Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Involvement of secondary metabolites in the pathogenesis of the American foulbrood of honey bees caused by Paenibacillus larvae
Sebastian
Müller
a,
Eva
Garcia-Gonzalez
b,
Elke
Genersch
*b and
Roderich D.
Süssmuth
*a
aInstitut für Chemie, Technische Universität Berlin, 10623, Berlin, Germany. E-mail: suessmuth@chem.tu-berlin.de
bInstitute for Bee Research, Department of Molecular Microbiology and Bee Diseases, 16540, Hohen Neuendorf, Germany. E-mail: elke.genersch@hu-berlin.de
First published on 23rd April 2015
Abstract
Covering: 2011 to end of 2014
The Gram-positive, spore-forming bacterium Paenibacillus larvae (P. larvae) is the causative agent of the epizootic American Foulbrood (AFB), a fatal brood disease of the western honey bee (Apis mellifera). AFB is one of the most destructive honey bee diseases since it is not only lethal for infected larvae but also for the diseased colonies. Due to the high impact of honey bees on ecology and economy this epizootic is a severe and pressing problem. Knowledge about virulence mechanisms and the underlying molecular mechanisms remain largely elusive. Recent genome sequencing of P. larvae revealed its potential to produce unknown secondary metabolites, like nonribosomal peptides and peptide–polyketide hybrids. This article highlights recent findings on secondary metabolites synthesized by P. larvae and discusses their role in virulence and pathogenicity towards the bee larvae.
Introduction
Western honey bees (Apis mellifera) are the most widely used managed pollinators for insect pollination-dependent agricultural crop and fruit cultures worldwide. Therefore, honeybees are seen as among the most important productive livestock.1 Agricultural pollination relies strongly (about 90%) on managed honey bees and the demand for honey bee pollination is still increasing.2,3 A significant reduction of the honey bee population will lead to serious consequences for most ecosystems under severe concomitant economic losses, not only in apiculture, but also in the entire agriculture worldwide. Honey bees are threatened by numerous pathogens, including viruses (e.g. deformed wing virus, DWV, transmitted by the mite Varroa destructor), fungi (e.g. Ascosphaera apis or Nosema spp.) and bacteria like Melissococcus plutonius (causing European Foulbrood) and Paenibacillus larvae (causing American Foulbrood).4 These pathogens pose various threats of different severity and some of them are even able to destroy entire colonies.Paenibacillus larvae and the American foulbrood
A deadly bee pathogen is the Gram-positive, facultative anaerobic, spore-forming bacterium Paenibacillus larvae (P. larvae), the etiological agent of the American Foulbrood (AFB).5 AFB is occurring worldwide and leads to massive losses of entire bee colonies every year. AFB is highly contagious and spreads very rapidly. In many countries, AFB is a notifiable disease and infected bee hives have to be burned to contain the disease. An effective treatment of AFB does still not exist, since the underlying molecular characteristics of the P. larvae infection remain largely elusive. Therefore, it is difficult to develop efficient approaches to fight this severe epizootic.Based on repetitive element PCR (repPCR) using enterobacterial repetitive intergenic consensus sequence (ERIC) primers,6 the species P. larvae has been classified into four different genotypes (ERIC I–IV) which could recently be confirmed by multi locus sequence typing (MLST) of hundreds of P. larvae isolates from all over the world.5,7 These genotypes also differ phenotypically with respect to spore and colony morphology, metabolism, and most importantly virulence.5,8,9 Therefore, the classification of the species into ERIC genotypes is of biological relevance. The genotypes ERIC I and ERIC II are regularly isolated from infected colonies worldwide, whereas ERIC III and ERIC IV are only represented by few historical isolates in type culture collections.10 Hence, only P. larvae ERIC I and ERIC II are of practical importance and will be covered in this review. Comparative exposure bioassays have demonstrated that the genotype ERIC II is more virulent on the larval level than ERIC I. It commonly kills bee larvae within 6–7 days, while ERIC I strains need up to 12 days to kill all infected larvae.11 These differences on the individual larval level translate to differences in virulence on the colony level because the earlier larvae die the more efficiently they can be removed by nurse bees engaged in brood hygiene as part of the social immune response of honey bees.12 The social immune response is better adapted to contain ERIC II infections than ERIC I infections because most ERIC II-infected larvae die early and, hence, these are more efficiently removed from the colony than the slower dying ERIC I-infected larvae. This leads to the paradoxical situation that P. larvae ERIC II is less virulent on the colony level than ERIC I.8
From the bacterium P. larvae only the spores are infectious and only young larvae (up to the age of 36 hours after egg hatching) are susceptible to infection.4,13 Hence adult honey bees do not become infected by the uptake of P. larvae endospores.14,15 Although only bee larvae are killed by P. larvae, the resulting lack of offspring ultimately leads to the collapse of the entire colony. In consequence weakened and collapsing colonies are likely robbed by strong colonies. Because the honey of P. larvae infected colonies is contaminated by P. larvae spores, bees robbing the food stores of such colonies efficiently transmit the disease to their own colonies.
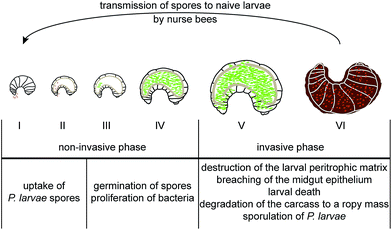
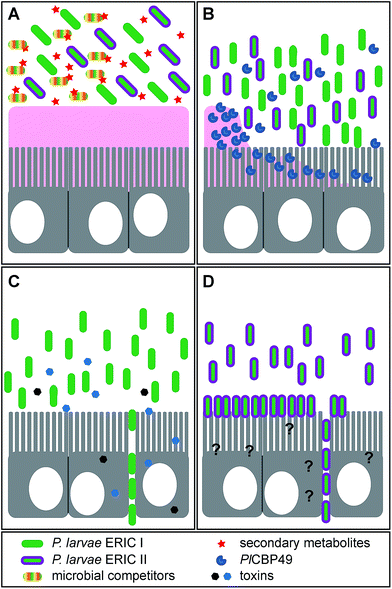
The infection is initiated as soon as P. larvae spores are taken up by larvae via spore contaminated larval food and germinate in the larval midgut (Fig. 1). The vegetative bacteria then massively proliferate in the midgut lumen. During this non-invasive phase of infection, no damage to the epithelial cell layer can be observed16 although the protective peritrophic matrix (PM) might already be affected (Fig. 2A and B).17P. larvae is able to metabolize sugars18 and crystalline chitin,17 hence, the larval food and chitin containing larval structures like the PM can serve as nutrition during this phase of infection (Fig. 2B).17 The invasive phase of the infection starts when the PM is degraded17,19 and the epithelial layer is attacked and penetrated by P. larvae (Fig. 2C and D).16 The bacteria invade the larval haemocoel (Fig. 1); a step that coincides with larval death.16 These characteristics of infection are shared by all P. larvae genotypes and the degradation of the larval PM has been shown to be a key step in AFB pathogenesis.17,19 The chitin-degrading protein PlCBP49 is responsible for PM degradation and is a unifying feature of pathogenesis reported to be expressed by both genotypes, ERIC I and ERIC II (Fig. 2B).19 In addition, genotype-specific virulence factors have been identified and it was proposed that P. larvae ERIC I and ERIC II follow different strategies when finally attacking and killing the infected larva (Fig. 2C and D).9,20 The ERIC I-genome harbors several functional toxin genes which are disrupted and, hence, presumably non-functional in the ERIC II-genome.9 Two of the ERIC I-specific toxins (Plx1, Plx2) have been characterized in more detail (Fig. 2C).21 Plx1 and Plx2 are novel AB-toxins [for reviews on this class of toxins see Falnes and Sandvig22 and Simon et al.23] and their expression could be linked to virulence.21 Plx1 is a single-chain AB-toxin belonging to an enigmatic AB-toxin family21 which also comprises MTX1 expressed by the entomopathogen Lysinibacillus sphaericus and the pierisin-like toxins expressed by some butterflies of the family Pieridae (Lepidoptera).24 Plx2 is a binary AB-toxin and its A-moiety resembles C3-like exoenzymes.21 The cellular targets of these novel AB-toxins still await identification.21 The ERIC II-genome harbors a functional S-layer protein gene (splA) which is mutated and, hence, non-functional in the ERIC I-genome.25 Accordingly, expression of S-layer proteins could only be demonstrated in P. larvae ERIC II.26 The P. larvae protein SplA was shown to mediate adhesion of P. larvae to primary bee midgut cells and expression of SplA could be linked to virulence of P. larvae ERIC II (Fig. 2D).25 The steps following P. larvae ERIC II adhesion to the midgut epithelium and those enabling the bacteria to penetrate the epithelial cell layer still remain elusive (Fig. 2D).25,26 These functionally characterized, genotype-specific virulence factors and genomic differences between P. larvae ERIC I and ERIC II clearly point to different virulence strategies pursued by ERIC I and ERIC II.9,20 However, irrespectively of the strategy, all P. larvae infections result in larval death and in the larval cadaver being converted in the brood cell to a ropy mass while non-infected, healthy larvae undergo metamorphosis and finally emerge as adult bees from the brood cell. Interestingly, this ropy mass is usually a pure culture of P. larvae,27,28 suggesting that P. larvae has some mechanism to outcompete other bacteria and especially saprophytes that have been reported to be present in the larval gut.29 Therefore the recent identification of secondary metabolite gene clusters in the genome of P. larvae may contribute to facilitate interactions with potential competitors (Fig. 2A).
