profiles were observed that are potentially responsible for nest mate discrimination.94 However, Z9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C29 was found via nMDS as conserved fertility signal in queens across populations, a compound that was already recognized earlier. Social parasites such as the bumblebees Bombus bohemicus and B. rupestris use similar nest-specific signals (worker trail pheromones) to find their hosts.95 Inter- and intraspecific odor variation of 45 hydrocarbons of nests of potential hosts were examined via GC-MS. PCA and discriminant function analysis (DFA) revealed interspecific and intercolonial differences in the odor bouquets of the host that were predominantly due to different patterns of alkenes. In Y-olfactometer choice tests B. rupestris showed a clear preference for the scent of its host proving that volatile signals enable parasite females to discriminate between potential host species.
C29 was found via nMDS as conserved fertility signal in queens across populations, a compound that was already recognized earlier. Social parasites such as the bumblebees Bombus bohemicus and B. rupestris use similar nest-specific signals (worker trail pheromones) to find their hosts.95 Inter- and intraspecific odor variation of 45 hydrocarbons of nests of potential hosts were examined via GC-MS. PCA and discriminant function analysis (DFA) revealed interspecific and intercolonial differences in the odor bouquets of the host that were predominantly due to different patterns of alkenes. In Y-olfactometer choice tests B. rupestris showed a clear preference for the scent of its host proving that volatile signals enable parasite females to discriminate between potential host species.
In all these examples the power of statistical analysis of complex metabolic signatures could be demonstrated. While separation of the hydrocarbons and testing is tedious and would risk overlooking effects of specific mixtures, the recognition and evaluation of metabolic patterns helps to unravel the complex language of CHCs.
2.5 MS/MS networking in comparative metabolomics
The capacity to generate vast data sets also challenges the ability for downstream processing.96 In the above mentioned examples the major goal was to eliminate signals of potentially irrelevant metabolites, but approaches to use MS-based molecular networking or other elaborate algorithms also allow to sketch more complex interaction networks by comprehensively evaluating metabolic patterns.97 An emerging tool is MS/MS networking, a visualization method for MS/MS data sets where molecules are clustered with regard to similarities in their molecular ion mass and fragmentation pattern.98 MS/MS spectra of complex mixtures of molecules are compared pair wise and their similarity is expressed as cosine of the normalized ratio of relative ion intensities (cosine similarity score). As a result structurally similar compounds with similar fragmentation patterns are located close to each other thus forming sub clusters of “molecular families” that share certain chemical properties. For identification of single nodes or whole sub clusters the network can be supplemented with standards and database entries.98 This identification differs from traditional approaches as MS/MS spectra are matched against other MS/MS spectra instead of matching them against protein sequences or molecular structures. Related or identical compounds are therefore detected before considering their possible identification. Instead of the traditional peak-by-peak identification consensus identifications from sets of spectra of related compounds are determined allowing a novel approach for classification.96,97,99 Common metabolites in data sets of interaction studies as well as the global molecular diversity of single species or certain taxa can be easily visualized. Also biosynthetic considerations can be taken into account during data evaluation.100–104 The software to generate MS/MS networks is freely available105,106 and even quantitative data sets can be considered.107To demonstrate the applicability of the MS/MS networking approach, several bacterial colonies were analyzed in situ on agar plates using nano desorption electrospray MS (DESI-MS).98 Networks of metabolites from Bacillus subtilis, Streptomyces coelicolor, S. marcescens, Mycobacterium smegmatis and Pseudomonas aeruginosa demonstrated the ability to visualize species-dependent molecular diversity. But also time-dependent networks of B. subtilis colonies could be generated to gain insights into bacterial developmental processes. Most important in the context of this review is the fact that also microbial interactions could be visualized as shown for S. coelicolor and B. subtilis (Fig. 4). Molecular networking was able to confirm earlier reports that B. subtilis PY79 elicits pigment production in S. coelicolor, whereas S. coelicolor increases production of the cannibalistic factors SKF and SDP in PY79 in the region of interaction.98
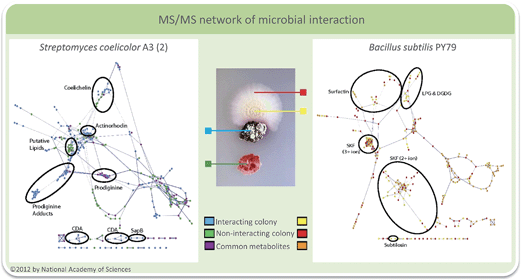 | ||
| Fig. 4 Interacting S. coelicolor (blue) and B. subtilis (yellow) in comparison to samples from noninteracting bacteria (red and green). The molecular network of S. coelicolor is shown on the left and that of B. subtilis on the right. With permission from the National Academy of Sciences USA.98 | ||
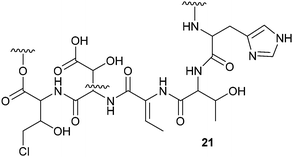
The antifungal activity of Pseudomonas sp. SH-C52 – a strain that protects sugar beet plants from soil born fungi108 – could be tentatively assigned to thanamycin 21 by generating a comparative MS/MS network of the wild type strain and two mutants. A partial structure of thanamycin 21 could be generated based on MS/MS data and analysis of the gene cluster. Since this initial report, MS/MS network-guided genome mining has been applied several times to facilitate the identification of non-ribosomal peptides (NRPs) and their associated gene clusters.99,104
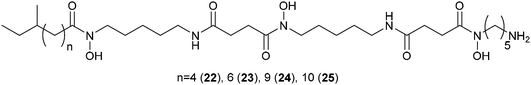
The antifungal activity of the octocoral symbiont Bacillus amyloliquefaciens against 13 different marine and terrestrial fungi was investigated using matrix-assisted laser desorption/ionization-imaging mass spectrometry (MALDI-IMS).109 In side-by-side interactions of B. amyloliquefaciens with Aspergillus fumigatus and A. niger potential antifungal metabolites were measured and visualized using seeded MS/MS networking. Within the network members of the iturin-family were spotted that proved to be active in antifungal bioassays. The excreted metabolites of Streptomyces coelicolor during interaction with five other actinomycetes showed a high specificity with regard to the interacting partner.110 By combining nano DESI-MS and MALDI-IMS one of the most comprehensive sets of molecules involved in microbial interactions to date was collected. Among the metabolites specific for the interaction a family of desferrioxamines 22–25 with acyl side chains of various lengths was triggered by siderophores from the neighboring actinomycetes. Altogether about 40% of all nodes within the spectral network were associated with the acyl-desferrioxamines 22–25 clearly characterizing the response of S. coelicolor. Besides the implications for the understanding of the chemical ecology of these strains, the study also outlines the importance of surveyed co-culturing in the search for new metabolites with potential pharmacological activity.
