Open Access Article
Open Access ArticleProbing the effect of arm length and inter- and intramolecular interactions in the formation of Cu(II) complexes of Schiff base ligands derived from some unsymmetrical tripodal amines†
Hassan
Keypour
*a,
Maryam
Shayesteh
a,
Sadegh
Salehzadeh
a,
Sébastien
Dhers
b,
Farahnaz
Maleki
a,
Hüseyin
Ünver
c and
Nefise
Dilek
d
aFaculty of Chemistry, Bu-Ali Sina University, Hamedan 65174, Iran. E-mail: haskey1@yahoo.com; Fax: +98 8118 380709; Tel: +98 9188131117
bDepartment of Chemistry and The MacDiarmid Institute for Advanced Materials and Nanotechnology, University of Otago, P. O. Box 56, Dunedin 9054, New Zealand
cDepartment of Physics, Faculty of Science, Ankara University, TR-06100 Tandogan, Ankara, Turkey
dDepartment of Physics, Arts and Sciences Faculty, Aksaray University, TR-68100, Aksaray, Turkey
First published on 30th July 2015
Abstract
The syntheses of two previously known, 2-((2-aminoethyl)(pyridin-2-ylmethyl)amino)ethanol (1) and 2-((3-aminopropyl)(pyridin-2-ylmethyl)amino)ethanol (2), and four new unsymmetrical N-capped tripodal amines, 2-((4-aminobutyl)(pyridin-2-ylmethyl)amino)ethanol (3), 3-((2-aminoethyl)(pyridin-2-ylmethyl)amino)propan-1-ol (4), 3-((3-aminopropyl)(pyridin-2-ylmethyl)amino)propan-1-ol (5) and 3-((4-aminobutyl)(pyridin-2-ylmethyl)amino)propan-1-ol (6), are reported. The ligands (3–4) feature a longer arm, 3-hydroxypropyl or butylamino, than in the analogues previously employed (2-hydroxyethyl arm, ethylamino-arm or propylamino-arm in 1 and 2). All six tripodal amines, 1–6, are equipped with a 2-methylpyridyl-arm and either an ethylamino-arm (1 and 4), propylamino-arm (2 and 5) or butylamino-arm (3 and 6). The new amines, 3–6, have been employed in one pot condensation reactions with 2-hydroxy-1-naphthaldehyde and salicylaldehyde (and its derivatives) in the presence of Cu(II) metal ions to generate a series of new mononuclear complexes, [MIILaldi](ClO4) as well as new dinuclear complexes [CuIILaldi]2(ClO4)2 of new ligands Laldi. Four monomeric complexes and one dimeric complex have been characterised by single crystal X-ray diffraction, revealing a distorted square-pyramidal copper(II) ion. A general comparison between these structures shows that the number and types of chelate ring sequences around the metal ions are important in the formation of structures. Theoretical studies show that the 3-hydroxypropyl arm in these complexes is a weak coordinating group and it can readily be removed from the coordination sphere of metal ions, resulting in a dimerised four coordinate complex. Calculations show that the interaction between the two monomeric fragments is very weak.
Introduction
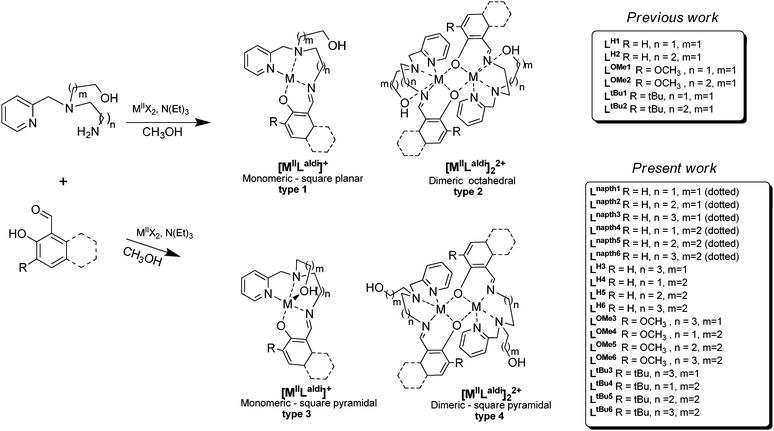
Recently, research on tripodal ligands and their related complexes has been an expanding field and is the subject of numerous reports.1 Transition metal complexes synthesised with this type of ligand display special physical, chemical or structural properties, such as unusual conformation, high thermodynamic stability and virtual kinetic inertness.2 These tripodal ligands can also serve as precursors for the synthesis of interesting macrobicyclic compounds3,4 which usually requires high dilution techniques5 or the use of metal ions as templates.6,7 The synthesis of model complexes mimicking the spectroscopic and structural properties of metalloprotein active sites can be undertaken by employing multidentate tripodal ligands, most of which possess aromatic donor functions like pyridyl and/or phenolic groups.8 However, the chemistry of asymmetric N-capped tripodal ligands which possess three pendant arms with different donor groups has not been well explored.9,10 This type of tripodal ligand is of particular interest in the context of modeling the asymmetric active metal sites such as those found in nitrile hydratase11 and horse liver alcohol dehydrogenase.12 Among these asymmetric N-capped tripodal ligands, our group is particularly interested in unsymmetric N-capped tripodal ligands with two different aliphatic arm lengths and one aromatic pendant arm. However these tripodal ligands bearing one pyridine arm are scarce.13 As part of a program to explore the coordination chemistry of partially unsymmetric tripodal N3O2 ligands, we herein report the synthesis and characterisation of new Cu complexes and compare them with our previous work. A direct influence of the lengths of the alkyl chains between central and terminal donor functions on the complex geometry was observed for the cadmium(II), nickel(II) and copper(II) complexes of these unsymmetrical tripodal ligands featuring different spacer lengths (Fig. 1). The coordination behavior towards Cd(II), Ni(II) and Cu(II) was investigated for ligands Laldi combining pyridine, amine, and alkoxy donor functions on ethylene, propylene and butylene spacers.14In previous work, we reported the synthesis of two new unsymmetrical triamines, 1 and 2, both of which feature three different arms: 2-methylpyridyl, either ethylamino- or propyl-amino, and ethanol (Fig. 1).14 Reaction of 1 or 2 with salicylaldehyde and its analogues gave a wide selection of compounds with which to probe the effects of strain in the resulting cadmium(II) and nickel(II) Schiff base complexes (Fig. 1).14 When the shorter ethylene linker was used between the tertiary and primary amine nitrogen atoms, a mononuclear Schiff base complex was structurally characterised in the case of [NiIILOMe1]+ whereas the more flexible propylene linker (LOMe2 and LH2 ligands) gave dinuclear complexes, [NiIILOMe2]22+ and [CdIILH2]22+, which were structurally characterised. The nickel(II) centre in [NiIILOMe1]+ has a distorted square planar geometry, whereas in [NiIILOMe2]22+ the geometry is distorted octahedral, as with the cadmium(II) ion in [CdIILH2]22+.14 The structural types observed to date are summarised in Fig. 1.
In this paper the effect of employing a 3-hydroxypropyl arm in place of the 2-hydroxyethyl arm, and a butylamino arm in place of the ethylamino or propylamino arm on the outcome of one pot condensations with various salicylaldehydes in the presence of copper(II) ions has been studied. Hence the synthesis of four new unsymmetrical tripodal triamines, 2-((4-aminobutyl)(pyridin-2-ylmethyl)amino)ethanol (3), 3-((2-aminoethyl)(pyridin-2-ylmethyl)amino)propan-1-ol (4), 3-((3-aminopropyl)(pyridin-2-ylmethyl)amino)propan-1-ol (5) or 3-((4-aminobutyl)(pyridin-2-ylmethyl)amino)propan-1-ol (6), is reported (Fig. 1). The synthesis and physical properties of the new Cu(II) complexes of the new ligands, HLH3–6, HLOMe3–6 and HLtBu3–6 formed in situ from the condensation of 2-hydroxybenzaldehyde with 3–6 (HLH3–6), 2-hydroxy-3-methoxy-benzaldehyde with 3–6 (HLOMe3–6), 3,5-di-tert-butylsalicylaldehyde with 3–6 (HLtBu3–6) and, for the first time, 2-hydroxy-1-naphthaldehyde with 1–6 (HLnapht1–6), are reported here. In addition, the X-ray crystal structures of [CuIILnapht2](ClO4), [CuIILOMe3](ClO4), [CuIILH3](ClO4), [CuIILH4](ClO4) and dimeric [CuIILH5]2(ClO4)2 are described.
Results and discussion
Four new unsymmetrical tripodal amines, 2-((4-aminobutyl)(pyridin-2-ylmethyl)amino)ethanol (3), 3-((2-aminoethyl)(pyridin-2-ylmethyl)amino)propan-1-ol (4), 3-((3-aminopropyl)(pyridin-2-ylmethyl)amino)propan-1-ol (5) and 3-((4-aminobutyl)(pyridin-2-ylmethyl)amino)propan-1-ol (6), were prepared in high yields. Amines 3–6 differ from the previously used amines, 1 and 2, in that 4 and 5 feature a 3-hydroxypropyl arm in place of the 2-hydroxyethyl arm, 3 features a butylamino arm in place of the ethylamino/propylamino arm and 6 features a 3-hydroxypropyl arm in place of the 2-hydroxyethyl arm and also a butylamino arm in place of the ethylamino/propylamino arm (Fig. S1–S12, ESI†). Subsequently, one pot reactions of amines 1–6 with 2-hydroxy-1-naphthaldehyde and salicylaldehyde derivatives in the presence of a Cu(II) metal salt were employed to generate new Cu(II) complexes of Schiff-base ligands Laldi, where aldi is H3–6, OMe3–6, tBu3–6 and napht1–6 (Fig. 1). Recrystallisation of powders obtained from the reaction mixture by vapour diffusion of diethyl ether (see the Experimental section below) gave either purified powders or single crystals which were analysed by single crystal X-ray diffraction, showing four monomeric and one dimeric compounds (vide infra).The infrared spectra of all complexes (Fig. S13–S30, ESI†) show a band at ca. 1613–1632 cm−1, attributable to the imine groups, and no bands due to ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) vibrations. Medium to strong bands at ca. 1596–1612 and 1437–1462 cm−1 are present in all cases, and correspond to the two highest energy ring vibrations of the coordinated pyridine.14,15 Absorptions attributable to the perchlorate ions are seen at approximately 1051–1088 and 619–625 cm−1. The lack of splitting suggests that they are not coordinated.