The species Paenibacillus larvae has originally been classified as Bacillus larvae.28 However, in 1991 the genus Bacillus was analyzed by comparative small-subunit rRNA (16S rRNA) sequence analysis and it was demonstrated that this genus comprised five phyletic lines, rRNA group 1–5 bacilli.30 A subsequent extensive taxonomic revision revealed that the rRNA group 3 bacilli including Bacillus larvae were considerably distinct from the other Bacilli and, hence, they were reclassified as the new genus Paenibacillus.31 The genus Paenibacillus comprises several bacterial species formerly known as Bacilli like P. alvei and P. polymyxa. As the prefix Paeni reflects, Paenibacilli are ‘almost’ Bacilli and they share several features and properties with Bacilli. Bacilli are known to synthesize various secondary metabolites, such as lanthipeptides, lipopeptides, polyketides and other small molecules.32 Representatives of such compounds known for decades are the cyclic peptide antibiotics gramicidin S and tyrocidine, the lipopeptides surfactin and mycosubtilin and the siderophore bacillibactin.33–36 Like the genus Bacillus, Paenibacillus also seems to be an interesting source of bioactive natural products, although to date only few examples exist: P. polymyxa is known to produce polymyxin and fursaricidin37,38 and Paenibacillus elgii B69 produces the catechol-type siderophore paenibactin, which is closely related to the Bacillus siderophore bacillibactin.39 Furthermore, Paenibacillus sp. F6-B70 was described to produce the macrolide antibiotic paenimacrolidin, which is active against methicillin-resistant Staphylococcus aureus.40 A cyclic lipopeptide, called battacin (octapeptin B5), was isolated from a Paenibacillus tianmuensis soil isolate and showed antibacterial activity against multidrug-resistant Gram-negative bacteria.41
Secondary metabolites from P. larvae
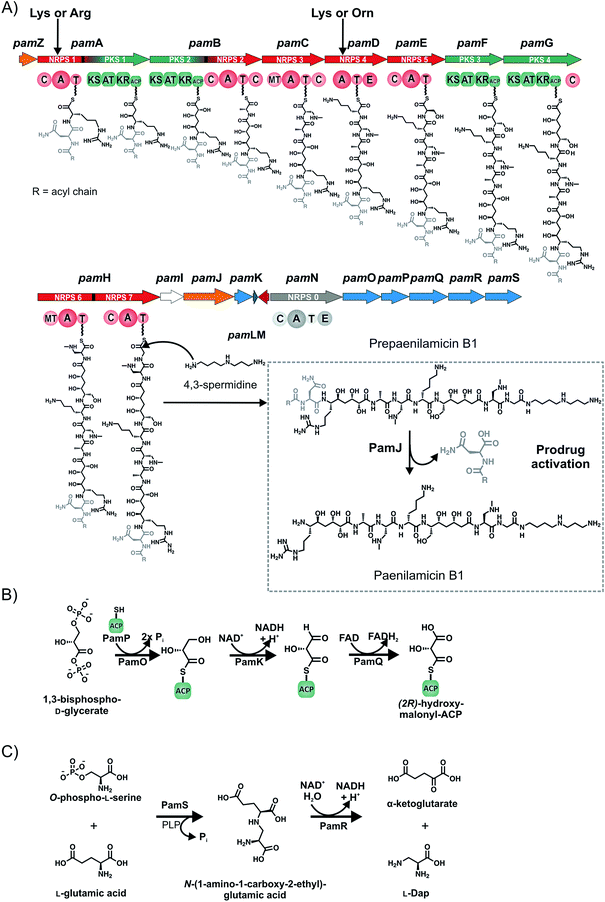
It was not until the entire genome sequences of P. larvae ERIC I and ERIC II were available9 that the complex giant gene clusters coding for secondary metabolites in the genomes of P. larvae ERIC I and ERIC II could be identified and correctly assembled (Fig. 3).9 Sequencing and annotation of the genomes of P. larvae strain DSM25430 (genotype ERIC II; 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 056
056![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 006 bp) and P. larvae strain DSM25719 (genotype ERIC I; 4
006 bp) and P. larvae strain DSM25719 (genotype ERIC I; 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 579
579![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 589 bp) facilitated the bioinformatics analysis of the genes, particularly of those being involved in pathogenesis and secondary metabolite production (Fig. 3 and 4).9 One NRPS was suggested to have similarities to the biosynthetic machinery of the siderophore bacillibactin and one NRPS/PKS hybrid gene cluster was suggested to encode the biosynthesis of a lipopeptide from the iturin family. An entirely cryptic cluster was a trimodular NRPS gene cluster which showed no apparent similarity to known gene clusters of other bacteria. A complex NRPS/PKS hybrid gene cluster showed similarities to the zwittermicin and xenocoumacin gene clusters as well as to other gene clusters dedicated to be responsible for the biosynthesis of NRP prodrugs. We set out for a genetic and biochemical characterization of these gene clusters followed by analytical characterization of their biosynthetic products as basis for elucidating their biological role in AFB pathogenesis.
589 bp) facilitated the bioinformatics analysis of the genes, particularly of those being involved in pathogenesis and secondary metabolite production (Fig. 3 and 4).9 One NRPS was suggested to have similarities to the biosynthetic machinery of the siderophore bacillibactin and one NRPS/PKS hybrid gene cluster was suggested to encode the biosynthesis of a lipopeptide from the iturin family. An entirely cryptic cluster was a trimodular NRPS gene cluster which showed no apparent similarity to known gene clusters of other bacteria. A complex NRPS/PKS hybrid gene cluster showed similarities to the zwittermicin and xenocoumacin gene clusters as well as to other gene clusters dedicated to be responsible for the biosynthesis of NRP prodrugs. We set out for a genetic and biochemical characterization of these gene clusters followed by analytical characterization of their biosynthetic products as basis for elucidating their biological role in AFB pathogenesis.