3 The systems biology approach
In contrast to the above mentioned reduction of complexity by pre-selecting specific groups of metabolites and by identifying single metabolites of relevance, systems biology approaches aim for a most comprehensive data collection and interpretation involving multiple-omics techniques.111,112 Within this section the combination of metabolomics with transcriptomics or genomics to gather a more complex picture of the chemical ecology of organismal interactions will be presented. Such approaches can aim towards a full understanding of the biological processes connected to the production or perception of compounds involved in chemical ecology. This is a complex task, which not only depends on the identification of the compound itself but also on the investigation of the involved biochemical pathways and physiological responses on a gene, transcript, protein, and metabolite level using the entire omics-toolbox. Additionally, a link to ecology that can be provided in lab or field experiments is required to determine the role of the identified principles. Combining all these techniques is challenging but enables the unbiased generation of novel hypotheses and opens up new research avenues.113Due to the advent of next generation sequencing technologies, genomic resources with a high degree of standardization can easily be obtained. However, the functional gene annotation that is relevant for the interconnection with metabolomics is lagging behind. In chemical ecology the understanding of dynamic metabolic networks is essential and it is therefore important to gather quantitative information using downstream-omics tools on the transcript, the protein or the metabolite level. A first challenging aspect for such an endeavor is the experimental design. Especially when taking the ecological situation of an organism into account elaborate incubation and sampling protocols are of key importance. Ideally, sampling should be performed in one step for all subsequent omics-technologies. In cases where time series are important this sampling should be minimal invasive to avoid disturbing the system. Substantial progress has been made and a recent protocol describes how one single sample can be used for analysis of metabolites, DNA, long and small RNA, and proteins via sequential isolation of those biomolecules.114 Since the available biomass might be limiting, such a protocol ideally needs only minute amounts of sample. The introduced protocol requires only 50 mg fresh weight for a full sample set and has been tested using the plant species Populus trichocarpa, Pinus radiata, and Arabidopsis thaliana, as well as the microalga Chlamydomonas reinhardtii. Successful implementation in four different laboratories with different background in handling of such protocols showed the robustness of the method. Data integration and analysis of the different –omics data sets are still a major challenge. Several approaches have been introduced to correlate the different data sets, but are highly challenging to apply.115,116 Thus often separate analysis of the respective data-sets and integration of the results is a more feasible approach as it is illustrated in the following examples.
The mode of infection and resistance of microalgae against a marine virus was addressed by Rosenwasser and collegues.116 Those viruses can control some of the largest oceanic algal blooms of the coccolithophore Emiliania huxleyi and thereby deeply influence organismal interactions in the plankton. The interaction of the algal host and its specific double stranded DNA virus was observed in a set of infection experiments involving both, lytic and non-lytic virus strains. Global host and virus transcriptome profiling was conducted simultaneously with metabolome analyses to unravel cellular pathways affected by the viral infections. The transcript analysis proposed the regulation of several biochemical pathways of both the host and the virus. Also distinct shifts in metabolite composition during lytic and non-lytic infection were observed by comparative metabolic profiling.116,117 In a combined weighted correlation network analysis that had to take into account a delay in metabolic compared to transcriptomic response, clusters from transcriptomics were correlated to metabolite levels. As many metabolites were correlated to one specific transcript expression pattern, integrated metabolic maps could be constructed. Especially during the early stages of viral infection a dynamic modulation of the host metabolism was observed. Based on the metabolic maps, a viral up regulation of algal fatty acid biosynthesis to help viral assembly, and a down regulation of host sphingolipid biosynthesis was observed. Viral influence on sphingolipids was already earlier identified in this system, thereby confirming the validity of the approach.118 But in addition, several terpenes including a group of sterols were substantially down regulated in agreement with a reduction of all host genes related to the production of isopentenyl-pyrophosphate in the mevalonate pathway. This down regulation could be assigned to an antiviral response mechanism that was further confirmed by inhibition experiments. Subsequent targeted metabolic analyses revealed that the suppression of steroid formation interferes with viral replication that requires these metabolites. Viral replication in E. huxleyi thus depends on the host metabolic machinery to provide sterols, fatty acids and sphingolipids as building blocks for viral progeny formation.
Plankton interactions were also the subject of another recent study that combined metabolome and proteome analyses during a competition situation.113 The sublethal allelopathic effects of the red-tide dinoflagellate Karenia brevis on two diatom competitors were addressed.119 Therefore Asterionellopsis glacialis, a diatom species that co-occurs with the red tide dinoflagellate and Thalassiosira pseudonana which may not have evolved resistance were challenged with K. brevis in co-cultivation experiments. In both diatoms the presence of the dinoflagellate caused changes in the metabolic and proteomic profiles. Up- and down regulated proteins and metabolites were identified tentatively and related to putative cell functions. Combining the proteomics and metabolomics results a molecular network was derived from which it was concluded that the naturally co-occurring species A. glacialis shows a more robust metabolism with only slight changes in co-cultivation. In contrast, allelopathy disrupted energy metabolism (glycolysis, photosynthesis) and impaired cellular protection mechanisms including cell membrane maintenance, osmoregulation and oxidative stress response in T. pseudonana (Fig. 5).113 This study provided several novel hypotheses for the underlying mechanisms in sub lethal allelopathy that will surely be the subject of testing in the near future.
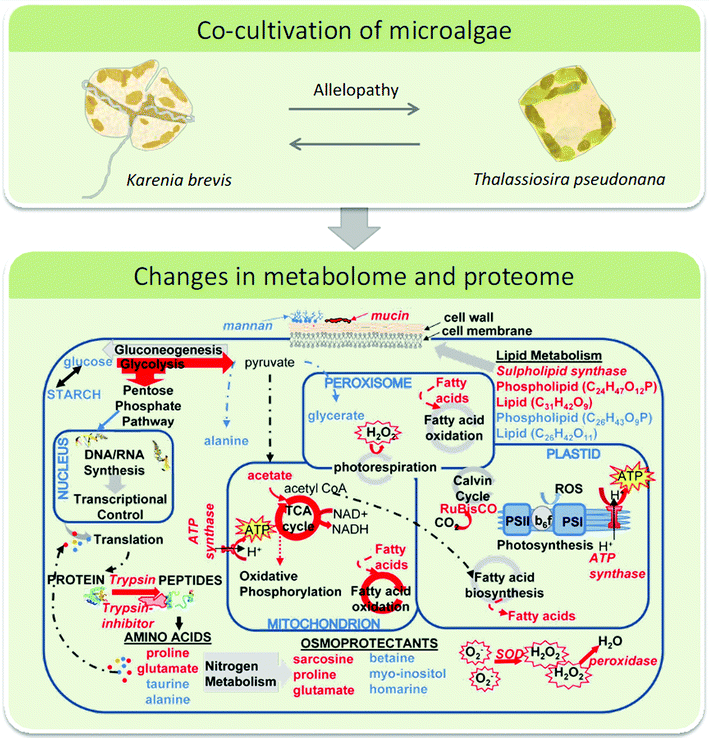 | ||
| Fig. 5 Top: interacting microalgae K. brevis and T. pseudonana were investigated using a metabolomics/proteomics approach. Bottom: the network of cellular pathways, enzymes, and metabolites in T. pseudonana under the influence of K. brevis are depicted. Pathways and metabolites enhanced by allelopathy are indicated by red arrows and compound names, respectively and those suppressed by blue labels. Below panel is reproduced with permission from the National Academy of Sciences.113 | ||
Successful combination of –omics approaches are also reported from the chemical ecology of higher plants.120,121 The majority of trees form beneficial relationships with ectomycorrhizal (ECM) fungi.122 After invasion an ECM-root tip is formed that can be considered as a type of symbiotic organ. The processes during invasive colonization of Populus root tips by the fungus Laccaria bicolor was the subject of a metabolomics/transcriptomics study by Tschaplinski et al.120 This study could benefit from the availability of a genomic and transcriptomic data repository of the fungus. In addition, both, a compatible host Populus trichocarpa and a recalcitrant host P. deltoides were accessible. The metabolic reprogramming that favors the formation of the ECM-root tip hybrid tissue in compatible interactions or that prevents the formation of such tissue in the recalcitrant host could be addressed by a combination of metabolomics and transcriptomics. In this case GC-MS proved to be the method of choice since data base supported evaluation of spectra allowed the tentative identification of regulated metabolites that could be brought into accordance with regulated pathways from transcriptomics.120 The symbiotic metabolome is altered throughout the colonization process with shifts in aromatic acids and fatty acid metabolism. The mutualistic ECM relationship might require such suppression or moderation of host defenses, allowing for the fungus to integrate into the root tissues. Explorative mapping of the observed metabolic changes in the benzoate degradation pathway using the KEGG database123,124 lead to a targeted transcriptome analysis. The results are in accordance with the fungus being the driver of metabolic reprogramming in the early stages of colonization. The challenge of ECM root tips of the recalcitrant P. deltoides with the fungus led to less pronounced metabolic shifts but plant defense related compounds such as tryptophan or salicin were elevated in the incompatible interaction. Targeted degradation assays further showed the capacity of the fungus to metabolize secreted defensive compounds. Thus diversity and half-life of plant defense compounds seem to be driving forces of the interaction.