O) vibrations. Medium to strong bands at ca. 1596–1612 and 1437–1462 cm−1 are present in all cases, and correspond to the two highest energy ring vibrations of the coordinated pyridine.14,15 Absorptions attributable to the perchlorate ions are seen at approximately 1051–1088 and 619–625 cm−1. The lack of splitting suggests that they are not coordinated.
The positive ion electrospray mass spectra of all complexes (Fig. S31–S48, ESI†) show a common peak, which is the fragment [CuIILaldi]+ associated with the loss of the ClO4− anion. The four copper(II) complexes using (Lald5)− as a ligand appear to be dimeric complexes [CuIILald5]2(ClO4)2 as the mass spectra exhibit peaks of very weak intensity consistent with the presence of a dication [CuLald5]22+. In all copper(II) complexes the most intense peaks are for the mononuclear [CuIILaldi]+ species which indicates that the dimer is unsurprisingly broken apart. On the other hand, a peak with a very weak intensity corresponding to the [MLaldi]2+ fragment is observed in most of the mononuclear complexes in present work and also in our previous work,14 even when their X-ray crystal structures show that they are mononuclear complexes. Thus it seems that the [MLaldi]2+ fragment observed in the mass spectra of the mononuclear complexes is formed due to a very small dimerization occurring in the mass spectrometer.
UV-Vis spectra of the fourtheen Cu(II) complexes in CH3CN solution showed a broad low-intensity absorption band occurring in the range 574 nm < λmax < 630 nm with molar extinction coefficient ranging between 92 M−1 cm−1 < ε < 174 M−1 cm−1. This is assigned to a d–d transition and is characteristic of five-coordinate copper(II) complexes with square pyramidal or distorted square pyramidal geometries, which generally exhibit a band in the 550–660 nm range (dxz, dyz → dx2–y2).16–24 In the case of four Cu complexes [CuIILaldi]ClO4 (i = 3), the respective λmax values in the range 600 nm < λmax < 633 nm and 121 M−1 cm−1 < ε < 157 M−1 cm−1 (each with a shoulder at 761–819 nm) are also indicative of square-pyramidal coordination according to the literature.25,26 In addition, a few absorption bands are found in the range 205–406 nm for all Cu(II) complexes, due to either charge transfer or π–π* transitions.19,21,27–29 Although the UV-Vis spectra of complexes with polydentate Schiff base ligands are not generally good indicators of geometry, the evidence gathered helps to support this geometry.
Room temperature magnetic moments were obtained for all mononuclear Cu(II) complexes. The magnetic moment values for these complexes lie in the 1.82–1.95 BM range. These values are close to the expected spin only magnetic moment value (1.73 BM) for the d9 Cu(II) system30 with single unpaired electron. For the four dinuclear copper complexes, the observed values of magnetic moment lie in the 1.32–1.66 BM range per Cu atom.
Crystal structures of [CuIILnapht2](ClO4), [CuIILH3](ClO4), [CuIILOMe3](ClO4) and [CuIILH4](ClO4)
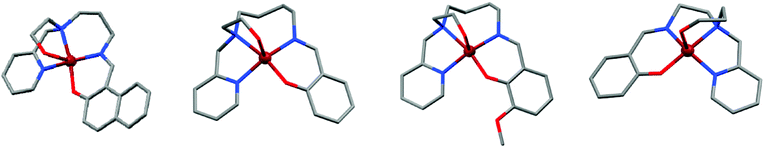
Green single crystals of [CuIILnapht2](ClO4), [CuIILH3](ClO4), [CuIILOMe3](ClO4) and [CuIILH4](ClO4) suitable to be studied by X-ray diffraction were obtained by slow diffusion of diethyl ether into a solution of the complex in MeOH. Crystal data and structure refinement are given in Table 1. These complexes are monometallic but differ in the space group adopted (P21/c, C2/c, P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) and I2/a, respectively). The molecular structures as well as selected bond lengths and angles are given in Fig. 2 and Table 2, respectively, and a comparison with the literature is also shown. The X-ray crystal structures of these complexes consist of [CuIILnapht2]+, [CuIILH3]+, [CuIILOMe3]+ and [CuIILH4]+ cations and perchlorate anions. The Cu(II) ion displays a distorted square pyramidal coordination, involving three N atoms and two O atoms. In comparison to the mononuclear [NiIILOMe1](ClO4) complex reported in our previous work,14 in which the hydroxyl group is not coordinated, in these mononuclear copper(II) complexes it is coordinated to the apical site of the approximate square pyramidal copper(II) ion (Table 2). As expected, this axially bound O donor atom makes a bond that is slightly longer than bond distances in the basal plane (∼2 Å). Among the Cu–N bonds, those involving the tertiary amine nitrogen atoms are the longest in all Cu complexes. The second longest Cu–N bond formed in both mononuclear and dinuclear complexes involves the Cu–Npy bonds. Comparison of the same bond lengths of square pyramidal Cu(II) complexes reported here with related reports in the literature is summarised in Table 2.14,19,31–38 The X-ray crystal structure analysis shows that in the case of [CuIILOMe3](ClO4), two [CuIILOMe3]+ cations are bonded through hydrogen bonding. Indeed, the hydrogen atom of the hydroxyl group of one cation is engaged in hydrogen bonding with the phenolic oxygen atom of the adjacent cation and vice versa (Fig. 5a). It seems that these intermolecular interactions between two molecules of such five coordinate complexes in [CuIILOMe3](ClO4) are relatively strong and prevent the formation of dinuclear compounds (see the Theoretical studies section). Note that the dataset for [CuIILOMe3](ClO4) was particularly bad, leading to a high R1 factor, this despite our best efforts to grow better crystals, the results presented here are from the best dataset obtained.
and I2/a, respectively). The molecular structures as well as selected bond lengths and angles are given in Fig. 2 and Table 2, respectively, and a comparison with the literature is also shown. The X-ray crystal structures of these complexes consist of [CuIILnapht2]+, [CuIILH3]+, [CuIILOMe3]+ and [CuIILH4]+ cations and perchlorate anions. The Cu(II) ion displays a distorted square pyramidal coordination, involving three N atoms and two O atoms. In comparison to the mononuclear [NiIILOMe1](ClO4) complex reported in our previous work,14 in which the hydroxyl group is not coordinated, in these mononuclear copper(II) complexes it is coordinated to the apical site of the approximate square pyramidal copper(II) ion (Table 2). As expected, this axially bound O donor atom makes a bond that is slightly longer than bond distances in the basal plane (∼2 Å). Among the Cu–N bonds, those involving the tertiary amine nitrogen atoms are the longest in all Cu complexes. The second longest Cu–N bond formed in both mononuclear and dinuclear complexes involves the Cu–Npy bonds. Comparison of the same bond lengths of square pyramidal Cu(II) complexes reported here with related reports in the literature is summarised in Table 2.14,19,31–38 The X-ray crystal structure analysis shows that in the case of [CuIILOMe3](ClO4), two [CuIILOMe3]+ cations are bonded through hydrogen bonding. Indeed, the hydrogen atom of the hydroxyl group of one cation is engaged in hydrogen bonding with the phenolic oxygen atom of the adjacent cation and vice versa (Fig. 5a). It seems that these intermolecular interactions between two molecules of such five coordinate complexes in [CuIILOMe3](ClO4) are relatively strong and prevent the formation of dinuclear compounds (see the Theoretical studies section). Note that the dataset for [CuIILOMe3](ClO4) was particularly bad, leading to a high R1 factor, this despite our best efforts to grow better crystals, the results presented here are from the best dataset obtained.