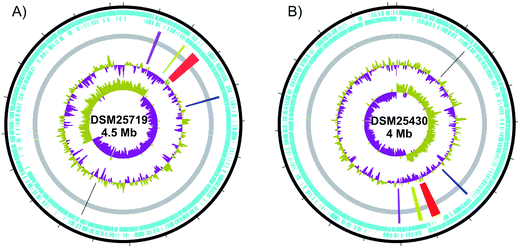 | ||
| Fig. 3 Graphical circular maps of the chromosome of Paenibacillus larvae. (A) ERIC I DSM25719 and (B) ERIC II DSM25430. Open reading frames on the forward and reverse strand are shown in blue. The inner circles show first GC% content and GC skew [(G – C)/(G + C)] purple and olive indicating higher and lower values than average, respectively. NRPS and PKS gene clusters are highlighted from the CDS grey bar. In ERIC II (B) Black: monomodular NRPS, blue: sevadicin gene cluster, red: paenilamicin gene cluster, yellow: paenilarvin gene cluster and purple: bacillibactin gene cluster. In ERIC I (A) the correspondent gene clusters are shown in the same colors. Circular maps were drawn using dnaplotter.81 | ||
Bacillibactin – occurrence of siderophores in P. larvae
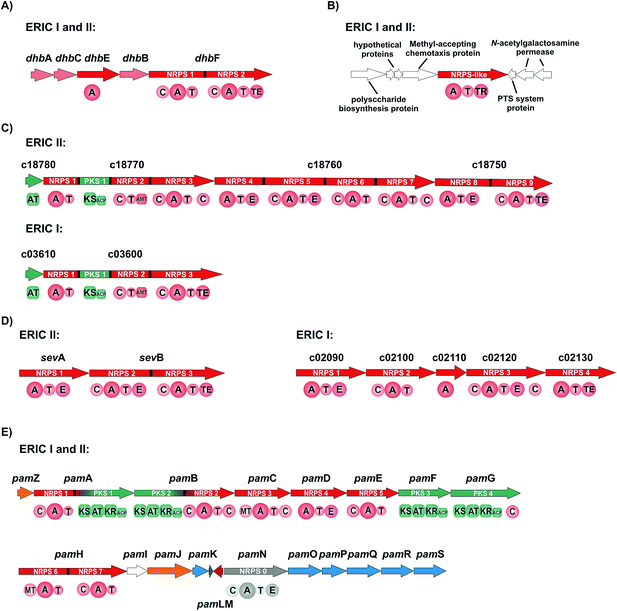
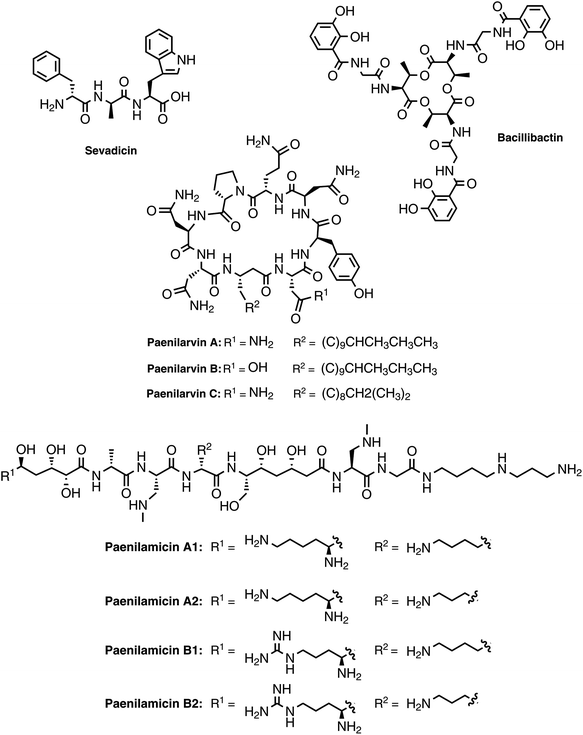
The gene cluster of P. larvae coding for the siderophore biosynthesis shows strong homology to the bacillibactin biosynthesis gene clusters of the Bacillus cereus sensu lato group and of Bacillus subtilis as well as to the paenibactin biosynthesis gene cluster of Paenibacillus elgii 69.42 Likewise to the bacillibactin gene cluster the P. larvae gene cluster was named dhb cluster (Fig. 4A).42 The dhb cluster is about 11 kbp in size and consists of five genes (dhbA, dhbC, dhbE, dhbB, and dhbF), which were annotated to encode for a 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase (DhbA), an isochorismate synthase (DhbC), a DHB-AMP-ligase (DhbE), an isochorismatase (DhbB), and a dimodular peptide synthetase (DhbF), respectively. On the protein level DhbACEBF of P. larvae and DhbACEBF of B. cereus show a high similarity between 70–79%. Proteins DhbA, B and C are predicted to synthesize 2,3-dihydroxybenzoate (DHB) from chorismate. The 60 kDa DHB-AMP-ligase DhbE with a similarity of 73% to DhbE of B. cereus is a stand-alone A domain, predicted to activate DHB. DhbF, with a similarity of 71% to DhbF of B. cereus, is a 267 kDa dimodular NRPS. The specificity-conferring amino acid sequence (DILQVGLIWK) of the A-domain is identical to DhbF1 of B. subtilis which activates Gly.35 The second A domain (DFWNIGMVHK) is identical to DhbF2 of B. subtilis and activates Thr.35 Both NRPSs DhbF1 and DhbF2 show a typical arrangement with a condensation (C) domain, an adenylation (A) domain and a thiolation (T) domain. The biosynthesis works in an iterative manner of three cycles and in the final stage, the C-terminal thioesterase (TE) domain of DhbF2 performs cleavage of the final peptide from the NRPS machinery under cyclisation to the macrolacton.35 Initially bioinformatic studies suggested the dhb cluster of P. larvae to encode for the NRPS machinery to synthesize paenibactin, a close bacillibactin homolog with Ala instead of Gly.39 For P. larvae strain DSM25430 (ERIC II) and strain DSM25719 (ERIC I) a combination of gene inactivation and mass spectrometric fragmentation analyses performed by our groups revealed that the NRPS machinery synthesizes the catechol-type siderophore bacillibactin and not paenibactin as expected before.42 Bacillibactin is the cyclic trimeric lactone of 2,3-dihydroxybenzoyl-Gly-L-Thr,35 which was assigned based on bioinformatics analysis and comparison with a reference compound from B. subtilis (Fig. 4). In general the dedicated function of siderophores is to secure iron supply under low iron conditions.43,44 For most pathogenic bacteria iron is a limiting factor and both, pathogen and host are competing for iron. Therefore, many pathogens, but also non-pathogenic bacteria synthesize and secrete siderophores as Fe3+ scavengers in iron limiting conditions. Comparison of wild-type strains and the appropriate gene inactivation strains (ΔdhbF) of both genotypes in larval infection assays however revealed no significant differences neither in total larval mortality nor in disease progression (cumulative larval mortality).42 Therefore, bacillibactin cannot be regarded as a virulence factor of P. larvae. In literature both is described, siderophores that act as virulence factors and siderophores not involved in pathogenicity. Recently, it could be shown that a B. cereus strain, unable to produce bacillibactin, showed an attenuated bacterial pathogenesis in insects.45 However, for a B. anthracis strain, insufficient in bacillibactin production, no phenotypical differences could be detected compared to the wild-type strain, suggesting that bacillibactin has no role in virulence.46Paenilarvins – iturin-like lipopeptide biosynthesis
As mentioned above, genotype ERIC II encodes a ∼37 kbp hybrid PKS/NRPS gene cluster responsible for the biosynthesis of iturin-like lipopeptides, the paenilarvins (Fig. 4B and 5).47 The iturins are structurally characterized by a macrocyclic heptapeptide core with an N-terminal β-amino fatty acid of various chain lengths.48 The β-amino group forms the macrolactam with the C-terminus of the heptapeptide. ERIC II contains one gene cluster with four ORFs of the same transcriptional directionality (Fig. 4B). The gene cluster arrangement is strongly reminiscent of the gene clusters responsible for the biosynthesis of the Bacillus lipopeptides iturin A and mycosubtilin.34,49 The first gene (c18780) encodes a trans-acting acyltransferase, which shows 63% sequence identity with ItuD from the iturin A biosynthesis. The initial PKS/NRPS encoded by c18770 shows 71% identity to the PKS/NRPS MycA of the mycosubtilin biosynthesis. Gene c18770 encodes the biosynthesis machinery for acylation and PKS-mediated extension of the acyl chain under involvement of the trans-acting acyltransferase and the reductive amination of the lipid chain by the class-III aminotransferase domain. Furthermore, the NRPS module for condensation of the first amino acid is encoded by c18770. Four additional A domains are found in c18760, whereas NRPS4 and 5 harbor epimerization domains for the stereochemical conversion of the activated L-amino acid to the cognate D-amino acid. On the protein level c18760 shows 72% identity with the NRPS MycB from Bacillus atrophaeus. Gene c18750 encodes for the NRPS responsible for the incorporation of the last two amino acids of the lipopeptide, whereas NRPS8 harbors an epimerization (E) domain. Additionally, the final NRPS contains a thioesterase domain, responsible for cyclisation and release of the lipopeptide.The isolation and structure elucidation was achieved for three new iturin-family lipopeptides named paenilarvins accomplished from the P. larvae strain DSM25430.47 Four additional derivatives of the paenilarvins were identified present in low amounts, but were not further characterized. Mass spectrometric data led to suggest variations in chain length and stereochemistry in the β-amino fatty acid side chain.47 Paenilarvin A shows an identical amino acid sequence as the recently identified mojavensin A from Bacillus mojavensis B0621A, but differs in the β-amino fatty acid side chain (Fig. 5).50 In paenilarvin B, Asn at position 2 is exchanged by Asp (Fig. 4). Paenilarvin C is a paenilarvin A analogue shortened by two methylene groups in the β-amino fatty acid side chain (Fig. 5). The biosynthesis begins with the activation of a fatty acid, which is likely derived from the primary metabolism. Subsequently, the fatty acid is loaded onto the first T domain, followed by a chain extension with malonyl-CoA catalyzed by PKS module and the trans-acting acyltransferase (c18780). The action of the aminotransferase (AMT) domain converts the extended fatty acid into the β-amino fatty acid by reductive transamination. The β-amino fatty acid is then condensed with Asx, catalyzed by the condensation domain of the first NRPS module. The next four NRPS modules, all expressed by the same gene (c18760), catalyze the activation and condensation of Tyr, Asn, Gln and Pro, respectively. Hereby, the first two NRPS harbor epimerization domains, resulting in D-Tyr and D-Asn in the final peptide. The last gene of the cluster (c18750) encodes the final two NRPS, responsible for the activation and condensation of two Asn, whereas the first Asn is converted to D-Asn by the epimerization domain. The last domain of the c18750 gene product is a thioesterase domain, which catalyzes the cyclisation and release of the paenilarvins.