An important tool for the deciphering of herbivory-induced gene-to-metabolite dynamics was introduced by Gulati et al.125 The novel transcript and metabolite data classification tool for experiments with multiple factors such as treatment, tissue, genetic context, or time-series was tested on Nicotiana attenuata using 3 tissues, 2 stressors and a 6-point-time-series for transcriptome and metabolome data. The value of the approach was demonstrated by revisiting well-characterized changes of regulatory genes and the oxylipin pathway. But the dimensionality reduction approach is also applicable to unravel rewiring of gene and metabolite networks in an explorative manner. Dynamic metabolite–metabolite correlation networks were created to cluster biochemically connected metabolites (e.g. shikimate pathway derived amino acids correlated with phenylpropanoid pathway metabolites). This study underlines the high plasticity of herbivore responses and provides a nice functional genomics tool to find novel genes and metabolites involved in herbivore response as shown for oxylipin signaling and the 17-hydroxygeranyllinalool diterpene glycoside (17-HGL-DTG) pathway. The root metabolome of N. attenuata under the influence of leaf herbivory was addressed using this approach.126 A multivariate time-series data analysis was used to evaluate leaf herbivory-elicited transcriptional and metabolic dynamics in the roots. Transient systemic responses in roots were detected. The semi-diurnal transcript oscillation was inverted and its major amplitude effects translated into root-specific secondary metabolite shifts.126 Using the multifactorial approach described in Gulati et al.125 tissue and treatment effects in a time-course data set could be analyzed that form the basis for further functional studies on the role of above ground herbivory on root metabolism.
The above examples might be considered as starting points of combined –omics studies in chemical ecology. These few publications that combined metabolomics with other –omics data sets to reveal basic principles of chemically mediated species interactions were all published within the last two years, thereby representing the start of a novel era in chemical ecology research. In other research areas relevant for the field, such as plant physiology, combined –omics approaches are also increasingly applied and several reviews already discussed the use of metabolomics, proteomics, transcriptomics and metabolic flux analyses as functional genomics tools.127–130
4 The comprehensive metabolome approach
This chapter will focus on the use of comprehensive metabolomics data of model species in chemical ecology and present an outline about the potential power of comprehensive data repositories. In the studies discussed so far the metabolome – the collection of all metabolites of an organism – has been covered only to some extent. Based on the hypothesis to be answered the focus of the study was on a certain subset of the metabolome. In such cases the exclusion of all other metabolites is crucial to increase the detection limit of the metabolites in focus. However, if the global metabolome response to species interactions is in focus of the study a comprehensive approach covering all present metabolites is needed.116 Such comprehensive metabolomes are challenging since no protocol exists so far that enables the extraction and analysis of the chemical diverse array of metabolites at once. However, for certain model organisms, comprehensive collections of metabolites with the aim to obtain complete metabolomes are currently assembled. The field of chemical ecology could benefit from such collections since they would define the metabolic potential and answer the question of what is possible in chemical signaling. Another issue, not only in metabolomics but also in all other –omics fields,121 is the identification and functional characterization of detected features, such as metabolites.120,132 Here chemical ecology can provide novel structures and hints towards their function off the beaten track of primary metabolism. There is clearly a need for public large spectral data bases that not only cover metabolites of key species to support such initiatives. This also calls for the deposition of own compounds with the according metadata to allow for knowledge-sharing between laboratories.It is estimated that about 5 million putative metabolites exist in the plant kingdom.133 Besides the structural diversity, also spatial and temporal variability as well as broad concentration ranges limit a comprehensive coverage of the metabolome. Therefore it has been recommended repeatedly to combine knowledge obtained with different extraction protocols and analytical tools such as GC(× GC)-MS, LC-MS, NMR or FT-IR.127,130 Standardized protocols addressing such combined approaches are however still under development.134–136 For certain model organisms quite comprehensive annotated data repositories exist as it is the case for Arabidopsis thaliana,137Escherichia coli,138 and Saccharomyces cerevisiae.139 Those repositories can serve as reference database not only for these species themselves but also for related organisms that might be of more interest in chemical ecology. http://PlantMetabolomics.org (PM) has been the first available online data repository covering over 3.100 compounds from A. thaliana wild type and 50 mutant lines that have been solely analyzed to accomplish this database.137 The aim is to annotate each metabolite with its experimental metadata and abundance following standardizing procedures (MSI,131 ArMet,140 MIAMET141). PM is further linked to several other databases including KEGG and PubChem to allow searching for genetic, chemical and biosynthetic pathway information. Additionally, protocols, tutorials as well as diverse statistical tools are provided. The Yeast Metabolome Database (YMDB) provides over 2000 metabolites linked to 995 genes or proteins, 66 biochemical pathways and 916 chemical reactions.139 1540 NMR- and 951 MS spectra are linked to 750 compounds. Intra- and extracellular concentrations are given for 627 metabolites. This data has been collected from books, articles, other databases and own measurements, and is linked to metadata, images, references and other databases. The E. coli metabolome database (ECMDB) is the most recent annotated data repository for E. coli (primarily strain K-12).138 It provides over 2600 metabolite entries with links to 1500 genes or proteins, 125 biochemical pathways and 2800 chemical reactions. It is also supplemented with 775 NMR spectra and 4035 MS spectra as well as intracellular concentration data for 800 compounds. But since S. cervesiae and E. coli serve mainly as model organisms in molecular biology and are rarely considered in chemical ecology these powerful data repositories address rather molecular biologists and system biologists.
Those data repositories are all species-specific, which is considered to be essential for the field of metabolomics as each organism has its own unique set of metabolites.138 Facing this dilemma it remains to be answered how to integrate the diverse variety of species that are interesting for chemical ecologists. The closest related model organism with its associated database could be used but much of the power of the repositories would be lost. A remarkable concept for a general purpose, cross-species, cross-platform data repository for metabolomics data that can overcome these limitations is realized in the MetaboLights platform.142,143 First results from studies in chemical ecology are already included71 and a broad usage would open up vast possibilities for the search of secondary metabolites in organismal interactions.