| Compound | [CuIILnapht2](ClO4) | [CuIILH3](ClO4) | [CuIILOMe3](ClO4) | [CuIILH4](ClO4) | [CuIILH5]2(ClO4)2 |
|---|---|---|---|---|---|
| Formula | C22H24ClCuN3O6 | C19H24ClCuN3O6 | C44H62Cl2Cu2N6O15 | C18H22ClCuN3O6 | C38H48Cl2Cu2N6O12 |
| Molecular weight (g mol−1) | 525.43 | 488.39 | 1112.98 | 475.38 | 978.80 |
| T (K) | 100(2) | 296 | 100(2) | 89(2) | 100(2) |
| Crystal system | Monoclinic | Monoclinic | Triclinic | Monoclinic | Orthorhombic |
| Space group | P21/c | C2/c |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
I2/a | Pbca |
| Z | 4 | 8 | 4 | 8 | 4 |
| a (Å) | 13.1270(2) | 11.5021(3) | 13.9094(3) | 19.0697(4) | 12.2844(2) |
| b (Å) | 13.7157(2) | 18.7307(5) | 18.2391(4) | 10.6833(2) | 14.1806(2) |
| c (Å) | 12.1763(2) | 20.7277(7) | 20.6775(6) | 18.9974(3) | 23.1490(3) |
| α (°) | 90 | 90 | 90.039(2) | 90 | 90 |
| β (°) | 103.203(2) | 95.504(1) | 91.901(2) | 95.766(2) | 90 |
| γ (°) | 90 | 90 | 111.884(2) | 90 | 90 |
| V (Å3) | 2134.34(6) | 4445.0(2) | 4864.6(2) | 3850.71(12) | 4032.56(10) |
| Density (g cm−3) | 1.635 | 1.460 | 1.520 | 1.640 | 1.612 |
| R 1 | 0.0308 | 0.045 | 0.1565 | 0.0382 | 0.0878 |
| wR2 | 0.0809 | 0.138 | 0.3908 | 0.1218 | 0.2657 |
| [CuIILnapht2](ClO4) | [CuIILH3](ClO4) | [CuIILOMe3](ClO4) | [CuIILH4](ClO) | [CuIILH5]2(ClO)2 | Sq pyr Cu(II) in the literature | Ref. | |
|---|---|---|---|---|---|---|---|
| Bond length [Å] | |||||||
| M(1)–N(imine) | 1.9486(15) | 1.989(3) | 2.008(10) | 1.929(2) | 1.969(7) | 1.923–1.969 | 14 and 31 |
| M(1)–N(py) | 2.0122(16) | 2.025(3) | 2.014(11) | 1.979(2) | 2.006(7) | 1.925–2.006 | 31 and 32 |
| M(1)–N(amine) | 2.0910(15) | 2.125(2) | 2.148(9) | 2.062(2) | 2.057(7) | 1.979–2.062 | 19 and 32–34 |
| M(1)–O(phenolic) | 1.9234(12) | 1.921(2) | 1.927(8) | 1.9172(17) | 1.937(5) | 1.917–2.096 | 19, 31 and 35–38 |
| M(1)⋯O(hydroxyalkyl) | 2.3802(13) | 2.241(2) | 2.222(9) | 2.2195(18) | 5.415 | 1.916–2.339 | 19 |
| M(1)–O(2)#1 | 2.407(6) | 2.423 | 38 | ||||
| M(1)⋯M(1) | 3.240 | 2.2729–3.001 | 19 and 31 | ||||
| Bond angle [°] | |||||||
| O(phenolic)–M(1)–N(imine) | 91.89(6) | 90.19(9) | 91.0(4) | 95.54(8) | 92.6(3) | ||
| O(phenolic)–M(1)–N(py) | 90.98(6) | 88.29(10) | 86.4(4) | 94.89(8) | 90.5(3) | ||
| N(imine)–M(1)–N(py) | 164.51(6) | 159.96(11) | 160.0(4) | 155.64(9) | 172.0(3) | ||
| O(phenolic)–M(1)–N(amine) | 170.51(6) | 166.57(10) | 164.0(4) | 179.08(8) | 165.4(3) | ||
| N(imine)–M(1)–N(amine) | 96.21(6) | 102.86(11) | 104.0(4) | 86.37(9) | 94.2(3) | ||
| N(py)–M(1)–N(amine) | 82.53(6) | 80.54(11) | 81.3(4) | 84.27(8) | 81.2(3) | ||
| O(phenolic)–M(1)–O(hydroxyalkyl) | 91.77(5) | 94.75(9) | 94.3(3) | 90.62(7) | |||
| N(imine)–M(1)–O(hydroxyalkyl) | 109.15(5) | 99.15(10) | 97.1(4) | 95.53(8) | |||
| N(py)–M(1)–O(hydroxyalkyl) | 85.96(5) | 100.92(10) | 102.9(4) | 106.77(8) | |||
| N(amine)–M(1)–O(hydroxyalkyl) | 80.92(5) | 80.2(9) | 78.6(3) | 89.27(7) | |||
| O(phenolic)–M(1)–O(2)#1 | 84.2(2) | ||||||
| N(imine)–M(1)–O(2)#1 | 91.0(2) | ||||||
| N(py)–M(1)–O(2)#1 | 96.7(2) | ||||||
| N(amine)–M(1)–O(2)#1 | 108.6(2) | ||||||
Variation of the length of the ligand arms leads to different sizes of chelate rings. These tripodal ligands are capable of forming both five and six membered chelate rings incorporating the copper ion in [CuIILnapht2](ClO4), [CuIILH4](ClO4) and [CuIILH5]2(ClO4)2 and also five, six and seven membered chelate rings in [CuIILH3](ClO4) and [CuIILOMe3](ClO4). For all complexes, the Namine–Cu–Npy angles are smaller than 90° [80.5–84.3°] for five membered chelate rings. The larger six-membered chelate rings lead to Ophenolic–Cu–Nimine angles that are all larger than 90° [90.2–94.5°]. A similar relationship between the Namine–Cu–Ohydroxyalkyl and also Namine–Cu–Nimine angles and the different chelate ring sizes is described. In the case of [CuIILnapht2](ClO4), [CuIILH3](ClO4) and [CuIILOMe3](ClO4) complexes, involving the 2-hydroxyethyl arm, the Namine–Cu–Ohydroxyalkyl angles are smaller than 90° [78.6–80.9°] for five membered chelate rings, whilst in the [CuIILH4](ClO4) complex involving the 3-hydroxypropyl arm, the Namine–Cu–Ohydroxyalkyl angle is ∼90° [89.27°] for the six-membered ring. The Namine–Cu–Nimine angle in [CuIILH4](ClO4) involving the ethylamine chain [86.37°] is smaller than 90° for the five membered chelate ring, in [CuIILnapht2](ClO4) and [CuIILH5]2(ClO4)2 complexes, involving the propylamine chain [94.2–96.2°] and also in [CuIILH3](ClO4) and [CuIILOMe3](ClO4) complexes involving the butylamine chain [102.9–104.0°] is larger than 90° for six and seven membered chelate rings, respectively (Table 2). The square pyramid in Cu complexes is somewhat trigonally distorted, as shown by the degree of trigonality, (τ),39–41 for Cu(1) being 0.10, 0.11, 0.066 and 0.39 for [CuIILnapht2](ClO4), [CuIILH3](ClO4), [CuIILOMe3](ClO4) and [CuIILH4](ClO4), respectively.
Crystal structure of [CuIILH5]2(ClO4)2
Green single crystals of [CuIILH5]2(ClO4)2 were obtained by slow diffusion of diethyl ether into a solution of the complex dissolved in a mixture of CH3CN and CH3OH, and crystallise in the orthorhombic crystal system and the Pbca space group. The molecular structure and selected bond lengths and bond angles related to the coordination environment of the metal and also bond lengths related to similar compounds are given in Fig. 3 and Table 2, respectively. The structure is a dinuclear, comprising two similar Cu(II) centers. Each copper atom has a pentacoordinate square-pyramidal geometry. N(1), N(2), N(3) and O(2) of a deprotonated Schiff base bind four coordination sites of Cu(1). Similarly, Cu(2) is coordinated by N(1), N(2), N(3) and O(2) of another deprotonated Schiff base. The fifth, apical, coordination site of each Cu(1) is occupied by O(2) from another ligand, thereby forming a di-phenoxido-bridged dimer, while the hydroxypropyl arm (O(1)) of the ligand remains uncoordinated (Fig. 3). The charge distribution was assigned based on the presence of only two ClO4− anions which make the complex a dication with two deprotonated ligands LH5−. Each phenoxy oxygen atom is bridging two complexes in an antisymmetric fashion giving Cu–Ophenoxy bond lengths of 2.407 and 1.937 Å – which are in the range of previously reported structures for the copper(II) dimeric complex having a phenoxy bridge.36–38 The coordination geometry around the copper centers is best described by the use of the τ-criterion,37 indicating that the coordination geometry in [CuIILH5]2+ is only slightly from square-pyramidal distorted (τ = 0.11). In this compound, N(1), N(2), N(3) and O(2) related to the same ligand occupy the equatorial positions and O(2)′, from the second ligand, occupies the apical position. The τ value for both Cu(1) in [CuIILH5]2(ClO4)2 is 0.11, which is similar to the value of Cu(II) in the monomers. Thus the geometries of both the copper centers in [CuIILH5]2(ClO4)2 are distorted square pyramidal. A 5,6,6-chelate ring sequence is observed in the dimer [CuIILH5]2+, with the expected square-pyramidal coordination geometry for both Cu(II). | ||
| Fig. 3 Perspective of [CuIILH5]2(ClO4)2. Hydrogen atoms and anions are omitted for clarity. Cu, C, N and O are represented in dark red, gray, blue and red, respectively. | ||
In contrast to coordinated ligands Lnapht2, LH3, LOMe3 and LH4 in [CuIILnapht2](ClO4), [CuIILH3](ClO4), [CuIILOMe3](ClO4) and [CuIILH4](ClO4) complexes, respectively, the ligand anion in complex [CuIILH5]2+ uses only four of the five donor groups binding to the copper atom, in which the hydroxypropyl arm remains uncoordinated and the phenolic oxygen atom bridges two Cu(II) atoms resulting in a Cu2O2 ring. This is not exclusively due to the steric situation in [CuIILH5]2(ClO4)2, but to the low stability of the six-membered chelate ring which would have been formed with the hydroxypropyl ligand arm: a similar behavior of related aliphatic tripodal ligands10 and also asymmetric tripodal ligands with two aliphatic and one aromatic arms has been already reported.26 In addition, the stability of the copper complex with the ligand trpn, which exclusively forms six-membered chelate rings, is shown to be about 105 times lower than the stability of the corresponding tren complex that contains only five-membered chelate rings.9c Examination of the Cu(II) complexes in the present study and the literature shows that Cu(II) atoms in tripodal complexes have a great flexibility in adoption of the number of chelate ring sequences around the metal atom to form a square pyramidal geometry around the central ion as observed for [CuIILnapht2](ClO4), 5,5,6,6, τ = 0.10; [CuIILH3] (ClO4), 5,5,7,6, τ = 0.11; [CuIILOMe3](ClO4), 5,5,7,6, τ = 0.066; [CuIILH4](ClO4), 5,6,5,6, τ = 0.39 and [CuIILH5]2(ClO4)2, 5,6,6, τ = 0.11, but it should be noted that Cu(II) in these types of complexes is not stable against the high number of six-membered chelate rings around the metal.9c For the dinuclear complex [CuIILH5]2(ClO4)2 the hypothetical mononuclear [CuIILH5](ClO4) would possess a 5,6,6,6 chelate ring sequence around the Cu(II) atom which, due to the high number of six-membered chelate rings, would be unstable. In order to form the stable structure with square pyramidal geometry, the hypothetical mononuclear complex would prefer to form a dinuclear structure with a 5,6,6 chelate ring sequence with a second identical ligand. Table 3 shows the comparison of the structural parameter (τ-value) for Cu(II) complexes characterized here.