Most members of the iturin family lipopeptides show strong antifungal and cytolytic activity by disruption of the plasma membrane, whereas the fatty acid chain length seems to be important for the bioactivity.51,52 In contrast to their good antifungal properties, most iturin-type lipopeptides show no or only weak antibacterial activity.48 The same was observed for the paenilarvins: neither paenilarvin A, nor paenilarvin B showed antibacterial activity against Gram-positive or Gram-negative bacteria, but both compounds were active against various filamentous fungi and yeasts: Mucor hiemalis, Aspergillus clavatus, Botryotinia fuckeliana, Hormoconis resinae, Penicillium capsulatum, Saccharomyces cerevisiae, Rhodotorula glutinis, Candida albicans, Wickerhamomyces anomalus, Nematospora coryli, Trichosporon oleaginous, Debaryomyces hansenii and Pichia membranifaciens. Paenilarvin A was significantly more active (MIC = 2.1–4.2 μg mL−1) than Paenilarvin B (MIC = 8.3–33.3 μg mL−1). In addition to the antimicrobial assays, the paenilarvins were also tested in a cell culture assay against the mouse fibroblast cell line L929. Thereby, paenilarvin A and B both showed also significant cytotoxicity (paenilarvin A: IC50 = 4 μg mL−1; paenilarvin B: IC50 > 10 μg mL−1). The authors were further interested in the effects of paenilarvin A and B against bee larva. Therefore, bee larvae were fed with paenilarvin A or B (5 μg) at days 4, 5 and 6 and larval mortality was recorded for 22 days. For both compounds a significant activity against bee larvae could be demonstrated: for paenilarvin A the mortality was 25% higher and for paenilarvin B 35% higher compared to the larval control group. In this study, paenilarvin C was not tested in any of the assays, whereas the effects of the shortened lipid chain for the increased bioactivity seem interesting. The authors estimate their studies on bee larvae as preliminary data, which have to be confirmed with different concentrations of compounds and with larvae at different developmental stages. The question whether paenilarvins are required by P. larvae to fight fungi as ecological niche competitors or to kill bee larva could not be answered in this study. The authors discussed a further hypothesis for the role of the paenilarvins: at subinhibitory concentrations they could act as signaling molecules, which is a hypothesis already suggested previously for other systems.53,54 In contrast, the corresponding gene cluster from P. larvae DSM25719 (genotype ERIC I) is much smaller (∼12.5 kbp) and contains only two genes (Fig. 4B). In this context only the biosynthetic machinery for the acylation, PKS-mediated extension and amidation as well as the NRPS for the incorporation of the first amino acid are present (Fig. 4B). The NRPS shows homology to NRPS9 of ERIC II, encoded by gene c18750. Accordingly, the ERIC I gene cluster could be responsible for the biosynthesis of another not yet identified (lipo)peptide or might be even inactive. However, no studies on the ERIC I gene cluster were published yet.
Sevadicin – A nonribosomal tripeptide
The genome of strain DSM25430 (genotype ERIC II) contains an 11.7 kbp gene cluster (sev cluster) consisting of only two genes (sevA and sevB) and coding for three NRPS modules without the presence of genes coding for tailoring enzymes or transport (Fig. 4C). In the NRPS genes epimerization (E) domains that catalyze the inversion of the α-stereocenter of amino acids are located in the first two modules while a TE domain was located at the C-terminal part of module 3. The predictions for A domain specificities were ambiguous, such that sequence details as well as peptide size were elusive. Help came from gene inactivation strains, incapable of sevadicin production. Subsequent analytical characterization rendered a linear peptide, called sevadicin with the sequence D-Phe–D-Ala–Trp (Fig. 5).55 While for strain DSM25430 (ERIC II) a peptide could be identified, strain DSM25719 (ERIC I) contains an NRPS gene cluster of five genes with four NRPS modules (Fig. 2C). The product of gene c02110 is annotated as a lone-standing A domain, predicted to activate Cys. At the protein level SevA and the product of c02090 are highly homologous, whereas the ERIC I NRPS is slightly shorter (927 aa) compared to SevA (1080 aa). Comparison of the ten highly conserved core motifs56 of the adenylation domains revealed that the ERIC I A domain lack the core motifs A1 and A2, suggesting an inactive module. Hereby, the involvement of the lone-standing A domain (gene c02110) as a trans-acting A domain could be hypothesized. Due to our inspection, the NRPS module encoded by gene c02100 seems to be active and is predicted to activate Val. The NRPS module encoded by c02120 shows a high homology to NRPS2 of SevB, with the exception of the first C domain. However, the C-terminal C domain is homologous to the C domain of NRPS3 of SevB. NRPS4 (encoded by c02130) is identical to the A–T–TE domains of SevB. Nevertheless, until to date no peptide was identified in ERIC I secretome.Testing the secretome of the sev gene inactivation strain showed that the antibacterial activity was clearly decreased while not completely lost compared to the wild-type secretome. Synthetic sevadicin showed only weak antibacterial activity against B. megaterium, suggesting alternative functions of sevadicin. These may be signaling networks, as it has recently shown for the aureusimines: nonribosomally synthesized diketopiperazine-like compounds produced by Staphylococcus aureus,57 which might be involved in regulating the expression of genes associated with virulence and with regulatory and redox-associated genes,58 suggesting a potential function for regulating a metabolic switch of S. aureus. Interestingly, P. larvae previously has also been hypothesized to undergo a metabolic switch during pathogenesis.16 In the larval gut during the non-invasive phase, P. larvae mainly takes its nutrients from the larval food ingested by larvae.18 Subsequently during the invasive phase, P. larvae switches to consuming larval structures, first the peritrophic membrane protecting the gut,17,19 then totally decomposing all larval tissues yielding a ropy mass consisting of a pure P. larvae culture.