Nevertheless, also chemical ecology can focus on model organisms, and several experiments on the chemical ecology of A. thaliana, Caenorhabditis elegans or Drosophila melanogaster have been conducted that can benefit from and contribute to the resources of joint metabolomics initiatives. Methods for the generation of comprehensive metabolomes of e.g. D. melanogaster144 or C. elegans,135 or at least standardized metabolomics methods for e.g. Bacillus subtilis134 or Staphylococcus aureus145 are suggested and it can be observed that even organisms that generated interest in the field of (chemical) ecology were promoted as models as it is the case with the tobacco plant Nicotiana attenuata or the unicellular diatom Skeletonema costatum. To assist metabolomics in chemical ecology standard operation procedures for such species with clear definition of specific protocols are needed as organism-specific characteristics can influence the extraction efficiency.146 For example for C. elegans tissue disruption is a crucial step.135 Tissue selection has also to be considered for more complex organisms since whole organism homogenates can be insufficient as was shown for D. melanogaster.144 Sample collection, preparation, storage, analysis and data handling can thus introduce a variability that interferes with comparative efforts between laboratories. At the time being it is an unrealistic goal to reach a comprehensive coverage of the ‘metabolome’ for every organism interesting in chemical ecology. But intelligent use of existing repositories147 normalized procedures and ethics of reporting and contributing to data bases148 will help to make metabolomics an even more powerful tool. Of particular importance is the metabolomics standards initiative (MSI) that was conceived in 2005 with the goal to develop universal minimum standard requirements for metabolomics analyses. Primary minimal standards were published 2007 encompassing five different areas of metabolomics analyses.131 Recommendations addressing the reporting of biological samples from mammals,149 plants,150 microbes,151 and of environmental samples152 were given, as well as for chemical analyses,153 data processing,154 data exchange structures,155 and ontology.156 To benefit from such initiatives, metabolomics researchers, also in chemical ecology, should stick wherever possible to these recommendations to make their work comparable.131 Due to lacking open source file formats the COordination of Standards in MetabOlomicS (COSMOS) was initiated subsequently and might prove as a powerful tool to handle and compare data also in the field of chemical ecology.142,157
5 Conclusions
From the above discussed examples it can be concluded that metabolomics represents an emerging technique in chemical ecology that has the power to shape the discipline in several ways. For one, the identification of signaling molecules will be greatly facilitated by at least partially eliminating the need of bioassay-guided fractionation for the identification of active metabolites. In a systems biology context, metabolomics enables connecting chemically mediated interactions to regulative events on a protein or transcriptome level. With the emerging possibilities of data repositories and the categorization of metabolites from ever more model species, the discipline of metabolomics will become more mature and tasks such as de-replication, metabolite assignment and even structure elucidation that are tedious today will become routine. There is still a long way to go to establish metabolomics as standard technique in labs of biologists, but interdisciplinary approaches involving analytical chemists are already in place. In addition to the above outlined avenues, metabolomics also opens up new fields of research in chemical ecology. Thus, e.g. imaging techniques will allow to spot the point of interaction in microbial communities,98,109 on surfaces and in the immediate surroundings of plants and algae,158–160 as well as in insects.161 But also the metabolome of single cells might become of interest, especially when aspects of cooperative interactions or cheaters in microbial communities are concerned.162–164 However the still limited capabilities of the available imaging techniques lead to smaller data-sets compared to other metabolomics techniques and quantification is still problematic. Novel concepts in metabolomics have the potential to allow the design of fundamentally new experiments in chemical ecology that will allow monitoring the ecological interactions of organisms in hitherto unrecognized complexity. One potential direction to go would be the space- and time resolved monitoring and manipulation of complex communities with the aim to identify the “chemical role” of each partner in set-ups that are a lot closer to the true ecology of the species than traditional, often over-simplified co-culturing approaches.6 Acknowledgements
We thank the Volkswagen Foundation, the German Research Foundation (DFG) within the framework “Collaborative Research Centre CRC1127 Chemical Mediators in complex Biosystems” and the Jena School for Microbial Communication for funding.References
- http://www.chemecol.org .
- S. M. Cook, Z. R. Khan and J. A. Pickett, in Annual Review of Entomology, Annual Reviews, Palo Alto, 2007, vol. 52, pp. 375–400 Search PubMed.
- A. Butenandt, R. Beckmann and E. Hecker, Hoppe-Seyler's Z. Physiol. Chem., 1961, 324, 71–83 CrossRef CAS.
- E. K. Prince and G. Pohnert, Anal. Bioanal. Chem., 2010, 396, 193–197 CrossRef CAS PubMed.
- J. Stokl, A. T. Dandekar and J. Ruther, J. Chem. Ecol., 2014, 40, 159–168 CrossRef PubMed.
- T. Alborn, T. C. J. Turlings, T. H. Jones, G. Stenhagen, J. H. Loughrin and J. H. Tumlinson, Science, 1997, 276, 945–949 CrossRef.
- A. Eberhard, A. L. Burlingame, C. Eberhard, G. L. Kenyon, K. H. Nealson and N. J. Oppenheimer, Biochemistry, 1981, 20, 2444–2449 CrossRef CAS.
- D. B. Wilburn, K. E. Bowen, P. W. Feldhoff and R. C. Feldhoff, J. Chem. Ecol., 2014, 40, 928–939 CrossRef CAS PubMed.
- K. Gase and I. T. Baldwin, Plant Ecol. Divers., 2012, 5, 485–490 CrossRef.
- C. Tittiger, J. Chem. Ecol., 2004, 30, 2335–2358 CrossRef CAS PubMed.
- M. Dicke, J. Plant Physiol., 1994, 143, 465–472 CrossRef CAS.
- W. Boland, J. Hopke, J. Donath, J. Nuske and F. Bublitz, Angew. Chem., Int. Ed., 1995, 34, 1600–1602 CrossRef CAS PubMed.
- O. Fiehn, Plant Mol. Biol., 2002, 48, 155–171 CrossRef CAS.
- G. J. Patti, O. Yanes and G. Siuzdak, Nat. Rev. Mol. Cell Biol., 2012, 13, 263–269 CrossRef CAS PubMed.
- B. B. Aldridge and K. Y. Rhee, Curr. Opin. Microbiol., 2014, 19, 90–96 CrossRef CAS PubMed.
- E. M. Lenz and I. D. Wilson, J. Proteome Res., 2007, 6, 443–458 CrossRef CAS PubMed.
- C. Junot, F. Fenaille, B. Colsch and F. Becher, Mass Spectrom. Rev., 2014, 33, 471–500 CrossRef CAS PubMed.
- M. Ernst, D. B. Silva, R. R. Silva, R. Z. N. Vencio and N. P. Lopes, Nat. Prod. Rep., 2014, 31, 784–806 RSC.
- K. Bingol and R. Bruschweiler, Anal. Chem., 2014, 86, 47–57 CrossRef CAS PubMed.
- S. L. Robinette, R. Bruschweiler, F. C. Schroeder and A. S. Edison, Acc. Chem. Res., 2012, 45, 288–297 CrossRef CAS PubMed.
- R. R. Forseth and F. C. Schroeder, Curr. Opin. Chem. Biol., 2011, 15, 38–47 CrossRef CAS PubMed.
- K. H. Liland, Trends Anal. Chem., 2011, 30, 827–84123 CrossRef CAS PubMed.
- E. Saccenti, H. C. J. Hoefsloot, A. K. Smilde, J. A. Westerhuis and M. M. W. B. Hendriks, Metabolomics, 2013, 10, 361–374 CrossRef.
- N. Gehlenborg, S. I. O'Donoghue, N. S. Baliga, A. Goesmann, M. A. Hibbs, H. Kitano, O. Kohlbacher, H. Neuweger, R. Schneider, D. Tenenbaum and A. C. Gavin, Nat. Methods, 2010, 7, S56–S68 CrossRef CAS PubMed.
- J. Sardans, J. Penuelas and A. Rivas-Ubach, Chemoecology, 2011, 21, 191–225 CrossRef CAS PubMed.
- D. D. Cheng, H. Kirk, P. P. J. Mulder, K. Vrieling and P. G. L. Klinkhamer, New Phytol., 2011, 192, 1010–1023 CrossRef CAS PubMed.
- M. Deicke, J.-P. Bellenger and T. Wichard, J. Chromatogr. A, 2013, 1298, 50–60 CrossRef CAS PubMed.