| 5-Coordinated complexes | Chelate ring sequence | τ-Value |
|---|---|---|
| [CuIILnapht2](ClO4) | 5,5,6,6 | 0.10 |
| [CuIILH3](ClO4) | 5,5,7,6 | 0.11 |
| [CuIILOMe3](ClO4) | 5,5,7,6 | 0.066 |
| [CuIILH4](ClO4) | 5,6,5,6 | 0.39 |
| [CuIILH5]2(ClO4)2 | 5,6,6 | 0.11 |
Theoretical studies
As described above, the present study brought us three interesting results. Firstly, we observed that the mononuclear five-coordinate complexes can be formed from most of ligands but dinuclear complexes only with a few of them. Secondly, X-ray crystal structures showed that hydrogen bonding can exist between two mononuclear five-coordinate complexes. Thirdly, mass spectra of both mononuclear and dinuclear complexes always show two characteristic peaks, one corresponding to a mononuclear fragment and one corresponding to a dinuclear fragment. In order to understand these results theoretical calculations were undertaken. The strength of interaction between two mononuclear four-coordinated fragments in one dinuclear complex and hydrogen bonding between two mononuclear five-coordinated complexes were evaluated (see Fig. 4). From this study it is clear that one of the coordinated arms in the mononulear complex leaves the metal ion and then the resulting four-coordinated complex can be dimerized. As can be seen in Fig. 3 and 4(a) in these dinuclear complexes, the hydroxyl group of the hydroxypropoyl arm is not coordinated to the metal ion. Thus the strength of the interaction between the hydroxyl group and the metal ion was evaluated, to figure out why it leaves the metal ion. However, the value of interaction energy between the metal ion and the whole ligand in these complexes was calculated first. As can be seen in Table 4, the values of interaction energies are relatively large and are in the range 629–634 kcal mol−1. Thus in all mononuclear complexes the Schiff base ligands are tightly bonded to the central metal ion. However, the data show that the interaction energy between two mononuclear fragments in the dinuclear complex [CuIILH5]2(ClO4)2 is very small and only −4.24 kcal mol−1 (see Table 5). Such a small interaction energy is not surprising as the interacting fragments are both cations. This explains why even in the mass spectra of the dinuclear complexes the major peak corresponds to a mononuclear complex. Indeed, inside the mass spectrometer the dinuclear complex readily breaks into two mononuclear complexes. Two forms, I and II, were considered for one of the mononuclear fragments of the dinuclear complex [CuIILH5]2(ClO4)2 and the geometry was optimised for both of them (see Fig. 5e and f). The difference between the above forms is that in form I the hydroxyl group is not coordinated to the metal ion but is indeed coordinated in form II. The data showed that the energy difference between the above two forms is only −2.57 kcal mol−1. Thus the interaction between the hydroxyl group and the central metal ion seems to be very weak. This means that the hydroxyl group simply leaves the metal ion, resulting in four-coordinated copper complexes which can be dimerized. This is the reason why in mass spectra of all complexes there is a very small peak corresponding to a dinuclear complex. | ||
| Fig. 4 Interacting four-coordinated fragments in dinuclear complex [CuIILH5]2(ClO4)2 (a) and hydrogen bonding between two [CuIILOMe3](ClO4) complexes (b). | ||
| Compound | E el (Hartree) | IE (kcal mol−1) | ||
|---|---|---|---|---|
| Cu2+ |
Lald−![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a a |
[CuLald]+ | ||
| a Frozen in the optimized geometry of the [CuLald]+ complex. | ||||
| [CuIILOMe3](ClO4) | −1639.151635 | −1165.9898167 | −2806.1509036 | −633.44 |
| [CuIILH3](ClO4) | −1639.151635 | −1051.6149949 | −2691.7726555 | −631.29 |
| [CuIILnapht2](ClO4) | −1639.151635 | −1165.7856412 | −2805.939729 | −629.05 |
| [CuIILH4](ClO4) | −1639.151635 | −1012.3607015 | −2652.5239637 | −634.81 |
Among the complexes synthesized here, both [CuIILH5]2(ClO4)2 and [CuIILH4](ClO4) complexes have the hydroxyl group at the end of a propyl chain. For all the other complexes the hydroxyl group is at the end of an ethyl chain. Indeed, only in the case of [CuIILH5]2(ClO4)2 and [CuIILH4](ClO4) we do observe an unstable six-membered chelate ring forming upon coordination of the hydroxyl group. For all other complexes the coordination of the hydroxyl group leads to the formation of a more stable five-membered chelate ring. Thus it seems that the formation of a dinuclear complex in which the hydroxyl group remained uncoordinated is quite expectable for both [CuIILH5]2(ClO4)2 and [CuIILH4](ClO4) complexes. However X-ray crystal structure analysis confirmed the formation of a dinuclear complex only in the case of the former complex. Thus two forms I and II for [CuIILH4](ClO4) were also considered and optimized (see Fig. 5c and d). As can be seen in Table 6, the data show that the energy difference between the above forms is about −5.32 kcal mol−1. Thus it seems that the interaction energy between the hydroxyl group and the metal ion in complex [CuIILH4](ClO4) is relatively large than that in complex [CuIILH5]2(ClO4)2. On the other hand, the interaction energy between two mononuclear fragments in the dinuclear complex [CuIILH5]2(ClO4)2 was only −4.24 kcal mol−1. The above interaction is larger than −2.57 kcal mol−1 and less than −5.3 kcal mol−1, calculated energy differences between forms I and II in complexes [CuIILH5]2(ClO4)2 and [CuIILH4](ClO4), respectively. This explains why the complex [CuIILH5]2(ClO4)2 is dimerized while [CuIILH4](ClO4) remained mononuclear. For [CuIILH5]2(ClO4)2, in contrast to [CuIILH4](ClO4), the value of interaction energy between two four coordinated fragments is larger than that between the hydroxyl group and the central metal ion.
| Compound | E el (Hartree) | ΔE (kcal mol−1) | |
|---|---|---|---|
| Form I | Form II | ||
| [CuIILH4](ClO4) | −2691.7690481 | −2691.7731458 | −5.32 |
| [CuIILH5]2(ClO4)2 | −2652.5239637 | −2652.5154793 | −2.57 |
In addition, we believe that the formation of hydrogen bonding between two mononuclear five-coordinated complexes prevents the formation of a dinuclear complex between two four-coordinated fragments. As seen in the previous section the X-ray crystal structure of the complex [CuIILOMe3](ClO4) showed that the hydrogen bonding is formed between two mononuclear complexes. Indeed the coordination of the hydroxyl group to the metal ion and then the formation of the hydrogen bond between two molecules of such a five-coordinated complex in [CuIILOMe3](ClO4) prevent the formation of a dinuclear complex which is formed in the case of [CuIILH5]2(ClO4)2. The value of the interaction between two mononuclear [CuIILOMe3](ClO4) complexes due to hydrogen bonding is about −7.28 kcal mol−1 (see Fig. 4). Interestingly, the latter value is larger than −4.24 kcal mol−1, and the interaction between two fragments in dinuclear complex [CuIILH5]2(ClO4)2 is larger than −2.57 and −5.3 kcal mol−1, the energy difference between forms I and II of complexes [CuIILH5]2(ClO4)2 and [CuIILH4](ClO4), respectively. Therefore these complexes are special cases in which the intermolecular interactions (herein hydrogen bonding) between two complexes can be stronger than some weak intramolecular metal–donor atom interactions. The formation of a mononuclear complex or a dinuclear one thus depends on the relative strength of inter- and intramolecular interactions.
Conclusion
In summary, we have reported the successful synthesis of 14 new mononuclear and a few dinuclear Cu(II) complexes by condensation of amines (3–6) with related aldehydes in the presence of Cu(II) metal ions. X-ray crystal structure determinations of [CuIILnapht2](ClO4), [CuIILH3](ClO4), [CuIILOMe3](ClO4) and [CuIILH4](ClO4) revealed them to be monomeric, except for the dimeric [CuIILH5]2(ClO4)2. There is a distorted square pyramidal environment around the central ion in both mononuclear and dinuclear complexes. In this work, emphasis has been put on the examination of structural relationships in the complexation behavior. By comparing crystal structures of the complexes identified to date, it is found that all of them use four donor groups of the ligand (three nitrogen atoms and phenoxy oxygen atoms) to bind the metal atom and the differences observed are related to hydroxyalkyl arms. These arms are not coordinated to the metal ion in all complexes but in both the coordinated and non-coordinated state they remain protonated. Examination of X-ray crystal structures of Cu(II) complexes in the present study shows that in the Cu(II) complexes with an ethyl-amino arm, the hydroxypropyl group is bound to the Cu(II) atom, while in the Cu(II) complexes with a propyl-amino arm, the hydroxypropyl arm remains uncoordinated and a dinuclear Cu(II) complex is formed. According to the theoretical study, the interaction between the hydroxyl group and the central metal ion seems to be very weak thus the hydroxyl group can leave the metal ion, yielding four-coordinated Cu(II) complexes which can be dimerized. The theoretical study also showed that the formation of a mononuclear complex or a dinuclear one depends on the relative strength of inter- and intramolecular interactions. In Cu(II) complexes with a hydroxyethyl arm, there is a stronger interaction between the hydroxyl group and the central metal ion due to the formation of a stable five-membered chelate ring upon coordination of the hydroxyl group to the metal ion. In the case of the Cu(II) complexes with a hydroxypropyl arm, an unstable six-membered chelate ring forms upon coordination of the hydroxyl group – the formation of dinuclear structures is more expectable in this case. Note that both ligands LH5 and LH4 have a hydroxypropyl arm, but the X-ray crystal structure analysis confirmed the formation of a dinuclear copper complex only in the case of LH5. By comparison of the structures, it seems that both aliphatic linkages are effective on the structure of dimeric complexes. According to theoretical studies, the value of interaction energy between two four coordinate fragments in the dinuclear [CuIILH5]2(ClO4)2 complex, in contrast to [CuIILH4](ClO4), is larger than that between the hydroxyl group and the central metal ion. Thus we can assume that a mononuclear five coordinated complex forms first, and in a second time the hydroxyl group leaves the metal ion, resulting in a four-coordinated complex which can be dimerized. In addition to intramolecular interactions, intermolecular interactions are also important in the formation of a mononuclear complex or a dinuclear one. The theoretical study also supported our assumption that the formation of hydrogen bonds between two mononuclear five-coordinated complexes prevents the formation of a dinuclear complex between two four coordinate fragments.Experimental
General remarks
Pyridine 2-carbaldehyde, 2-hydroxy-3-methoxybenzaldehyde, 2-hydroxybenzaldehyde, 2-hydroxy-1-naphthaldehyde and metal salt were obtained from Aldrich and used without further purification. 3,5-Di-tert-butyl-2-hydroxybenzaldehyde was synthesised according to the literature procedure.42 All other chemicals and solvents were of reagent grade and were used as received except for methanol that was dried (Mg) before use.Caution!