Paenilamicins – complex peptide–polyketide hybrids
A remarkably complex NRPS/PKS hybrid gene cluster (pam gene cluster) of about 60 kbp, which shows no greater similarities to any known gene clusters, is found in both genotypes ERIC I and ERIC II, respectively (Fig. 4D). The biggest part of the gene cluster consists of five NRPS (pamC-E, pamH and pamN), two PKS (pamF and G) and two PKS/NRPS hybrid genes (pamA and B).59 Six genes are involved in precursor biosynthesis (pamK and pamO-S). Two genes have a likely function in transport and/or resistance (pamZ and pamJ). PamM is most probably responsible for transcriptional regulation, whereas PamL is a lone-standing acyl-carrier protein (ACP) and PamI shows weak homology to BtrH-like proteins.The considerably challenging structure elucidation was achieved by isolating four derivatives, named paenilamicins (Fig. 4).60 Based on the structures of paenilamicin A1/A2 and paenilamicin B1/B2 a comprehensive biosynthetic picture could be drawn (Fig. 6A): the A domain of PamA shows relaxed substrate specificity and must be capable of activating either Lys or Arg. The biosynthesis continues with a PKS1-mediated Claisen condensation with malonyl-CoA and a ketoreductase-catalyzed reduction of the amino acid carbonyl to yield a secondary alcohol and the generation of a cadaverine (Cad) or agmatine (Agm) residue, respectively. A subsequent Claisen condensation with an unusual (2R)-hydroxymalonyl is then performed by the PKS part of the PKS/NRPS hybrid PamB, followed by another ketoreduction. (2R)-Hydroxymalonyl-ACP is most likely provided by the catalytic action of PamKOPQ, proteins that show high similarity to (2R)-hydroxymalonyl-ACP biosynthesis proteins known from the zwittermicin biosynthesis.61,62 PamK shows 78% similarities to the 3-hydroxyacyl-CoA-dehydrogenease (ZmaG), PamO shares 88% similarities with the FkbH-like protein (ZmaN) and PamP is annotated as an ACP with high similarity (86%) to ZmaD/H, whereas PamQ is an acyl-CoA dehydrogenase with 89% similarity to ZmaE. The proposed biosynthesis of (2R)-hydroxymalonyl-ACP requires glycolysis intermediate 1,3-bisphospho-D-glycerate as a substrate (Fig. 6B).61 The 1,3-bisphospho-D-glycerate is tethered to PamP and dephosphorylated by PamO. The glycerate-ACP is subsequently oxidized by PamK and PamQ, respectively yielding in (2R)-hydroxymalonyl-ACP (Fig. 6B). Next, a D-Ala is condensed to the building block by NRPS2. Hereby, D-Ala is not stereochemically converted by an epimerization domain of NRPS2 (no E domain could be detected by sequence analysis). Because of the occurrence of a gene coding for an alanine racemase it is proposed that D-Ala is provided by this enzyme and directly activated by the A domain, as it was shown for cyclosporin and HC toxin.63,64 The biosynthesis continues with the condensation of the amino acids N-methyl-diaminopropionic acid (mDap), D-Lys/D-Orn, and Ser. Thereby, Dap is provided by PamS and PamR from the substrates O-phospho-L-Ser and Glu (Fig. 6C).65 PamS is a 2,3-diaminopropionate biosynthesis protein (SbnB-like, 80% similarity) and PamR is a pyridoxal-5'-phosphate-dependent protein subunit β (SbnA-like, 80% similarity), which are known from the Dap biosynthesis of the staphyloferrin B biosynthesis (Fig. 6C).65 The presence of N-methyl-Dap is supported by an N-methylation domain in the NRPS PamC. In contrast to common N-methylations occurring at the peptide bond, this N-methylation is located in the amino acid side-chain. As already described for the A domain of PamA, the A domain of PamD shows also a relaxed substrate specificity and is capable to activate Lys and Orn, resulting in another two paenilamicin derivatives. The biosynthesis proceeds with two consecutive condensations of malonyl-CoA with followed ketoreductions catalyzed by PKS3 (PamF) and 4 (PamG). Together, PamE-G proteins are responsible for the biosynthesis of the structural motif galantinic acid (Gla). Interestingly, the same structural motif is found in the siderophor anachelin produced by the cyanobacterium Anabaena cylindrical, suggesting the same biosynthetic setup.66 Catalyzed by the bimodular NRPS PamH, another mDap and a Gly are attached. An interesting hypothesis is provided for the attachment of the C-terminal 4,3-spermidine. The protein PamI shows homology to the BtrH protein family. A functional classification for a BtrH protein was given for the biosynthesis of butirosin, where the BtrH protein is described as an aminoglycoside 1-N-acyltransferase responsible for the ligation of the aminoglycoside side chain of butirosin.67 At the C-terminus of PamH (after the T domain of the Gly specific NRPS) a similar domain could be identified and the authors hypothesized that the domains could somehow be responsible for the directional attachment of 4,3-spermidine to the paenilamicins and thereby release of the peptide from the NRPS.
Another interesting aspect is the suggested production of a prodrug called prepaenilamicin, which parallels the (pre)xenocoumacins, compounds from Xenorhabdus nematophila described by Bode and coworkers.68,69 Therein two genes seem to be conserved: a NRPS specific for the activation of (acyl)-D-Asn or (acyl)-D-Gln and a D-Asn specific carboxypeptidase. For the pam system the A domain of NRPS8 (PamN) is predicted to activate Asn or Asx by three different prediction tools and harbors an E domain responsible for transformation of L-Asx into D-Asx. Moreover, PamN is homologous to the starting module XcnA of the xenocoumacin biosynthesis gene cluster as well as to starting modules of other biosynthesis gene clusters responsible for the biosynthesis of nonribosomal prodrugs like amicoumacin (B. pumilus), zwittermicin (B. thuringiensis), colibactin (E. coli), and other similar NRPS/PKS clusters with unknown products that all are predicted to activate Asn or Asx.68,69
Bode and coworkers proposed two different architectures of the peptidases: type I with XcnG as the model peptidase comprises a signal peptide followed by a periplasmatic D-Asn specific carboxypeptidase and three C-terminal transmembrane helices. Type II has a signal peptide followed by a peptidase domain, nine transmembrane helices and an ABC transporter.68 The peptidase subunit is responsible for the specific cleavage of the acylated D-Asp moiety of the prexenocoumacins, resulting in the bioactive xenocoumacins.68 The protein PamJ was found to be homologous to XcnG suggesting that PamJ could be a peptidase (Fig. 6A). Although PamJ shows structural differences (type II architecture) to XcnG (type I architecture) a comparable biosynthesis and pro-drug activation mechanism seems to be likely (Fig. 6A). The described pro-drug mechanism is a strategy of the producer strain to avoid self-damaging.
For the paenilamicins antibacterial activity against various Gram-positive strains (P. alvei, B. subtilis, B. licheniformis, and B. megaterium) was found, which are all strains with a potential association with honey bees.59 Beside the antibacterial activity the paenilamicins exhibit also antifungal activity against yeasts (P. pastoris and S. cerevisiae) and the plant pathogenic filamentous fungus F. oxysporum.59 Cytotoxic effects found in vitro against the Trichoplusia ni (Lepidotera-derived) insect cell line Tn5, suggested that paenilamicin could act as a toxin against bee larvae. However bee larva infection assays with a P. larvae strain unable to synthesize paenilamicin (P. larvae ΔpamA) showed no significant differences in total mortality of bee larvae.59 These results show that in the P. larvae/honey bee larvae system it is obviously not always possible to infer from in vitro data on in vivo function. Nevertheless, the authors also analyzed the time course of infection revealing that the curve for cumulative mortality was shifted for the larval groups infected with the mutant strain (ΔpamA), indicating a significantly delayed onset of mortality in comparison to the groups infected with wild-type P. larvae. Accordingly, the LT100 (lethal time) also significantly differed: wild-type P. larvae required about 8 days, whereas the mutant strain needed 12 days to accomplish killing of all infected larvae.59 The proposed main function of paenilamicin was shown in a recently published study:60 in the corresponding assay larvae were co-infected with both P. larvae and the saprophytic bacterium P. alvei. After larval death dead larvae were examined for P. alvei survival by determination of P. alvei growth on agar medium. This assay showed that only 11% vital P. alvei could be recovered from larvae infected with wild-type P. larvae. Interestingly, in co-infection assays where larvae were infected with P. alvei and P. larvae ΔpamA the recovery rate of P. alvei was significantly higher (about 62%). The results of the larvae co-infection assays show, that a main function of the paenilamicins is outcompeting competitors in the ecological niche larval gut and may explain why the ropy mass that remains after larval death is a pure culture of P. larvae (Fig. 2A).