- M. Deicke, J. F. Mohr, J.-P. Bellenger and T. Wichard, Analyst, 2014, 139, 6096–6099 RSC.
- O. Baars, F. M. M. Morel and D. H. Perlman, Anal. Chem., 2014, 86, 11298–11305 CrossRef CAS PubMed.
- E. Croisier, M. Rempt and G. Pohnert, Phytochemistry, 2010, 71, 574–580 CrossRef CAS PubMed.
- M. Rempt and G. Pohnert, Angew. Chem., Int. Ed., 2010, 49, 4755–4758 CrossRef CAS PubMed.
- N. S. Cortina, D. Krug, A. Plaza, O. Revermann and R. Müller, Angew. Chem., Int. Ed., 2012, 51, 811–816 CrossRef CAS PubMed.
- D. Krug and R. Müller, Nat. Prod. Rep., 2014, 31, 768–783 RSC.
- M. Kutyniok and C. Müller, J. Exp. Bot., 2012, 63, 6199–6210 CrossRef CAS PubMed.
- J. Hofmann, N. El Ashry Ael, S. Anwar, A. Erban, J. Kopka and F. Grundler, Plant J., 2010, 62, 1058–1071 CAS.
- M. Kutyniok and C. Müller, Oecologia, 2013, 173, 1367–1377 CrossRef PubMed.
- M. Kutyniok, M. Persicke and C. Müller, J. Chem. Ecol., 2014, 40, 118–127 CrossRef CAS PubMed.
- S. G. Kim, F. Yon, E. Gaquerel, J. Gulati and I. T. Baldwin, PLoS One, 2011, 6, e26214 CAS.
- M. Stitz, I. T. Baldwin and E. Gaquerel, PLoS One, 2011, 6, e25925 CAS.
- E. Gaquerel, H. Kotkar, N. Onkokesung, I. Galis and I. T. Baldwin, PLoS One, 2013, 8, e62336 CAS.
- N. Onkokesung, E. Gaquerel, H. Kotkar, H. Kaur, I. T. Baldwin and I. Galis, Plant Physiol., 2012, 158, 389–407 CrossRef CAS PubMed.
- G. Glauser, G. Marti, N. Villard, G. A. Doyen, J.-L. Wolfender, T. C. J. Turlings and M. Erb, Plant J., 2011, 68, 901–911 CrossRef CAS PubMed.
- G. Marti, M. Erb, J. Boccard, G. Glauser, G. R. Doyen, N. Villard, C. A. M. Robert, T. C. J. Turlings, S. Rudaz and J.-L. Wolfender, Plant, Cell Environ., 2013, 36, 621–639 CrossRef CAS PubMed.
- A. E. Rodriguez Estrada, A. Hegeman, H. Corby Kistler and G. May, Fungal Genet. Biol., 2011, 48, 874–885 CrossRef CAS PubMed.
- W. Jonkers, A. E. Estrada, K. Lee, A. Breakspear, G. May and H. C. Kistler, Appl. Environ. Microbiol., 2012, 78, 3656–3667 CrossRef CAS PubMed.
- K. Lee, J. J. Pan and G. May, FEMS Microbiol. Lett., 2009, 299, 31–37 CrossRef CAS PubMed.
- A. Steinbrenner, S. Gómez, S. Osorio, A. Fernie and C. Orians, J. Chem. Ecol., 2011, 37, 1294–1303 CrossRef CAS PubMed.
- S. Gomez, A. D. Steinbrenner, S. Osorio, M. Schueller, R. A. Ferrieri, A. R. Fernie and C. M. Orians, Entomol. Exp. Appl., 2012, 144, 101–111 CAS.
- A. Combès, I. Ndoye, C. Bance, J. Bruzaud, C. Djediat, J. Dupont, B. Nay and S. Prado, PLoS One, 2012, 7, e47313 Search PubMed.
- C. A. Smith, E. J. Want, G. O'Maille, R. Abagyan and G. Siuzdak, Anal. Chem., 2006, 78, 779–787 CrossRef CAS PubMed.
- M. Scognamiglio, V. Fiumano, B. D'Abrosca, A. Esposito, Y. H. Choi, R. Verpoorte and A. Fiorentino, Phytochemistry, 2014, 106, 69–85 CrossRef CAS PubMed.
- R. Schweiger, M. C. Baier, M. Persicke and C. Müller, Nat. Commun., 2014, 5, 3886 CAS.
- G. M. Nylund, F. Weinberger, M. Rempt and G. Y. N. Pohnert, PLoS One, 2011, 6, e29359 CAS.
- E. K. Prince, K. L. Poulson, T. L. Myers, R. D. Sieg and J. Kubanek, Harmful Algae, 2010, 10, 39–48 CrossRef CAS PubMed.
- C. Paul, A. Barofsky, C. Vidoudez and G. Pohnert, Mar. Ecol.: Prog. Ser., 2009, 389, 61–70 CrossRef.
- C. Paul, M. Mausz and G. Pohnert, Metabolomics, 2013, 9, 349–359 CrossRef CAS.
- J. Gillard, J. Frenkel, V. Devos, K. Sabbe, C. Paul, M. Rempt, D. Inzé, G. Pohnert, M. Vuylsteke and W. Vyverman, Angew. Chem., Int. Ed., 2013, 52, 854–857 CrossRef CAS PubMed.
- A. Ianora, M. Boersma, R. Casotti, A. Fontana, J. Harder, F. Hoffmann, H. Pavia, P. Potin, S. A. Poulet and G. Toth, Estuaries Coasts, 2006, 29, 531–551 CrossRef CAS.
- E. Van Donk, A. Ianora and M. Vos, Hydrobiologia, 2011, 668, 3–19 CrossRef CAS.
- S. Goulitquer, P. Potin and T. Tonon, Mar. Drugs, 2012, 10, 849–880 CrossRef CAS PubMed.
- K. A. Fredrickson and S. L. Strom, J. Plankton Res., 2009, 31, 135–152 CrossRef PubMed.
- G. Pohnert and W. Boland, Nat. Prod. Rep., 2002, 19, 108–122 RSC.
- J. R. Seymour, R. Simo, T. Ahmed and R. Stocker, Science, 2010, 329, 342–345 CrossRef CAS PubMed.
- B. Vanelslander, C. Paul, J. Grueneberg, E. K. Prince, J. Gillard, K. Sabbe, G. Pohnert and W. Vyverman, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 2412–2417 CrossRef CAS PubMed.
- C. Beemelmanns, A. Woznica, R. A. Aegado, A. M. Cantley, N. King and J. Clardy, J. Am. Chem. Soc., 2014, 136, 10210–10213 CrossRef CAS PubMed.
- C. Dreanno, K. Matsumura, N. Dohmae, K. Takio, H. Hirota, R. R. Kirby and A. S. Clare, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 14396–14401 CrossRef CAS PubMed.
- J. Gillard, J. Frenkel, V. Devos, K. Sabbe, C. Paul, M. Rempt, D. Inze, G. Pohnert, M. Vuylsteke and W. Vyverman, Angew. Chem., Int. Ed., 2013, 52, 854–857 CrossRef CAS PubMed.
- J. Frenkel, W. Vyverman and G. Pohnert, Plant J., 2014, 79, 632–644 CrossRef CAS PubMed.
- K. L. Poulson, R. D. Sieg and J. Kubanek, Nat. Prod. Rep., 2009, 26, 729–745 RSC.
- R. J. M. Weber, E. Selander, U. Sommer and M. R. Viant, Mar. Drugs, 2013, 11, 4158–4175 CrossRef CAS PubMed.