Whilst no problems were encountered in the course of this work, perchlorate mixtures are potentially explosive and should therefore be handled with appropriate care.Infrared spectra were obtained between 4000 and 400 cm−1 on a Bruker Alpha FT-ATR IR spectrometer with a diamond anvil Alpha-P module for all complexese. UV-Vis spectra were recorded on a Jasco V550 spectrophotometer. ESI mass spectra were recorded at the University of Otago on a Bruker MicrOTOFQ spectrometer exception for [CuIILH3](ClO4), [CuIILOMe3](ClO4) and [CuIILtBu3](ClO4) complexes that the spectra were recorded using a Kratos-MS-50T spectrometer. Room temperature magnetic moments were determined using a Johnson Matthey MSB-MK1 magnetic susceptibility balance. Standard microanalysis for all complexes was carried out using a Perkin-Elmer, CHNS/O elemental analyzer model 2400. 1H and 13C NMR spectra were taken in CDCl3 on a Jeol 90 MHz spectrometer using Si(CH3)4 as an internal standard. Crystals suitable for X-ray diffraction were obtained by slow diffusion of diethyl ether vapor into methanol or a mixture of methanol and acetonitrile. Single crystal X-ray crystallographic data were collected at 100 K for [CuIILnapht2](ClO4), [CuIILOMe3](ClO4) and [CuIILH5]2(ClO4)2 (λ = 1.54184) and also at 89 K for [CuIILH4](ClO4) (λ = 0.71073) on a Bruker Kappa APEX II area detector diffractometer (University of Otago), using graphite monochromatic Mo-Kα radiation. In the case of the [CuIILH3](ClO4) complex, single crystal X-ray crystallographic data were collected at 296 K (λ = 1.54184) on a Bruker SMART BREEZE CCD diffractometer using APEX2 software.43 The data were collected for Lorentz and polarization effects and semi-empirical absorption corrections (SCALE) were applied. The structures were solved by direct or Patterson methods (SHELXS-97)44 and refined against all F2 data (SHELX-97).45 All non-hydrogen atoms were modelled anisotropically except where noted. Unless otherwise specified, hydrogen atoms were inserted at calculated positions and rode on the atoms to which they were attached. In the case of this complex, absorption correction was applied to collected data using multi-scan, SADAPS V2012/1 software.43 The title compound was solved by direct methods46 using SHELXS-97 and refined using SHELXL-97.46 The weighted R-factor, wR and goodness of fit S are based on F2. The threshold expression of F2 > 2 sigma (F2) is used only for calculating R-factors. All estimated standard deviations (e.s.ds) are estimated using the full covariance matrix. The cell e.s.ds are taken into account individually in the estimation of e.s.d.s in distances, angles, and torsion angles; correlations between e.s.d.s in cell parameters are only used when they are defined by crystal symmetry. All non-hydrogen atoms were refined anisotropically and hydrogen atoms were added according to the theoretical model. In the case of complex [CuIILOMe3](ClO4), we were not able to refine the X-ray crystal structure of this complex exactly.
Computational details
The crystallographic structure of all [CuLaldi]+ complexes was fully optimized using the def2-SVP basis set at the M0647 level of theory. The calculated bond lengths and bond angles in optimized complexes were in good agreement with corresponding experimental data. Calculated root mean squares (RMSs) for metal–ligand bond distances were less than 0.055 (see Table 7). The interaction energy between the Cu2+ metal ion and the anionic ligand in mononulear complexes was calculated with the following equation:| ΔE = EAB − (EABA + EABB), |
| Compounds | Cu–Namine | Cu–Npy | Cu–Nimine | Cu–O− | Cu–OH | RMS |
|---|---|---|---|---|---|---|
| a The data obtained at the M06/def2-SVP level are given as a plain text and experimental data are in italic. b For this complex this is the distance between Cu(1)–O-(2) bonds. | ||||||
| [CuIILOMe3](ClO4) | 2.190 | 2.045 | 2.009 | 1.944 | 2.209 | 0.041 |
| 2.136 | 2.039 | 2.013 | 2.013 | 2.232 | ||
| [CuIILH3](ClO4) | 2.227 | 2.046 | 1.985 | 1.916 | 2.298 | 0.054 |
| 2.125 | 2.025 | 1.990 | 1.921 | 2.236 | ||
| [CuIILnapht2](ClO4) | 2.140 | 2.030 | 1.940 | 1.916 | 2.339 | 0.030 |
| 2.091 | 2.012 | 1.949 | 1.923 | 2.380 | ||
| [CuIILH4](ClO4) | 2.113 | 2.017 | 1.935 | 1.905 | 2.250 | 0.032 |
| 2.062 | 1.979 | 1.928 | 1.917 | 2.220 | ||
| [CuIILH5]2(ClO4)2 | 2.102 | 2.027 | 1.988 | 1.975 | 2.340b | 0.042 |
| 2.056 | 2.006 | 1.970 | 1.938 | 2.407 | ||
Synthesis and characterization
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)py. 1H NMR (CDCl3, ppm) δ = 1.36–1.55 (m, 4H); 2.33 (b, 3H), 2.55–2.72 (m, 6H); 3.58–3.60 (t, 2H); 3.78 (s, 2H); 7.14–7.17 (m, 1H); 7.14–7.17 (t, 1H); 7.29 (d, 1H); 7.62–7.66 (td, 1H); 8.52 (d, 1H). 13C NMR (CDCl3, ppm) δ = 24.057; 29.248; 40.432; 54.065; 56.110; 58.750; 59.688; 121.349, 122.493; 135.879; 148.314; 159.228.
N)py. 1H NMR (CDCl3, ppm) δ = 1.36–1.55 (m, 4H); 2.33 (b, 3H), 2.55–2.72 (m, 6H); 3.58–3.60 (t, 2H); 3.78 (s, 2H); 7.14–7.17 (m, 1H); 7.14–7.17 (t, 1H); 7.29 (d, 1H); 7.62–7.66 (td, 1H); 8.52 (d, 1H). 13C NMR (CDCl3, ppm) δ = 24.057; 29.248; 40.432; 54.065; 56.110; 58.750; 59.688; 121.349, 122.493; 135.879; 148.314; 159.228.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)py. 1H NMR (CDCl3, ppm) δ = 1.309–1.625 (m, 2H); 2.432–2.631 (m, 6H); 3.334–3.640 (m, 4H); 4.466 (s, 3H); 6.837–7.403 (m, 3H); 8.234 (d, 1H). 13C NMR (CDCl3, ppm) δ = 29.061; 38.186; 51.476; 54.671; 59.340; 59.826; 121.420, 122.367; 135.981; 148.220; 158.791.
N)py. 1H NMR (CDCl3, ppm) δ = 1.309–1.625 (m, 2H); 2.432–2.631 (m, 6H); 3.334–3.640 (m, 4H); 4.466 (s, 3H); 6.837–7.403 (m, 3H); 8.234 (d, 1H). 13C NMR (CDCl3, ppm) δ = 29.061; 38.186; 51.476; 54.671; 59.340; 59.826; 121.420, 122.367; 135.981; 148.220; 158.791.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)py. 1H NMR (CDCl3, ppm) δ = 1.497 (m, 4H); 2.309–2.546 (m, 6H); 3.482 (s, 4H); 4.436 (s, 3H); 6.856–7.505 (m, 3H); 8.299 (d, 1H). 13C NMR (CDCl3, ppm) δ = 25.886; 28.631; 39.169; 50.729; 51.830; 59.026; 59.715; 121.485, 122.507; 136.132; 148.245; 158.371.
N)py. 1H NMR (CDCl3, ppm) δ = 1.497 (m, 4H); 2.309–2.546 (m, 6H); 3.482 (s, 4H); 4.436 (s, 3H); 6.856–7.505 (m, 3H); 8.299 (d, 1H). 13C NMR (CDCl3, ppm) δ = 25.886; 28.631; 39.169; 50.729; 51.830; 59.026; 59.715; 121.485, 122.507; 136.132; 148.245; 158.371.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)py. 1H NMR (CDCl3, ppm) δ = 1.324–1.503 (m, 6H); 2.275–2.454 (m, 6H); 3.505 (s, 4H); 4.538 (s, 3H); 6.960–7.455 (m, 3H); 8.298 (s, 1H). 13C NMR (CDCl3, ppm) δ = 23.495; 28.598; 29.624; 40.283; 51.612; 53.237; 59.436; 60.711, 121.218; 122.362; 135.758; 148.031, 158.747.
N)py. 1H NMR (CDCl3, ppm) δ = 1.324–1.503 (m, 6H); 2.275–2.454 (m, 6H); 3.505 (s, 4H); 4.538 (s, 3H); 6.960–7.455 (m, 3H); 8.298 (s, 1H). 13C NMR (CDCl3, ppm) δ = 23.495; 28.598; 29.624; 40.283; 51.612; 53.237; 59.436; 60.711, 121.218; 122.362; 135.758; 148.031, 158.747.
General synthesis of the complexes
A solution of 0.5 mmol of appropriate aldehyde, 2-hydroxy-3-methoxy-benzaldehyde (0.08 g, 0.5 mmol), 2-hydroxybenzaldehyde (0.06 g, 0.5 mmol), 3,5-di-tert-butyl-2-hydroxybenzaldehyde (0.117 g, 0.5 mmol) or 2-hydroxy-1-naphthaldehyde (0.086 g, 0.5 mmol) in methanol (50 ml) and 7 drops of N(Et)3 were added dropwise to a refluxing solution of Cu(ClO4)2·6H2O (0.184 g, 0.5 mmol) and the corresponding amine, 1 (0.098 g, 0.5 mmol) or 2 (0.105 g, 0.5 mmol) or 3 (0.112 g, 0.5 mmol) or 4 (0.105 g, 0.5 mmol) or 5 (0.112 g, 0.5 mmol) or 6 (0.119 g, 0.5 mmol) in the same solvent (50 mL). After refluxing for 12 h, the solution was concentrated in a rotary evaporator (EXTREME CAUTION!) at room temperature to ca. 5–10 mL. A small volume of diethyl ether was added slowly, producing a powdery precipitate. The powdery Cu(II) products were filtered off, washed with cold diethyl ether and dried under vacuum. Crystalline or powdery compounds were obtained by slow diffusion of diethyl ether vapor into a solution of these compounds in methanol or a mixture of methanol and acetonitrile, as detailed below.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio via slow vapour diffusion of Et2O yields green powder (0.10 g, 79.8%). Anal. calc. for C21H22ClCuN3O6: C, 49.32; H, 4.34; N, 8.22. Found: C, 49.77; H, 4.55; N, 7.92%. IR (ATR, cm−1) 1616 ν(C
1 ratio via slow vapour diffusion of Et2O yields green powder (0.10 g, 79.8%). Anal. calc. for C21H22ClCuN3O6: C, 49.32; H, 4.34; N, 8.22. Found: C, 49.77; H, 4.55; N, 7.92%. IR (ATR, cm−1) 1616 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)imi, 1604, 1458 ν(C
N)imi, 1604, 1458 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)py and ν(C
N)py and ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1088, 620 ν(ClO4−). ESI-MS (MeOH, m/z+): 411.1 [CuIILnapht1]+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 228 (21
C), 1088, 620 ν(ClO4−). ESI-MS (MeOH, m/z+): 411.1 [CuIILnapht1]+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 228 (21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 097), 390 (2572), 572 (116). Magnetic moment: μeff = 2.2 BM [Gouy].