A cryptic monomodular NRPS-like gene cluster
The genotypes, ERIC I (gene c22070) and ERIC II (gene c04880) both contain a monomodular NRPS-like gene (Fig. 4E). It encodes for an A domain, a T domain and a C-terminal thioesterase reduction (TR) domain, but lacks a C domain. The A domain of the P. larvae NRPS-like protein is predicted to activate Phe and the module arrangement has similarities to the fungal NRPS LnaA (1042 aa) and LnbA (1007 aa) from Aspergillus flavus NRRL3357,70 although the putative NRPS-like protein from P. larvae (1239 aa) shares only weak homology to LnaA and LnbA, respectively. The A domains of LnaA and LnbA specifically activate Tyr and biosynthesis products are piperazine and pyrazine derivatives, as well as a hemiacetal-containing morpholine.70 In a biosynthesis model the authors suggest that Tyr is activated by the A domains of LnaA and/or LnbA, transferred to the T domains and subsequently reduced to yield a tyrosinal by the catalytic action of the TR domain. Further reduction and cyclisation is supposed to be catalyzed be the reductases LnaB and LnbB.70 In the context of the P. larvae cluster no additional reductases were identified, furthermore despite the NRPS gene no further genes show homologies to the lna or lnb cluster. The structures are reminiscent of aureusimines from S. aureus and diketopiperazines from Streptomyces although different biosynthesis mechanisms as bimodular NRPS and tRNA-dependent assembly are employed.57,71 Likewise to above mentioned bacteria, the diketopiperazines and the piperazine/pyrazine might play a role in a signaling networks.57,70,71 From the present data we hypothesize that P. larvae is able to produce related compounds which could have a similar function as quorum sensing molecules, which needs to be proven in further work.Gene clusters of ribosomally synthesized secondary metabolites
Apart from the above mentioned gene clusters encoding for nonribosomal peptides, the genotypes ERIC I and ERIC II contain also genes encoding for the synthesis of ribosomal peptides. The genomes of P. larvae strains DSM25430 and DSM25719 were analyzed with the antiSMASH72 online tool to assess the potential for the biosynthesis of bacteriocins and ribosomally synthesized and posttranslationally modified peptides (RiPPs).The manual assignment of genes rendered three putative bacteriocin and RiPP gene clusters for genotype ERIC II, thereof one lanthipeptide and two bacteriocins. For genotype ERIC I putative biosynthesis genes or gene cluster-like arrangements for eight bacteriocins and one lanthipeptide were found. In ERIC II the lanthipeptide cluster comprises a gene (c31290) coding for a putative lantibiotic dehydratase with a C-terminal thiopeptide-type bacteriocin biosynthesis domain. Gene c31300 encodes a LanC-like cyclase and c31310 for a truncated ABC transporter (183 aa). A protease was not found in the cluster, but is also not universally found in class I gene clusters,73,74 while however the assigned prepropeptide gene (c31270) has only weak sequence homology to known prepropeptides. A putative gene cluster for the biosynthesis of a bacteriocin and with homology to gene clusters of several B. amyloliquefaciens and B. subtilis species is gene c36580. NCBI's conserved domain search75 showed that the C-terminal part has homologies to class IId cyclic uberolysin-like bacteriocins.76 A putative ABC transporter of 233 aa (gene c36600), could be responsible for transport and self-resistance. Even if a large part of the genes of this cluster could not be assigned regarding their functions one could assume a functional gene cluster for the expression of a bacteriocin. Another uberolysin-like putative bacteriocin (73 aa) is encoded by gene c40350 with 49.3% identity and 69.9% similarity to uberolysin from Streptococcus uberis.77 The truncated gene product of c40340 (191 aa) shows only weak homology to the cyclizing protein UblB (535 aa),77 assuming that the P. larvae protein is non-functional. Three putative ABC transporters are encoded by the genes c40280, c40250 and c40240.
Probably an incomplete class II lanthipeptide gene cluster is spanning the region 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 806
806![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 134–1
134–1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 812
812![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 604, which contains a truncated lanM-like gene and a putative prepropeptide (c19180) with weak homology to known lanthipeptides and lacking characteristic features of precursor peptides. Likewise, bacteriocin gene clusters (c05230, c05830, c21080, c21140, c24550, c28660, c31740, c34160) putatively are disrupted or incomplete.
604, which contains a truncated lanM-like gene and a putative prepropeptide (c19180) with weak homology to known lanthipeptides and lacking characteristic features of precursor peptides. Likewise, bacteriocin gene clusters (c05230, c05830, c21080, c21140, c24550, c28660, c31740, c34160) putatively are disrupted or incomplete.
Summary and outlook
Genome sequencing of Paenibacillus larvae strains DSM25430 (genotype ERIC II) and DSM25719 (genotype ERIC I) revealed the presence of gene clusters coding for the synthesis of secondary metabolites. With a size of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 056
056![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 006 bp (ERIC II) and 4
006 bp (ERIC II) and 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 579
579![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 589 bp (ERIC I), 2.99% and 2.23% of the genomes are devoted to the biosynthesis of small molecules, respectively (Fig. 3).
589 bp (ERIC I), 2.99% and 2.23% of the genomes are devoted to the biosynthesis of small molecules, respectively (Fig. 3).
The gene clusters encode NRPS peptides and NRPS/PKS hybrid peptides of which four structural families have been identified (Fig. 4 and 5). Interestingly, exclusive polyketide synthases have not been identified. Furthermore, no gene clusters responsible for the biosynthesis of terpenes, aminoglycosides and other small molecules have been identified (own analysis). Putative clusters responsible for ribosomally synthesized peptides could be detected in both P. larvae genotypes, whereas antiSMASH analysis of genotype ERIC I showed more putative clusters (one lanthipeptide and eight bacteriocin clusters) compared to genotype ERIC II (one lanthipeptide, two bacteriocin clusters). Most clusters seem to be incomplete or exhibited only weak homologies to known gene clusters of ribosomally synthesized peptides.76,78 Nevertheless, some core precursor peptides showed homologies to known bacteriocins.79 Experimental data is necessary to prove which clusters are active and to elucidate the roles of bacteriocins and RiPPs in AFB.
In recent work four of the dedicated nonribosomal secondary metabolites were identified and their structures characterized. The siderophore bacillibactin is generally widespread among Bacillus species. Since gene inactivation studies of the gene cluster rendered no effect on virulence we hypothesize that the siderophore may be an evolutionary remnant still enabling P. larvae to sequester iron under certain circumstances. The other three compounds apparently have antimicrobial defense tasks: the nonribosomal tripeptide sevadicin shows antibacterial activity while paenilarvins, iturin-like lipopeptides, have antifungal activity against various filamentous fungi and yeasts. For iturin family lipopeptides fungicidal and strong cytolytic activity has been described while they display no or only weak antibacterial activities.48,51,52 Finally, paenilamicins show both, antibacterial as well as antifungal activity in addition to cytotoxic activity against insect cells.59 It is very likely, that the array of these compounds acts in a synergistic manner as this was already described for effects on pathogenic fungi by bacillomycin and fengycin.80 Exposure bioassays provided first evidence for the in vivo function of the paenilamicins during AFB pathogenesis: (i) by killing bacterial (and maybe fungal) competitors the paenilamicins enable P. larvae to fight its ecological niche and (ii) the paenilamicins influence the time-course of disease in bee larvae although the mechanism remains elusive.59 Therefore, paenilamicins act as virulence factors for P. larvae during larval infection. The high specialization of P. larvae on honey bees and the uniqueness of sevadicin and paenilamicin in this bacterium may indicate a more intricate role of these two compounds.
Still not clear is the cytotoxicity of compounds which becomes critical in the moment when P. larvae breaks through the midgut lumen. While chitinases and maybe also other enzymes dissolve cellular structures, secondary metabolites could have a function in protecting the larval cadaver against other bacteria and fungi that act as nutrient competitors. Another function, especially for sevadicin due to weak antibacterial activity, could be a role as a signaling molecule involved in the regulation of biological processes, as it could be shown for the aureusimines57,58 and other antibiotics.54
The described genomic differences between P. larvae ERIC I and ERIC II also concern the secondary metabolite gene clusters. The genome of the P. larvae genotype ERIC I harbors also the paenilamicin and the bacillibactin clusters, although paenilamicin could not be identified from fractionated secretomes yet. Additionally, ERIC I harbors two clusters that differ from homologous clusters from genotype ERIC II, and whose products are not identified yet. One of these clusters is homologous to the iturin gene cluster, which consists in contrast to the gene cluster of ERIC II, of only two genes. One of these genes is a trans-acting acyltransferase nearly identical to c18780 from ERIC II Ser103Ala; the other gene is a NRPS/PKS hybrid with high homology to gene c18770 from ERIC II, but harboring a TE domain at the C-terminus. This cluster could be responsible for the synthesis of another not yet identified (lipo)peptide or might be even an inactive gene cluster. Another gene cluster responsible for the biosynthesis of an unknown peptide from the genotype ERIC I is a homolog of the sev gene cluster. The ERIC I gene cluster consists of five genes (two genes in ERIC II) and likely includes a trans-acting A domain in the peptide synthesis. Close inspection of the gene cluster led us to suggest the synthesis of a tetrapeptide. Furthermore, a monomodular NRPS-like gene is harbored by both genotypes, whose compounds are not identified yet. NRPS-like proteins with the architecture A–T–TR are predominantly known from fungi. Such as the NRPS-like proteins from the fungus A. flavus, which are responsible for the biosynthesis of metabolites involved in a signaling network resulting in a metabolic switch. Interestingly, also P. larvae is undergoing a metabolic switch.16 Further work is necessary to identify all compounds produced by P. larvae and to illuminate their functions.