- S. H. von Reuss, N. Bose, J. Srinivasan, J. J. Yim, J. C. Judkins, P. W. Sternberg and F. C. Schroeder, J. Am. Chem. Soc., 2012, 134, 1817–1824 CrossRef CAS PubMed.
- A. Choe, T. Chuman, S. H. von Reuss, A. T. Dossey, J. J. Yim, R. Ajredini, A. A. Kolawa, F. Kaplan, H. T. Alborn, P. E. A. Teal, F. C. Schroeder, P. W. Sternberg and A. S. Edison, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 20949–20954 CrossRef CAS PubMed.
- F. C. Schroeder, Chem. Biol., 2015, 22, 7–16 CrossRef CAS PubMed.
- J. Srinivasan, F. Kaplan, R. Ajredini, C. Zachariah, H. T. Alborn, P. E. A. Teal, R. U. Malik, A. S. Edison, P. W. Sternberg and F. C. Schroeder, Nature, 2008, 454, 1115–1146 CrossRef CAS PubMed.
- J. Srinivasan, S. H. von Reuss, N. Bose, A. Zaslaver, P. Mahanti, M. C. Ho, O. G. O'Doherty, A. S. Edison, P. W. Sternberg and F. C. Schroeder, PLoS Biol., 2012, 10, e1001237 CAS.
- C. Pungaliya, J. Srinivasan, B. W. Fox, R. U. Malik, A. H. Ludewig, P. W. Sternberg and F. C. Schroeder, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 7708–7713 CrossRef CAS PubMed.
- Y. Izrayelit, J. Srinivasan, S. L. Campbell, Y. Jo, S. H. von Reuss, M. C. Genoff, P. W. Sternberg and F. C. Schroeder, ACS Chem. Biol., 2012, 7, 1321–1325 CrossRef CAS PubMed.
- N. Bose, J. M. Meyer, J. J. Yim, M. G. Mayer, G. V. Markov, A. Ogawa, F. C. Schroeder and R. J. Sommer, Curr. Biol., 2014, 24, 1536–1541 CrossRef CAS PubMed.
- A. Choe, T. Chuman, S. H. von Reuss, A. T. Dossey, J. J. Yim, R. Ajredini, A. A. Kolawa, F. Kaplan, H. T. Alborn, P. E. A. Teal, F. C. Schroeder, P. W. Sternberg and A. S. Edison, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 20949–20954 CrossRef CAS PubMed.
- P. Mahanti, N. Bose, A. Bethke, J. C. Judkins, J. Wollam, K. J. Dumas, A. M. Zimmerman, S. L. Campbell, P. J. Hu, A. Antebi and F. C. Schroeder, Cell Metab., 2014, 19, 73–83 CrossRef CAS PubMed.
- S. L. Robinette, R. Ajredini, H. Rasheed, A. Zeinomar, F. C. Schroeder, A. T. Dossey and A. S. Edison, Anal. Chem., 2011, 83, 1649–1657 CrossRef CAS PubMed.
- A.-M. Rottler, S. Schulz and M. Ayasse, J. Chem. Ecol., 2013, 39, 67–75 CrossRef CAS PubMed.
- A. Smith, W. Vanderpool, J. Millar, L. Hanks and A. Suarez, Chemoecology, 2014, 24, 29–34 CrossRef CAS PubMed.
- S. E. F. Evison, R. S. Ferreira, P. D'Ettorre, D. Fresneau and C. Poteaux, J. Chem. Ecol., 2012, 38, 1441–1449 CrossRef CAS PubMed.
- A. Van Oystaeyen, R. C. Oliveira, L. Holman, J. S. van Zweden, C. Romero, C. A. Oi, P. d'Ettorre, M. Khalesi, J. Billen, F. Wackers, J. G. Millar and T. Wenseleers, Science, 2014, 343, 287–290 CrossRef CAS PubMed.
- H. C. Kelstrup, K. Hartfelder, F. S. Nascimento and L. M. Riddiford, J. Exp. Biol., 2014, 217, 2399–2410 CrossRef PubMed.
- E. Amsalem, J. Kiefer, S. Schulz and A. Hefetz, J. Chem. Ecol., 2014, 40, 900–912 CrossRef CAS PubMed.
- F. Menzel, J. Orivel, M. Kaltenpoth and T. Schmitt, Chemoecology, 2014, 24, 105–119 CrossRef PubMed.
- W. Bonckaert, F. P. Drijfhout, P. d'Ettorre, J. Billen and T. Wenseleers, J. Chem. Ecol., 2012, 38, 42–51 CrossRef CAS PubMed.
- A. Sramkova, C. Schulz, R. Twele, W. Francke and M. Ayasse, Naturwissenschaften, 2008, 95, 515–522 CrossRef CAS PubMed.
- H. C. Kelstrup, K. Hartfelder, F. S. Nascimento and L. M. Riddiford, Front. Zool., 2014, 11, 78 CrossRef PubMed.
- A. A. Smith, B. Holldobler and J. Liebig, J. Chem. Ecol., 2008, 34, 1275–1282 CrossRef CAS PubMed.
- A. A. Smith, B. Holldober and J. Liebig, Curr. Biol., 2009, 19, 78–81 CrossRef CAS PubMed.
- A. A. Smith, J. G. Millar, L. M. Hanks and A. V. Suarez, J. Exp. Biol., 2013, 216, 3917–3924 CrossRef CAS PubMed.
- E. Bunk, A. Sramkova and M. Ayasse, Chemoecology, 2010, 20, 189–198 CrossRef CAS PubMed.
- A. Guthals, J. D. Watrous, P. C. Dorrestein and N. Bandeira, Mol. BioSyst., 2012, 8, 2535–2544 RSC.
- J. Y. Yang, L. M. Sanchez, C. M. Rath, X. Liu, P. D. Boudreau, N. Bruns, E. Glukhov, A. Wodtke, R. de Felicio, A. Fenner, W. R. Wong, R. G. Linington, L. Zhang, H. M. Debonsi, W. H. Gerwick and P. C. Dorrestein, J. Nat. Prod., 2013, 76, 1686–1699 CrossRef CAS PubMed.
- J. Watrous, P. Roach, T. Alexandrov, B. S. Heath, J. Y. Yang, R. D. Kersten, M. van der Voort, K. Pogliano, H. Gross, J. M. Raaijmakers, B. S. Moore, J. Laskin, N. Bandeira and P. C. Dorrestein, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, E1743–E1752 CrossRef CAS PubMed.
- D. D. Nguyen, C.-H. Wu, W. J. Moree, A. Lamsa, M. H. Medema, X. Zhao, R. G. Gavilan, M. Aparicio, L. Atencio, C. Jackson, J. Ballesteros, J. Sanchez, J. D. Watrous, V. V. Phelan, C. van de Wiel, R. D. Kersten, S. Mehnaz, R. De Mot, E. A. Shank, P. Charusanti, H. Nagarajan, B. M. Duggan, B. S. Moore, N. Bandeira, B. Ø. Palsson, K. Pogliano, M. Gutiérrez and P. C. Dorrestein, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, E2611–E2620 CrossRef CAS PubMed.
- K. A. Aliferis and S. Jabaji, PLoS One, 2012, 7, e42576 CAS.
- F. Jourdan, Metabolomics Coming of Age with Its Technological Diversity, in Advances in Botanical Res., ed. D. Rolin, 2013, vol. 67, pp. 557–591 Search PubMed.
- F. Jourdan, L. Cottret, L. Huc, D. Wildridge, R. Scheltema, A. Hillenweck, M. P. Barrett, D. Zalko, D. G. Watson and L. Debrauwer, Metabolomics, 2010, 6, 312–321 CrossRef CAS PubMed.