097), 390 (2572), 572 (116). Magnetic moment: μeff = 2.2 BM [Gouy].
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)imi; 1449 ν(C
N)imi; 1449 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) and ν(C
N) and ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1088, 619 ν(ClO4). ESI-MS (MeOH, m/z+): 425.1 [CuIILnapht2]+, 851.2 ([CuIILnapht2]+2 + H), 949.2 ([CuIILnapht2]+2)ClO4. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 298 (17
C), 1088, 619 ν(ClO4). ESI-MS (MeOH, m/z+): 425.1 [CuIILnapht2]+, 851.2 ([CuIILnapht2]+2 + H), 949.2 ([CuIILnapht2]+2)ClO4. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 298 (17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 419), 387 (6369), 583 (141). Magnetic moment: μeff = 1.72 BM [Gouy].
419), 387 (6369), 583 (141). Magnetic moment: μeff = 1.72 BM [Gouy].
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)imi; 1450 ν(C
N)imi; 1450 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C)py, 1080, 620ν(ClO4). ESI-MS (MeOH, m/z+): 389.1 [CuLH3]+, 777.2 ([CuLH3]2−H)+, 879.2 ([CuIILH3]2 + 2H)ClO4+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 224 (137
C)py, 1080, 620ν(ClO4). ESI-MS (MeOH, m/z+): 389.1 [CuLH3]+, 777.2 ([CuLH3]2−H)+, 879.2 ([CuIILH3]2 + 2H)ClO4+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 224 (137![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 236), 272 (25
236), 272 (25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 701), 374 (1377), 463 (314.12), 612 (128.46), 761 (76.53). Magnetic moment: μeff = 1.84 BM [Gouy].
701), 374 (1377), 463 (314.12), 612 (128.46), 761 (76.53). Magnetic moment: μeff = 1.84 BM [Gouy].
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio via slow vapour diffusion of Et2O yields green crystals (0.10 g, 80%). Anal. calc. for C20H26ClCuN3O7: C, 46.25; H, 5.05; N, 8.09. Found: C, 45.39; H, 5.29; N, 7.72%. IR (ATR, cm−1): 1615 ν(C
1 ratio via slow vapour diffusion of Et2O yields green crystals (0.10 g, 80%). Anal. calc. for C20H26ClCuN3O7: C, 46.25; H, 5.05; N, 8.09. Found: C, 45.39; H, 5.29; N, 7.72%. IR (ATR, cm−1): 1615 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)imi; 1600, 1454 ν(C
N)imi; 1600, 1454 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)py and ν(C
N)py and ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1078, 621 ν(ClO4). ESI-MS (MeOH, m/z+): 419.1 [CuIILOMe3]+, 837.3 ([CuIILOMe3]2 − H)+, 939.2 ([CuIILOMe3]2 + 2H)ClO4+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 238 (31
C), 1078, 621 ν(ClO4). ESI-MS (MeOH, m/z+): 419.1 [CuIILOMe3]+, 837.3 ([CuIILOMe3]2 − H)+, 939.2 ([CuIILOMe3]2 + 2H)ClO4+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 238 (31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 324), 281 (18
324), 281 (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 414), 384 (2537), 483 (303.65), 602 (133.7), 781 (94). Magnetic moment: μeff = 2 BM [Gouy].
414), 384 (2537), 483 (303.65), 602 (133.7), 781 (94). Magnetic moment: μeff = 2 BM [Gouy].
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)imi; 1441 ν(C
N)imi; 1441 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1085, 622 ν(ClO4). ESI-MS (MeOH, m/z+): 501.2 [CuIILtBu3]+, 1001.5 ([CuIILtBu3]2 − H)+, 1103.4 ([CuIILtBu3]2 + 2H)ClO4+. UV-Vis {λmax, nm (λmax, M−1 cm−1)} in CH3CN: 228 (15
C), 1085, 622 ν(ClO4). ESI-MS (MeOH, m/z+): 501.2 [CuIILtBu3]+, 1001.5 ([CuIILtBu3]2 − H)+, 1103.4 ([CuIILtBu3]2 + 2H)ClO4+. UV-Vis {λmax, nm (λmax, M−1 cm−1)} in CH3CN: 228 (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 389), 247 (14
389), 247 (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 363.4), 279 (9711.6), 320 (3988), 381 (3323), 486 (122), 633 (121), 819 (60). Magnetic moment: μeff = 1.88 BM [Gouy].
363.4), 279 (9711.6), 320 (3988), 381 (3323), 486 (122), 633 (121), 819 (60). Magnetic moment: μeff = 1.88 BM [Gouy].
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio via slow vapour diffusion of Et2O yields green powder (0.09 g, 68%). Anal. calc. for C23.25H27ClCuN3O6.25: C, 51.01; H, 4.97; N, 7.68. Found: C, 51.2; H, 4.92; N, 7.73%. IR (ATR, cm−1, cm−1) 1615 ν(C
1 ratio via slow vapour diffusion of Et2O yields green powder (0.09 g, 68%). Anal. calc. for C23.25H27ClCuN3O6.25: C, 51.01; H, 4.97; N, 7.68. Found: C, 51.2; H, 4.92; N, 7.73%. IR (ATR, cm−1, cm−1) 1615 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)imi; 1604, 1447 ν(C
N)imi; 1604, 1447 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)py and ν(C
N)py and ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1079, 620 ν(ClO4). ESI-MS (MeOH, m/z+): 439.1 [CuIILnapht3]+, 879.3 ([CuIILnapht3]2 + H)+, 979.2 ([CuIILnapht3]2 + 2H)ClO4+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 238 (35
C), 1079, 620 ν(ClO4). ESI-MS (MeOH, m/z+): 439.1 [CuIILnapht3]+, 879.3 ([CuIILnapht3]2 + H)+, 979.2 ([CuIILnapht3]2 + 2H)ClO4+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 238 (35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 335), 311 (17
335), 311 (17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 908), 398 (6036), 485 (sh), 600 (157), 770 (98). Magnetic moment: μeff = 2.01 BM [Gouy].
908), 398 (6036), 485 (sh), 600 (157), 770 (98). Magnetic moment: μeff = 2.01 BM [Gouy].
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio via vapour diffusion of Et2O yields green crystals (0.178 g, 75%). Anal. calc. for C18H22ClCuN3O6: C, 45.48; H, 4.66; N, 8.84. Found: C, 45.42; H, 4.93; N, 9.02%. IR (ATR, cm−1): 1632 ν(C
1 ratio via vapour diffusion of Et2O yields green crystals (0.178 g, 75%). Anal. calc. for C18H22ClCuN3O6: C, 45.48; H, 4.66; N, 8.84. Found: C, 45.42; H, 4.93; N, 9.02%. IR (ATR, cm−1): 1632 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)imi, 1600, 1445 ν(C
N)imi, 1600, 1445 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)py and ν(C
N)py and ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1071, 619 ν(ClO4). ESI-MS (MeOH, m/z+): 375.1 [CuIILH4]+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 222 (18
C), 1071, 619 ν(ClO4). ESI-MS (MeOH, m/z+): 375.1 [CuIILH4]+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 222 (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 271), 244 (17
271), 244 (17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 469), 266 (14
469), 266 (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 079), 371 (3402), 587 (169). Magnetic moment: μeff = 1.82 BM [Gouy].
079), 371 (3402), 587 (169). Magnetic moment: μeff = 1.82 BM [Gouy].
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)imi; 1444 ν(C
N)imi; 1444 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)py and ν(C
N)py and ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1068, 620 ν(ClO4). ESI-MS (MeOH, m/z+): 405.1 [CuIILOMe4]+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 205 (22
C), 1068, 620 ν(ClO4). ESI-MS (MeOH, m/z+): 405.1 [CuIILOMe4]+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 205 (22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 476), 240 (17
476), 240 (17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 281), 270 (12
281), 270 (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 011), 382 (2324), 575 (174). Magnetic moment: μeff = 1.90 BM [Gouy].
011), 382 (2324), 575 (174). Magnetic moment: μeff = 1.90 BM [Gouy].
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)imi; 1612, 1462 ν(C
N)imi; 1612, 1462 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)py and ν(C
N)py and ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1070, 620 ν(ClO4). ESI-MS (MeOH, m/z+): 487.2 [CuIILBu4]+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 227 (17
C), 1070, 620 ν(ClO4). ESI-MS (MeOH, m/z+): 487.2 [CuIILBu4]+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 227 (17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 049), 248 (14
049), 248 (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 636), 274 (10
636), 274 (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800), 383 (3105), 630 (155). Magnetic moment: μeff = 1.95 BM [Gouy].
800), 383 (3105), 630 (155). Magnetic moment: μeff = 1.95 BM [Gouy].
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio via slow vapour diffusion of Et2O yields green powder (0.191 g, 73%). Anal. calc. for C22H24ClCuN3O6: C, 50.29; H, 4.60; N, 8.00. Found: C, 50.67; H, 4.72; N, 7.73%. IR (ATR, cm−1): 1618 ν(C
1 ratio via slow vapour diffusion of Et2O yields green powder (0.191 g, 73%). Anal. calc. for C22H24ClCuN3O6: C, 50.29; H, 4.60; N, 8.00. Found: C, 50.67; H, 4.72; N, 7.73%. IR (ATR, cm−1): 1618 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)imi; 1607, 1437 ν(C
N)imi; 1607, 1437 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)py and ν(C
N)py and ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1072, 619 ν(ClO4−). ESI-MS (MeOH, m/z+): 425.1 [CuIILnapht4]+, 951.2 ([CuIILnapht4]2ClO4 + 2H)+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 226 (27
C), 1072, 619 ν(ClO4−). ESI-MS (MeOH, m/z+): 425.1 [CuIILnapht4]+, 951.2 ([CuIILnapht4]2ClO4 + 2H)+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 226 (27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 244), 237 (sh), 248 (sh), 287 (10
244), 237 (sh), 248 (sh), 287 (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 913), 313 (8927), 383 (4194), 574 (162). Magnetic moment: μeff = 1.86 BM [Gouy].