In conclusion entomopathogens appear as a rich and yet underinvestigated source of novel and structurally diverse secondary metabolites. The bee pathogen P. larvae is no exception to this rule and an in-depth study and characterization of the secondary metabolites produced by the honey bee pathogen P. larvae is highly relevant not only for bee pathologists but for a broader community interested in discovering and elucidating novel antibacterial substances. Hence, improving our knowledge of the secondary metabolites of P. larvae will allow a better understanding of roles of secondary metabolites in cell biology and pathogenesis, especially in AFB. In addition, the further characterization of novel metabolites, such as paenilamicin, could bring new resources in the drug development market.
Acknowledgements
The authors are grateful to the Ministries for Agriculture from Brandenburg and Sachsen-Anhalt, Germany, the German Research Foundation (Graduate School 1121), and the Cluster of Excellence “Unifying Concepts in Catalysis” (UniCat) coordinated by TU Berlin.Notes and references
- R. Morse and N. Calderone, Bee Cult., 2000, 128, 1–15 Search PubMed.
- M. A. Aizen, L. A. Garibaldi, S. A. Cunningham and A. M. Klein, Curr. Biol., 2008, 18, 1572–1575 CrossRef CAS PubMed.
- M. A. Aizen and L. D. Harder, Curr. Biol., 2009, 19, 915–918 CrossRef CAS PubMed.
- E. Genersch, Appl. Microbiol. Biotechnol., 2010, 87, 87–97 CrossRef CAS PubMed.
- E. Genersch, E. Forsgren, J. Pentikainen, A. Ashiralieva, S. Rauch, J. Kilwinski, I. Fries and J. Pentikäinen, Int. J. Syst. Evol. Microbiol., 2006, 56, 501–511 CrossRef CAS PubMed.
- J. Versalovic and M. Schneider, Methods Mol. Cell. Biol., 1994, 5, 25–40 CAS.
- B. J. Morrissey, T. Helgason, L. Poppinga, A. Fünfhaus, E. Genersch and G. E. Budge, Environ. Microbiol., 2014, 17, 1414–1424 CrossRef PubMed.
- S. Rauch, A. Ashiralieva, K. Hedtke and E. Genersch, Appl. Environ. Microbiol., 2009, 75, 3344–3347 CrossRef CAS PubMed.
- M. Djukic, E. Brzuszkiewicz, A. Fünfhaus, J. Voss, K. Gollnow, L. Poppinga, H. Liesegang, E. Garcia-Gonzalez, E. Genersch and R. Daniel, PLoS One, 2014, 9, e90914 Search PubMed.
- E. Genersch, J. Invertebr. Pathol., 2010, 103, S10–S19 CrossRef PubMed.
- E. Genersch, A. Ashiralieva and I. Fries, Appl. Environ. Microbiol., 2005, 71, 7551–7555 CrossRef CAS PubMed.
- N. Wilson-Rich, M. Spivak, N. H. Fefferman and P. T. Starks, Annu. Rev. Entomol., 2009, 54, 405–423 CrossRef CAS PubMed.
- L. Hasemann, Am. Bee J., 1961, 101, 298–299 Search PubMed.
- J. D. Hitchcock, A. Stoner, W. T. Wilson and D. M. Menapace, J. Kans. Entomol. Soc., 1979, 52, 238–246 Search PubMed.
- W. T. Wilson, J. Invertebr. Pathol., 1971, 17, 247–255 CrossRef CAS.
- D. Yue, M. Nordhoff, L. H. Wieler and E. Genersch, Environ. Microbiol., 2008, 10, 1612–1620 CrossRef CAS PubMed.
- E. Garcia-Gonzalez and E. Genersch, Environ. Microbiol., 2013, 15, 2894–2901 CAS.
- S. Neuendorf, K. Hedtke, G. Tangen and E. Genersch, Microbiology, 2004, 150, 2381–2390 CrossRef CAS PubMed.
- E. Garcia-Gonzalez, L. Poppinga, A. Fünfhaus, G. Hertlein, K. Hedtke, A. Jakubowska and E. Genersch, PLoS Pathog., 2014, 10, e1004284 Search PubMed.
- A. Fünfhaus, A. Ashiralieva, R. Borriss, E. Genersch and A. Funfhaus, Environ. Microbiol. Rep., 2009, 1, 240–250 CrossRef PubMed.
- A. Fünfhaus, L. Poppinga and E. Genersch, Environ. Microbiol., 2013, 15, 2951–2965 Search PubMed.
- P. O. Falnes and K. Sandvig, Curr. Opin. Cell Biol., 2000, 12, 407–413 CrossRef CAS.
- N. C. Simon, K. Aktories and J. T. Barbieri, Nat. Rev. Microbiol., 2014, 12, 599–611 CrossRef CAS PubMed.
- I. Carpusca, T. Jank and K. Aktories, Mol. Microbiol., 2006, 62, 621–630 CrossRef CAS PubMed.
- L. Poppinga, B. Janesch, A. Fünfhaus, G. Sekot, E. Garcia-Gonzalez, G. Hertlein, K. Hedtke, C. Schäffer and E. Genersch, PLoS Pathog., 2012, 8, e1002716 CAS.
- A. Fünfhaus and E. Genersch, Environ. Microbiol. Rep., 2012, 4, 194–202 CrossRef PubMed.
- E. C. Holst, Science, 1945, 102, 593–594 CAS.
- G. F. White, USDA Bur. Entomol. Tech. Ser., 1906, 14, 1–50 Search PubMed.
- S. Vojvodic, S. M. Rehan and K. E. Anderson, PLoS One, 2013, 8, e72106 CAS.
- C. Ash, J. A. Farrow, M. Dorsch, E. Stackebrandt and M. D. Collins, Int. J. Syst. Bacteriol., 1991, 41, 343–346 CrossRef CAS PubMed.
- C. Ash, F. G. Priest and M. D. Collins, Antonie van Leeuwenhoek, 1993, 64, 253–260 CrossRef CAS.
- T. Stein, Mol. Microbiol., 2005, 56, 845–857 CrossRef CAS PubMed.
- F. Lipmann, W. Gevers, H. Kleinkauf and R. Roskoski, Adv. Enzymol. Relat. Areas Mol. Biol., 1971, 35, 1–34 CAS.
- E. H. Duitman, L. W. Hamoen, M. Rembold, G. Venema, H. Seitz, W. Saenger, F. Bernhard, R. Reinhardt, M. Schmidt, C. Ullrich, T. Stein, F. Leenders and J. Vater, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 13294–13299 CrossRef CAS.
- J. J. May, T. M. Wendrich and M. A. Marahiel, J. Biol. Chem., 2001, 276, 7209–7217 CrossRef CAS PubMed.
- K. Arima, A. Kakinuma and G. Tamura, Biochem. Biophys. Res. Commun., 1968, 31, 488–494 CrossRef CAS.
- J. R. Cath, T. S. G. Jones and S. Wilkinson, Ann. N. Y. Acad. Sci., 1949, 51, 917–923 CrossRef PubMed.
- Y. Kajimura and M. Kaneda, J. Antibiot., 1996, 49, 129–135 CrossRef CAS.
- Y. Wen, X. Wu, Y. Teng, C. Qian, Z. Zhan, Y. Zhao and O. Li, Environ. Microbiol., 2011, 13, 2726–2737 CrossRef CAS PubMed.
- X.-C. Wu, C.-D. Qian, H.-H. Fang, Y.-P. Wen, J.-Y. Zhou, Z.-J. Zhan, R. Ding, O. Li and H. Gao, Microb. Biotechnol., 2011, 4, 491–502 CrossRef CAS PubMed.
- C.-D. Qian, X.-C. Wu, Y. Teng, W.-P. Zhao, O. Li, S.-G. Fang, Z.-H. Huang and H.-C. Gao, Antimicrob. Agents Chemother., 2012, 56, 1458–1465 CrossRef CAS PubMed.
- G. Hertlein, S. Müller, E. Garcia-Gonzalez, L. Poppinga, R. D. Süssmuth and E. Genersch, PLoS One, 2014, 9, e108272 Search PubMed.
- J. Y. Lee, K. D. Passalacqua, P. C. Hanna and D. H. Sherman, PLoS One, 2011, 6, e20777 CAS.
- B. Bister, D. Bischoff, G. J. Nicholson, M. Valdebenito, K. Schneider, G. Winkelmann, K. Hantke and R. D. Süssmuth, Biometals, 2004, 17, 471–481 CrossRef CAS.
- D. Segond, E. Abi Khalil, C. Buisson, N. Daou, M. Kallassy, D. Lereclus, P. Arosio, F. Bou-Abdallah and C. Nielsen Le Roux, PLoS Pathog., 2014, 10, e1003935 Search PubMed.