- M. Sugimoto, M. Kawakami, M. Robert, T. Soga and M. Tomita, Curr. Bioinf., 2012, 7, 96–108 CrossRef CAS PubMed.
- W.-T. Liu, A. Lamsa, W. R. Wong, P. D. Boudreau, R. Kersten, Y. Peng, W. J. Moree, B. M. Duggan, B. S. Moore, W. H. Gerwick, R. G. Linington, K. Pogliano and P. C. Dorrestein, J. Antibiot., 2014, 67, 99–104 CrossRef CAS PubMed.
- P. Shannon, A. Markiel, O. Ozier, N. S. Baliga, J. T. Wang, D. Ramage, N. Amin, B. Schwikowski and T. Ideker, Genome Res., 2003, 13, 2498–2504 CrossRef CAS PubMed.
- M. E. Smoot, K. Ono, J. Ruscheinski, P. L. Wang and T. Ideker, Bioinformatics, 2011, 27, 431–432 CrossRef CAS PubMed.
- J. R. Winnikoff, E. Glukhov, J. Watrous, P. C. Dorrestein and W. H. Gerwick, J. Antibiot., 2014, 67, 105–112 CrossRef CAS PubMed.
- R. Mendes, M. Kruijt, I. de Bruijn, E. Dekkers, M. van der Voort, J. H. M. Schneider, Y. M. Piceno, T. Z. DeSantis, G. L. Andersen, P. A. H. M. Bakker and J. M. Raaijmakers, Science, 2011, 332, 1097–1100 CrossRef CAS PubMed.
- W. J. Moree, J. Y. Yang, X. Zhao, W.-T. Liu, M. Aparicio, L. Atencio, J. Ballesteros, J. Sanchez, R. G. Gavilan, M. Gutierrez and P. C. Dorrestein, J. Chem. Ecol., 2013, 39, 1045–1054 CrossRef CAS PubMed.
- M. F. Traxler, J. D. Watrous, T. Alexandrov, P. C. Dorrestein and R. Kolter, mBio, 2013, 4, e00459-13 CrossRef PubMed.
- K.-C. Tan, S. V. S. Ipcho, R. D. Trengove, R. P. Oliver and P. S. Solomon, Mol. Plant Pathol., 2009, 10, 703–715 CrossRef CAS PubMed.
- D. Yang, X. Du, Z. Yang, Z. Liang, Z. Guo and Y. Liu, Eng. Life Sci., 2014, 14, 456–466 CrossRef CAS PubMed.
- K. L. Poulson-Ellestad, C. M. Jones, J. Roy, M. R. Viant, F. M. Fernandez, J. Kubanek and B. L. Nunn, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 9009–9014 CrossRef CAS PubMed.
- L. Valledor, M. Escandon, M. Meijon, E. Nukarinen, M. J. Canal and W. Weckwerth, Plant J., 2014, 79, 173–180 CrossRef CAS PubMed.
- P. Langfelder, B. Zhang and S. Horvath, Bioinformatics, 2008, 24, 719–720 CrossRef CAS PubMed.
- S. Rosenwasser, M. A. Mausz, D. Schatz, U. Sheyn, S. Malitsky, A. Aharoni, E. Weinstock, O. Tzfadia, S. Ben-Dor, E. Feldmesser, G. Pohnert and A. Vardi, Plant Cell, 2014, 26, 2689–2707 CrossRef CAS PubMed.
- M. A. Mausz and G. Pohnert, J. Plant Physiol., 2015, 172, 137–148 CrossRef CAS PubMed.
- A. Vardi, B. A. S. Van Mooy, H. F. Fredricks, K. J. Popendorf, J. E. Ossolinski, L. Haramaty and K. D. Bidle, Science, 2009, 326, 861–865 CrossRef CAS PubMed.
- E. K. Prince, T. L. Myers and J. Kubanek, Limnol. Oceanogr., 2008, 53, 531–541 CrossRef.
- T. J. Tschaplinski, J. M. Plett, N. L. Engle, A. Deveau, K. C. Cushman, M. Z. Martin, M. J. Doktycz, G. A. Tuskan, A. Brun, A. Kohler and F. Martin, Mol. Plant-Microbe Interact., 2014, 27, 546–556 CrossRef CAS PubMed.
- J. Gulati, I. T. Baldwin and E. Gaquerel, Plant J., 2014, 77, 880–892 CrossRef CAS PubMed.
- A. Zuccaro, U. Lahrmann and G. Langen, Curr. Opin. Plant Biol., 2014, 20, 135–145 CrossRef PubMed.
- http://www.genome.jp/kegg/ .
- P. E. Larsen, A. Sreedasyam, G. Trivedi, G. K. Podila, L. J. Cseke and F. R. Collart, BMC Syst. Biol., 2011, 5, 70 CrossRef PubMed.
- J. Gulati, S. G. Kim, I. T. Baldwin and E. Gaquerel, Plant Physiol., 2013, 162, 1042–1059 CrossRef CAS PubMed.
- J. Gulati, I. T. Baldwin and E. Gaquerel, Plant J., 2014, 77, 880–892 CrossRef CAS PubMed.
- W. Weckwerth, Annu. Rev. Plant Biol., 2003, 54, 669–689 CrossRef CAS PubMed.
- M. Macel, N. M. van Dam and J. J. B. Keurentjes, Mol. Ecol. Resour., 2010, 10, 583–593 CrossRef CAS PubMed.
- K. Saito and F. Matsuda, Annu. Rev. Plant Biol., 2010, 61, 463–489 CrossRef CAS PubMed.
- W. Weckwerth, Anal. Bioanal. Chem., 2011, 400, 1967–1978 CrossRef CAS PubMed.
- O. Fiehn, D. Robertson, J. Griffin, M. van der Werf, B. Nikolau, N. Morrison, L. W. Sumner, R. Goodacre, N. W. Hardy, C. Taylor, J. Fostel, B. Kristal, R. Kaddurah-Daouk, P. Mendes, B. van Ommen, J. C. Lindon and S. A. Sansone, Metabolomics, 2007, 3, 175–178 CrossRef CAS.
- R. Verpoorte, Y. H. Choi and H. K. Kim, Phytochem. Anal., 2010, 21, 2–3 CrossRef CAS PubMed.
- W. Weckwerth, Planta Med., 2011, 77, 1232–1232 Search PubMed.
- H. Meyer, H. Weidmann and M. Lalk, Microb. Cell Fact., 2013, 12, 13 CrossRef PubMed.
- F. M. Geier, E. J. Want, A. M. Leroi and J. G. Bundy, Anal. Chem., 2011, 83, 3730–3736 CrossRef CAS PubMed.
- A. D. Hegeman, Briefings Funct. Genomics, 2010, 9, 139–148 CrossRef CAS PubMed.
- P. Bais, S. M. Moon, K. He, R. Leitao, K. Dreher, T. Walk, Y. Sucaet, L. Barkan, G. Wohlgemuth, M. R. Roth, E. S. Wurtele, P. Dixon, O. Fiehn, B. M. Lange, V. Shulaev, L. W. Sumner, R. Welti, B. J. Nikolau, S. Y. Rhee and J. A. Dickerson, Plant Physiol., 2010, 152, 1807–1816 CrossRef CAS PubMed.
- A. C. Guo, T. Jewison, M. Wilson, Y. F. Liu, C. Knox, Y. Djoumbou, P. Lo, R. Mandal, R. Krishnamurthy and D. S. Wishart, Nucleic Acids Res., 2013, 41, D625–D630 CrossRef CAS PubMed.