913), 313 (8927), 383 (4194), 574 (162). Magnetic moment: μeff = 1.86 BM [Gouy].
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio via vapour diffusion of Et2O yields green crystals (0.342 g, 70%). Anal. calc. for C38H48Cl2Cu2N6O12: C, 46.63; H, 4.94; N, 8.59. Found: C, 46.72; H, 4.90; N, 9.01%. IR (KBr disc, cm−1): 1620 ν(C
1 ratio via vapour diffusion of Et2O yields green crystals (0.342 g, 70%). Anal. calc. for C38H48Cl2Cu2N6O12: C, 46.63; H, 4.94; N, 8.59. Found: C, 46.72; H, 4.90; N, 9.01%. IR (KBr disc, cm−1): 1620 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)imi; 1596, 1440 ν(C
N)imi; 1596, 1440 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)py and ν(C
N)py and ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1069, 619 ν(ClO4). ESI-MS (MeOH, m/z+): 389.1 [CuIILH5]+, 879.1 ([CuIILH5]2ClO4 + 2H)+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 257 (6806), 305 (2924), 404 (379), 598 (92). Magnetic moment: μeff = 1.58 B.M per Cu atom [Gouy].
C), 1069, 619 ν(ClO4). ESI-MS (MeOH, m/z+): 389.1 [CuIILH5]+, 879.1 ([CuIILH5]2ClO4 + 2H)+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 257 (6806), 305 (2924), 404 (379), 598 (92). Magnetic moment: μeff = 1.58 B.M per Cu atom [Gouy].
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio via slow vapour diffusion of Et2O yields green powder (0.348 g, 67%). Anal. calc. for C40H52Cl2Cu2N6O14: C, 46.25; H, 5.05; N, 8.09. Found: C, 46.17; H, 5.25; N, 8.4%. IR (ATR, cm−1): 1620 ν(C
1 ratio via slow vapour diffusion of Et2O yields green powder (0.348 g, 67%). Anal. calc. for C40H52Cl2Cu2N6O14: C, 46.25; H, 5.05; N, 8.09. Found: C, 46.17; H, 5.25; N, 8.4%. IR (ATR, cm−1): 1620 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)imi; 1444 ν(C
N)imi; 1444 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1078, 620 ν(ClO4). ESI-MS (MeOH, m/z+): 419.1 [CuIILOMe5]+, 939.2 ([CuIILOMe5]2 + 2H)ClO4+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 258 (9704), 313 (3700), 406 (754), 587 (122). Magnetic moment: μeff = 1.52 BM per Cu atom [Gouy].
C), 1078, 620 ν(ClO4). ESI-MS (MeOH, m/z+): 419.1 [CuIILOMe5]+, 939.2 ([CuIILOMe5]2 + 2H)ClO4+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 258 (9704), 313 (3700), 406 (754), 587 (122). Magnetic moment: μeff = 1.52 BM per Cu atom [Gouy].
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)imi; 1600, 1440 ν(C
N)imi; 1600, 1440 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)py and ν(C
N)py and ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1074, 620 ν(ClO4). ESI-MS (MeOH, m/z+): 501.2 [CuIILtBu5]+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 210 (127
C), 1074, 620 ν(ClO4). ESI-MS (MeOH, m/z+): 501.2 [CuIILtBu5]+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 210 (127![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 302), 229 (14
302), 229 (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 206), 248 (13
206), 248 (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 790), 275 (9945), 306 (4208), 383 (2838), 610 (166). Magnetic moment: μeff = 1.32 BM per Cu atom [Gouy].
790), 275 (9945), 306 (4208), 383 (2838), 610 (166). Magnetic moment: μeff = 1.32 BM per Cu atom [Gouy].
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)imi; 1602, 1449 ν(C
N)imi; 1602, 1449 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)py and ν(C
N)py and ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1079, 620 ν(ClO4−). ESI-MS (MeOH, m/z+): 439.1 [CuIILnapht5]+, 979.2 ([CuIILnapht5]2 + 2H)ClO4+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 224 (36
C), 1079, 620 ν(ClO4−). ESI-MS (MeOH, m/z+): 439.1 [CuIILnapht5]+, 979.2 ([CuIILnapht5]2 + 2H)ClO4+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 224 (36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 505), 322 (10
505), 322 (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 751), 382 (4649), 402 (sh), 599 (140). Magnetic moment: μeff = 1.66 BM Cu atom [Gouy].
751), 382 (4649), 402 (sh), 599 (140). Magnetic moment: μeff = 1.66 BM Cu atom [Gouy].
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)imi; 1446 ν(C
N)imi; 1446 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1049, 620 ν(ClO4). ESI-MS (MeOH, m/z+): 403.1326 [CuLH6]+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 223 (49
C), 1049, 620 ν(ClO4). ESI-MS (MeOH, m/z+): 403.1326 [CuLH6]+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 223 (49![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 480), 273 (15
480), 273 (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 334), 307 (5202), 371 (3433), 619 (136). Magnetic moment: μeff = 1.78 BM [Gouy].
334), 307 (5202), 371 (3433), 619 (136). Magnetic moment: μeff = 1.78 BM [Gouy].
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio via slow vapour diffusion of Et2O yields green powder (0.18 g, 68%). Anal. calc. for C21.25H29ClCuN3O7.25: C, 47.14; H, 5.40; N, 7.76. Found: C, 47.24; H, 5.39; N, 7.72%. IR (ATR, cm−1): 1615 ν(C
1 ratio via slow vapour diffusion of Et2O yields green powder (0.18 g, 68%). Anal. calc. for C21.25H29ClCuN3O7.25: C, 47.14; H, 5.40; N, 7.76. Found: C, 47.24; H, 5.39; N, 7.72%. IR (ATR, cm−1): 1615 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)imi; 1446 ν(C
N)imi; 1446 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1074, 625 ν(ClO4). ESI-MS (MeOH, m/z+): 433.1411 [CuIILOMe6]+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 204 (23
C), 1074, 625 ν(ClO4). ESI-MS (MeOH, m/z+): 433.1411 [CuIILOMe6]+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 204 (23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 919), 237 (21
919), 237 (21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 533), 281 (13
533), 281 (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 442), 381 (3564), 594 (145). Magnetic moment: μeff = 1.74 BM [Gouy].
442), 381 (3564), 594 (145). Magnetic moment: μeff = 1.74 BM [Gouy].
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)imi; 1460 ν(C
N)imi; 1460 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1081, 621 ν(ClO4). ESI-MS (MeOH, m/z+): 515.2532 [CuIILtBu6]+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 203 (19
C), 1081, 621 ν(ClO4). ESI-MS (MeOH, m/z+): 515.2532 [CuIILtBu6]+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 203 (19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 536), 228 (15
536), 228 (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 706), 248 (14
706), 248 (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 105), 280 (9412), 328 (3974), 384 (3135), 507 (165), 621 (164). Magnetic moment: μeff = 1.92 BM [Gouy].
105), 280 (9412), 328 (3974), 384 (3135), 507 (165), 621 (164). Magnetic moment: μeff = 1.92 BM [Gouy].
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio via slow vapour diffusion of Et2O yields green powder (0.19 g, 69%). Anal. calc. for C24H28ClCuN3O6: C, 52.08; H, 5.10; N, 7.59. Found: C, 52.42; H, 5.22; N, 7.33%. IR (ATR, cm−1, cm−1) 1622 ν(C
1 ratio via slow vapour diffusion of Et2O yields green powder (0.19 g, 69%). Anal. calc. for C24H28ClCuN3O6: C, 52.08; H, 5.10; N, 7.59. Found: C, 52.42; H, 5.22; N, 7.33%. IR (ATR, cm−1, cm−1) 1622 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)imi; 1597, 1445 ν(C
N)imi; 1597, 1445 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)py and ν(C
N)py and ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1051, 619 ν(ClO4). ESI-MS (MeOH, m/z+): 453.1437 [CuIILnapht6]+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 227 (26
C), 1051, 619 ν(ClO4). ESI-MS (MeOH, m/z+): 453.1437 [CuIILnapht6]+. UV-Vis {λmax, nm (εmax, M−1 cm−1)} in CH3CN: 227 (26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 485), 235 (26
485), 235 (26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 573), 311 (12
573), 311 (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 151), 383 (4209), 399 (4221), 588 (112). Magnetic moment: μeff = 2.02 BM [Gouy].
151), 383 (4209), 399 (4221), 588 (112). Magnetic moment: μeff = 2.02 BM [Gouy].
Acknowledgements
We are grateful to the Faculty of Chemistry of Bu-Ali Sina University, Ministry of Science, Research and Technology of Iran, the University of Otago and the MacDiarmid Institute for Advanced Materials and Nanotechnology for financial support (PhD scholarship for SD), and Professor Sally Brooker for hosting MS for a 6-month research visit.Notes and references
- (a) L. F. Szczepura, L. M. Witham and K. J. Takeuchi, Coord. Chem. Rev., 1998, 174, 5–32 CrossRef CAS; (b) J. M. Baumeister, R. Alberto, K. Ortner, B. Spingler, P. A. Schubiger and T. A. Kaden, J. Chem. Soc., Dalton Trans., 2002, 4143–4151 RSC; (c) I. Yoon, Y. W. Shin, J. Kim, K.-M. Park, S. B. Park and S. S. Lee, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 2002, 58, m165–m166 Search PubMed; (d) K. S. Choi, D. Kang, J.-E. Lee, J. Seo and S. S. Lee, Bull. Korean Chem. Soc., 2006, 27, 747–754 CrossRef CAS; (e) S. Chung, W. Kim, S. B. Park, I. Yoon, S. S. Lee and D. D. Sung, J. Chem. Soc., Chem. Commun., 1997, 965–966 RSC; (f) S. S. Lee, J. M. Park, D. Y. Kim, J. H. Jung and M. H. Cho, Chem. Lett., 1995, 1009–1010 CrossRef CAS; (g) Y.-S. Xie, H. Jiang, X.-T. Liu, Z.-Y. Zhou, Q.-L. Liu and X.-L. Xu, Collect. Czech. Chem. Commun., 2002, 67, 1647–1657 CrossRef CAS.