- S. Cendrowski, W. MacArthur and P. Hanna, Mol. Microbiol., 2004, 51, 407–417 CrossRef CAS PubMed.
- S. Sood, H. Steinmetz, H. Beims, K. I. Mohr, M. Stadler, M. Djukic, W. von der Ohe, M. Steinert, R. Daniel and R. Müller, Chembiochem, 2014, 15, 1947–1955 CrossRef CAS PubMed.
- R. Maget-Dana and F. Peypoux, Toxicology, 1994, 87, 151–174 CrossRef CAS.
- K. Tsuge, T. Akiyama and M. Shoda, J. Bacteriol., 2001, 183, 6265–6273 CrossRef CAS PubMed.
- Z. Ma, N. Wang, J. Hu and S. Wang, J. Antibiot., 2012, 65, 317–322 CrossRef CAS PubMed.
- R. H. Baltz, V. Miao and S. K. Wrigley, Nat. Prod. Rep., 2005, 22, 717–741 RSC.
- L. Thimon, F. Peypoux, J. Wallach and G. Michel, FEMS Microbiol. Lett., 1995, 128, 101–106 CrossRef CAS PubMed.
- D. Romero, M. F. Traxler, D. López and R. Kolter, Chem. Rev., 2011, 111, 5492–5505 CrossRef CAS PubMed.
- R. I. Aminov, Environ. Microbiol., 2009, 11, 2970–2988 CrossRef CAS PubMed.
- E. Garcia-Gonzalez, S. Müller, P. Ensle, R. D. Süssmuth and E. Genersch, Environ. Microbiol., 2014, 16, 1297–1309 CrossRef CAS PubMed.
- M. A. Marahiel, T. Stachelhaus and H. D. Mootz, Chem. Rev., 1997, 97, 2651–2674 CrossRef CAS PubMed.
- M. A. Wyatt, W. Wang, C. M. Roux, F. C. Beasley, D. E. Heinrichs, P. M. Dunman and N. A. Magarvey, Science, 2010, 329, 294–296 CrossRef CAS PubMed.
- M. A. Wyatt, W. Wang, C. M. Roux, F. C. Beasley, D. E. Heinrichs, P. M. Dunman and N. A. Magarvey, Science, 2011, 333, 1381 CAS.
- E. Garcia-Gonzalez, S. Müller, G. Hertlein, N. Heid, R. D. Süssmuth and E. Genersch, MicrobiologyOpen, 2014, 3, 642–656 CrossRef CAS PubMed.
- S. Müller, E. Garcia-Gonzalez, A. Mainz, G. Hertlein, N. C. Heid, E. Mösker, H. van den Elst, H. S. Overkleeft, E. Genersch and R. D. Süssmuth, Angew. Chem., Int. Ed., 2014, 53, 10821–10825 CrossRef PubMed.
- Y. A. Chan, M. T. Boyne, A. M. Podevels, A. K. Klimowicz, J. Handelsman, N. L. Kelleher and M. G. Thomas, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 14349–14354 CrossRef CAS PubMed.
- Y. A. Chan and M. G. Thomas, Biochemistry, 2010, 49, 3667–3677 CrossRef CAS PubMed.
- G. Weber, K. Schörgendorfer, E. Schneider-Scherzer and E. Leitner, Curr. Genet., 1994, 26, 120–125 CrossRef CAS.
- J. S. Scott-Craig, D. G. Panaccione, J. A. Pocard and J. D. Walton, J. Biol. Chem., 1992, 267, 26044–26049 CAS.
- M. J. Kobylarz, J. C. Grigg, S. J. Takayama, D. K. Rai, D. E. Heinrichs and M. E. P. Murphy, Chem. Biol., 2014, 21, 379–388 CrossRef CAS PubMed.
- H. Beiderbeck, K. Taraz, H. Budzikiewicz and A. E. Walsby, Z. Naturforsch., C: J. Biosci., 2000, 55, 681–687 CAS.
- N. M. Llewellyn, Y. Li and J. B. Spencer, Chem. Biol., 2007, 14, 379–386 CrossRef CAS PubMed.
- D. Reimer, K. M. Pos, M. Thines, P. Grün and H. B. Bode, Nat. Chem. Biol., 2011, 7, 888–890 CrossRef CAS PubMed.
- D. Reimer and H. B. Bode, Nat. Prod. Rep., 2014, 31, 154–159 RSC.
- R. R. Forseth, S. Amaike, D. Schwenk, K. J. Affeldt, D. Hoffmeister, F. C. Schroeder and N. P. Keller, Angew. Chem., Int. Ed., 2013, 52, 1590–1594 CrossRef CAS PubMed.
- P. Belin, M. Moutiez, S. Lautru, J. Seguin, J.-L. Pernodet and M. Gondry, Nat. Prod. Rep., 2012, 29, 961–979 RSC.
- K. Blin, M. H. Medema, D. Kazempour, M. A. Fischbach, R. Breitling, E. Takano and T. Weber, Nucleic Acids Res., 2013, 41, W204–W212 CrossRef PubMed.
- C. Corvey, T. Stein, S. Düsterhus, M. Karas and K.-D. Entian, Biochem. Biophys. Res. Commun., 2003, 304, 48–54 CrossRef CAS.
- M. C. Booth, C. P. Bogie, H. G. Sahl, R. J. Siezen, K. L. Hatter and M. S. Gilmore, Mol. Microbiol., 1996, 21, 1175–1184 CAS.
- A. Marchler-Bauer, S. Lu, J. B. Anderson, F. Chitsaz, M. K. Derbyshire, C. DeWeese-Scott, J. H. Fong, L. Y. Geer, R. C. Geer, N. R. Gonzales, M. Gwadz, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, F. Lu, G. H. Marchler, M. Mullokandov, M. V. Omelchenko, C. L. Robertson, J. S. Song, N. Thanki, R. A. Yamashita, D. Zhang, N. Zhang, C. Zheng and S. H. Bryant, Nucleic Acids Res., 2011, 39, D225–D229 CrossRef CAS PubMed.
- M. Maqueda, M. Sánchez-Hidalgo, M. Fernández, M. Montalbán-López, E. Valdivia and M. Martínez-Bueno, FEMS Microbiol. Rev., 2008, 32, 2–22 CrossRef CAS PubMed.
- R. E. Wirawan, K. M. Swanson, T. Kleffmann, R. W. Jack and J. R. Tagg, Microbiology, 2007, 153, 1619–1630 CrossRef CAS PubMed.
- P. G. Arnison, M. J. Bibb, G. Bierbaum, A. A. Bowers, T. S. Bugni, G. Bulaj, J. A. Camarero, D. J. Campopiano, G. L. Challis, J. Clardy, P. D. Cotter, D. J. Craik, M. Dawson, E. Dittmann, S. Donadio, P. C. Dorrestein, K.-D. Entian, M. A. Fischbach, J. S. Garavelli, U. Göransson, C. W. Gruber, D. H. Haft, T. K. Hemscheidt, C. Hertweck, C. Hill, A. R. Horswill, M. Jaspars, W. L. Kelly, J. P. Klinman, O. P. Kuipers, A. J. Link, W. Liu, M. A. Marahiel, D. A. Mitchell, G. N. Moll, B. S. Moore, R. Müller, S. K. Nair, I. F. Nes, G. E. Norris, B. M. Olivera, H. Onaka, M. L. Patchett, J. Piel, M. J. T. Reaney, S. Rebuffat, R. P. Ross, H.-G. Sahl, E. W. Schmidt, M. E. Selsted, K. Severinov, B. Shen, K. Sivonen, L. Smith, T. Stein, R. D. Süssmuth, J. R. Tagg, G.-L. Tang, A. W. Truman, J. C. Vederas, C. T. Walsh, J. D. Walton, S. C. Wenzel, J. M. Willey and W. A. van der Donk, Nat. Prod. Rep., 2013, 30, 108–160 RSC.
- H. Abriouel, C. M. A. P. Franz, N. Ben Omar and A. Gálvez, FEMS Microbiol. Rev., 2011, 35, 201–232 CrossRef CAS PubMed.
- X.-H. Chen, A. Koumoutsi, R. Scholz and R. Borriss, J. Mol. Microbiol. Biotechnol., 2009, 16, 14–24 CrossRef CAS PubMed.
- T. Carver, N. Thomson, A. Bleasby, M. Berriman and J. Parkhill, Bioinformatics, 2009, 25, 119–120 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2015 |