- T. Jewison, C. Knox, V. Neveu, Y. Djoumbou, A. C. Guo, J. Lee, P. Liu, R. Mandal, R. Krishnamurthy, I. Sinelnikov, M. Wilson and D. S. Wishart, Nucleic Acids Res., 2012, 40, D815–D820 CrossRef CAS PubMed.
- H. Jenkins, N. Hardy, M. Beckmann, J. Draper, A. R. Smith, J. Taylor, O. Fiehn, R. Goodacre, R. J. Bino, R. Hall, J. Kopka, G. A. Lane, B. M. Lange, J. R. Liu, P. Mendes, B. J. Nikolau, S. G. Oliver, N. W. Paton, S. Rhee, U. Roessner-Tunali, K. Saito, J. Smedsgaard, L. W. Sumner, T. Wang, S. Walsh, E. S. Wurtele and D. B. Kell, Nat. Biotechnol., 2004, 22, 1601–1606 CrossRef CAS PubMed.
- R. J. Bino, R. D. Hall, O. Fiehn, J. Kopka, K. Saito, J. Draper, B. J. Nikolau, P. Mendes, U. Roessner-Tunali, M. H. Beale, R. N. Trethewey, B. M. Lange, E. S. Wurtele and L. W. Sumner, Trends Plant Sci., 2004, 9, 418–425 CrossRef CAS PubMed.
- C. Steinbeck, P. Conesa, K. Haug, T. Mahendraker, M. Williams, E. Maguire, P. Rocca-Serra, S. A. Sansone, R. M. Salek and J. L. Griffin, Metabolomics, 2012, 8, 757–760 CrossRef CAS PubMed.
- K. Haug, R. M. Salek, P. Conesa, J. Hastings, P. de Matos, M. Rijnbeek, T. Mahendraker, M. Williams, S. Neumann, P. Rocca-Serra, E. Maguire, A. Gonzalez-Beltran, S. A. Sansone, J. L. Griffin and C. Steinbeck, Nucleic Acids Res., 2013, 41, D781–D786 CrossRef CAS PubMed.
- V. R. Chintapalli, M. Al Bratty, D. Korzekwa, D. G. Watson and J. A. T. Dow, PLoS One, 2013, 8, e78066 CAS.
- H. Meyer, M. Liebeke and M. Lalk, Anal. Biochem., 2010, 401, 250–259 CrossRef CAS PubMed.
- C. Vidoudez and G. Pohnert, Metabolomics, 2012, 8, 654–669 CrossRef CAS.
- R. M. Salek, K. Haug, P. Conesa, J. Hastings, M. Williams, T. Mahendraker, E. Maguire, A. N. Gonzalez-Beltran, P. Rocca-Serra, S. A. Sansone and C. Steinbeck, Database, 2013 Search PubMed , p.bat029.
- J. Griss, A. R. Jones, T. Sachsenberg, M. Walzer, L. Gatto, J. Hartler, G. G. Thallinger, R. M. Salek, C. Steinbeck, N. Neuhauser, J. Cox, S. Neumann, J. Fan, F. Reisinger, Q. W. Xu, N. del Toro, Y. Perez-Riverol, F. Ghali, N. Bandeira, I. Xenarios, O. Kohlbacher, J. A. Vizcaino and H. Hermjakob, Mol. Cell. Proteomics, 2014, 13, 2765–2775 CAS.
- J. L. Griffin, A. W. Nicholls, C. A. Daykin, S. Heald, H. C. Keun, I. Schuppe-Koistinen, J. R. Griffiths, L. L. Cheng, P. Rocca-Serra, D. V. Rubtsov and D. Robertson, Metabolomics, 2007, 3, 179–188 CrossRef CAS.
- O. Fiehn, L. W. Sumner, S. Y. Rhee, J. Ward, J. Dickerson, B. M. Lange, G. Lane, U. Roessner, R. Last and B. Nikolau, Metabolomics, 2007, 3, 195–201 CrossRef CAS.
- M. J. van der Werf, R. Takors, J. Smedsgaard, J. Nielsen, T. Ferenci, J. C. Portais, C. Wittmann, M. Hooks, A. Tomassini, M. Oldiges, J. Fostel and U. Sauer, Metabolomics, 2007, 3, 189–194 CrossRef CAS PubMed.
- N. Morrison, D. Bearden, J. G. Bundy, T. Collette, F. Currie, M. P. Davey, N. S. Haigh, D. Hancock, O. A. H. Jones, S. Rochfort, S. A. Sansone, D. Stys, Q. Teng, D. Field and M. R. Viant, Metabolomics, 2007, 3, 203–210 CrossRef CAS.
- L. W. Sumner, A. Amberg, D. Barrett, M. H. Beale, R. Beger, C. A. Daykin, T. W. M. Fan, O. Fiehn, R. Goodacre, J. L. Griffin, T. Hankemeier, N. Hardy, J. Harnly, R. Higashi, J. Kopka, A. N. Lane, J. C. Lindon, P. Marriott, A. W. Nicholls, M. D. Reily, J. J. Thaden and M. R. Viant, Metabolomics, 2007, 3, 211–221 CrossRef CAS PubMed.
- R. Goodacre, D. Broadhurst, A. K. Smilde, B. S. Kristal, J. D. Baker, R. Beger, C. Bessant, S. Connor, G. Calmani, A. Craig, T. Ebbels, D. B. Kell, C. Manetti, J. Newton, G. Paternostro, R. Somorjai, M. Sjostrom, J. Trygg and F. Wulfert, Metabolomics, 2007, 3, 231–241 CrossRef CAS.
- N. W. Hardy and C. F. Taylor, Metabolomics, 2007, 3, 243–248 CrossRef CAS.
- S. A. Sansone, D. Schober, H. J. Atherton, O. Fiehn, H. Jenkins, P. Rocca-Serra, D. V. Rubtsov, I. Spasic, L. Soldatova, C. Taylor, A. Tseng and M. R. Viant, Metabolomics, 2007, 3, 249–256 CrossRef CAS.
- R. M. Salek, K. Haug and C. Steinbeck, Gigascience, 2013, 2, 8 CrossRef PubMed.
- A. L. Lane, L. Nyadong, A. S. Galhena, T. L. Shearer, E. P. Stout, R. M. Parry, M. Kwasnik, M. D. Wang, M. E. Hay, F. M. Fernandez and J. Kubanek, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 7314–7319 CrossRef CAS PubMed.
- K. Grosser, L. Zedler, M. Schmitt, B. Dietzek, J. Popp and G. Pohnert, Biofouling, 2012, 28, 687–696 CrossRef CAS PubMed.
- D. Holscher, S. Dhakshinamoorthy, T. Alexandrov, M. Becker, T. Bretschneider, A. Buerkert, A. C. Crecelius, D. De Waele, A. Elsen, D. G. Heckel, H. Heklau, C. Hertweck, M. Kai, K. Knop, C. Krafft, R. K. Maddula, C. Matthaus, J. Popp, B. Schneider, U. S. Schubert, R. A. Sikora, A. Svatos and R. L. Swennen, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 105–110 CrossRef PubMed.
- F. Kaftan, V. Vrkoslav, P. Kynast, P. Kulkarni, S. Bocker, J. Cvacka, M. Knaden and A. Svatos, J. Mass Spectrom., 2014, 49, 223–232 CrossRef CAS PubMed.
- A. Svatos, Anal. Chem., 2011, 83, 5037–5044 CrossRef CAS PubMed.
- C.-J. Shih, P.-Y. Chen, C.-C. Liaw, Y.-M. Lai and Y.-L. Yang, Nat. Prod. Rep., 2014, 31, 739–755 RSC.
- B. B. Misra, S. M. Assmann and S. Chen, Trends Plant Sci., 2014, 19, 637–646 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2015 |