- S. Paul, A. K. Barik, R. J. Butcher and S. K. Kar, Polyhedron, 2000, 19, 2661–2666 CrossRef CAS.
- L. R. Gahan, T. M. Donlevy and T. W. Hambley, Inorg. Chem., 1990, 19, 1451–1454 CrossRef.
- A. M. Sargeson, Pure Appl. Chem., 1984, 56, 1603–1619 CrossRef CAS.
- K. Ziegler, Methodes der Organischen Chemie, Part 2, Houben-weyl, Georg-Thieme-Verlag, Stuttgart, 1955, vol. 4, p.729 Search PubMed.
- J. Hunter, J. Nelson, C. Harding, M. McCann and V. McKee, J. Chem. Soc., Chem. Commun., 1990, 1148–1151 RSC.
- T. M. Garrett, T. J. McMurry, M. W. Hosseini, Z. E. Reyes, F. E. Hahn and K. N. Raymond, J. Am. Chem. Soc., 1991, 113, 2965–2977 CrossRef CAS.
- L. Busetto and V. Zanotti, Inorg. Chim. Acta, 2008, 361, 3004–3011 CrossRef CAS PubMed.
- (a) R. L. Fanshawe and A. G. Blackman, Inorg. Chem., 1995, 34, 421–423 CrossRef CAS; (b) A. M. Dittler-Klingemann and F. E. Hahn, Inorg. Chem., 1996, 35, 1996–1999 CrossRef CAS; (c) A. M. Dittler-Klingemann, C. Orvig, F. E. Hahn, F. Thaler, C. Hubbard, R. van Eldik, S. Schindler and I. Fábián, Inorg. Chem., 1996, 35, 7798–7803 CrossRef CAS.
- C. Ochs, F. E. Hahn and R. Frohlich, Chem. – Eur. J., 2000, 6, 2193–2199 CrossRef CAS.
- (a) B. A. Brennan, G. Alms, M. J. Nelson, L. T. Durney and R. G. Scarrow, J. Am. Chem. Soc., 1996, 118, 9194–9195 CrossRef CAS; (b) R. G. Scarrow, B. S. Strickler, J. J. Ellison, S. C. Shoner, J. A. Kovacs, J. G. Cummings and M. J. Nelson, J. Am. Chem. Soc., 1998, 120, 9237–9245 CrossRef CAS; (c) L. Heinrich, Y. Li, J. Vaissermann, G. Chottard and J.-C. Chottard, Angew. Chem., Int. Ed. Engl., 1999, 38, 3526–3528 CrossRef CAS.
- M. T. Werth, S.-F. Tang, G. Formicka, M. Zeppezauer and M. K. Johnson, Inorg. Chem., 1995, 34, 218–228 CrossRef CAS.
- (a) Y.-H. Chiu, G. J. Gabriel and J. W. Canary, Inorg. Chem., 2005, 44, 40–44 CrossRef CAS PubMed; (b) G. J. Christian, A. Llobet and F. Maseras, Inorg. Chem., 2010, 49, 5977–5985 CrossRef CAS PubMed.
- H. Keypour, A. H. Jamshidi, M. Rezaeivala and L. Valencia, Polyhedron, 2013, 52, 872–878 CrossRef CAS PubMed.
- M. Kalanithi, M. Rajarajan, P. Tharmaraj and C. D. Sheela, Spectrochim. Acta, Part A, 2012, 87, 155–162 CrossRef CAS PubMed.
- G. A. McLachlan, G. D. Fallon, R. L. Martin and L. Spiccia, Inorg. Chem., 1995, 34, 254–261 CrossRef CAS.
- S. J. Brudenell, L. Spiccia and E. R. T. Tiekink, Inorg. Chem., 1996, 35, 1974–1979 CrossRef CAS; A. B. P. Lever, Studies in physical and theoretical chemistry 33: Inorganic electronic spectroscopy, Elsevier, Amsterdam, 1997 Search PubMed.
- B. Graham, M. T. W. Hearn, P. C. Junk, C. M. Kepert, F. E. Mabbs, B. Moubaraki, K. S. Murray and L. Spiccia, Inorg. Chem., 2001, 40, 1536–1543 CrossRef CAS PubMed.
- C. Ochs, F. E. Hahn and R. Fröhlich, Eur. J. Inorg. Chem., 2001, 2427–2436 CrossRef CAS.
- M. Du, Y.-M. Guo, S.-T. Chen, X.-H. Bu and J. Ribas, Inorg. Chim. Acta, 2003, 346, 207–214 CrossRef CAS.
- S. Bhattacharyya, S. B. Kumar, S. K. Dutta, E. R. T. Tiekink and M. Chaudhury, Inorg. Chem., 1996, 35, 1967–1973 CrossRef CAS.
- B. Sarkar, M. S. Ray, Y.-Z. Li, Y. Song, A. Figuerola, E. Ruiz, J. Cirera, J. Cano and A. Ghosh, Chem. – Eur. J., 2007, 13, 9297–9309 CrossRef CAS PubMed.
- B. Sarkar, S. Konar, C. J. Gomez-Garcia and A. Ghosh, Inorg. Chem., 2008, 47, 11611–11619 CrossRef CAS PubMed.
- B. J. Hathaway and A. A. G. Tomlinson, Coord. Chem. Rev., 1970, 5, 1–43 CrossRef CAS.
- M. Duggan, N. Ray, B. Hathaway, G. Tomlinson, P. Brint and K. Pelin, J. Chem. Soc., Dalton Trans., 1980, 1342–1348 RSC.
- N. J. Ray, L. Hulett, R. Sheahan and B. Hathaway, J. Chem. Soc., Dalton Trans., 1981, 1463–1469 RSC.
- E. Colacio, J. M. Domínguez-Vera, M. Gazi, R. Kivekäs, J. M. Moreno and A. Pajunen, J. Chem. Soc., Dalton Trans., 2000, 505–509 RSC.
- (a) M. J. Scott, S. C. Lee and R. H. Holm, Inorg. Chem., 1994, 33, 4651–4662 CrossRef CAS; (b) M. T. Gardner, G. Deinum, Y. Kim, G. T. Babcock, M. J. Scott and R. H. Holm, Inorg. Chem., 1996, 35, 6878–6884 CrossRef CAS PubMed; (c) R. T. Conley, Infrared Spectroscopy, Allyn & Bacon Inc., Boston, 1966 Search PubMed; (d) K. Nakamoto, Infrared and Raman Spectra of Inorganic and Coordination Compounds Part A and B, 5h edn, Wiley, New York, 1997 Search PubMed.
- N. Mondal, D. K. Dey, S. Mitra and V. Gramlich, Polyhedron, 2001, 20, 607–613 CrossRef CAS.
- A. Bottcher, H. Elias, E. Jager, H. Lanffelderora, M. Mazur, L. Mullor, H. Paulas, P. Pelikan, M. Rudolph and M. Valko, Inorg. Chem., 1993, 32, 4131–4138 CrossRef.
- R. N. Patel, S. P. Rawat, M. Choudhary, V. P. Sondhiya, D. K. Patel, K. K. Shukla, D. K. Patel, Y. Singh and R. Pandey, Inorg. Chim. Acta, 2012, 392, 283–291 CrossRef CAS PubMed.
- A. Kumar Sharma and R. Mukherjee, Inorg. Chim. Acta, 2008, 361, 2768–2776 CrossRef PubMed.
- E. V. Rybak-Akimova, A. Y. Nazarenko, L. Chen, P. W. Krieger, A. M. Herrera, V. V. Tarasov and P. D. Robinson, Inorg. Chim. Acta, 2001, 324, 1–15 CrossRef CAS.
- N. J. Lundin, I. G. Hamilton and A. G. Blackman, Polyhedron, 2004, 23, 97–102 CrossRef CAS PubMed.
- F. Tuna, G. I. Pascu, J. P. Sutter, M. Andruh, S. Golhen, J. Guillevic and H. Pritzkow, Inorg. Chim. Acta, 2003, 342, 131–138 CrossRef CAS.
- B. Sreenivasulu, M. Vetrichelvan, F. Zhao, S. Gao and J. J. Vittal, Eur. J. Inorg. Chem., 2005, 4635–4645 CrossRef CAS PubMed.
- Y. Yahsi and H. Kara, Inorg. Chim. Acta, 2013, 397, 110–116 CrossRef CAS PubMed.
- L. C. Nathan, J. E. Koehne, J. M. Gilmore, K. A. Hannibal, W. E. Dewhirst and T. D. Mai, Polyhedron, 2003, 22, 887–894 CrossRef CAS.
- A. W. Addison, T. N. Rao, J. Reedijk, J. van Rijn and G. C. Verschoor, J. Chem. Soc., Dalton Trans., 1984, 7, 1349–1356 RSC.
- K. P. Maresca, G. H. Bonavia, J. W. Babich and J. Zubieta, Inorg. Chim. Acta, 1999, 284, 252–257 CrossRef CAS.
- M. Li, A. Ellern and J. H. Espenson, Inorg. Chem., 2005, 44, 3690–3699 CrossRef CAS PubMed.
- J. F. Larrow and E. N. Jacobson, J. Org. Chem., 1994, 59, 1939–1942 CrossRef CAS.
- Bruker, APEX2, SAINT, SADABS and SHELXTL Bruker AXS Inc., Madison, Wisconsin, USA, 2012.
- (a) G. M. Sheldrick, Acta Crystallogr., Sect. A: Found. Crystallogr., 1990, 46, 467–473 CrossRef; (b) G. M. Sheldrick, Methods Enzymol., 1997, 276, 628–641 CAS.
- G. M. Sheldrick and T. R. Schneider, Methods Enzymol., 1997, 277, 319–343 CAS.
- G. M. Sheldrick, SHELXS97, University of Gottingen, Germany, 1997 Search PubMed.
- Y. Zhao and D. G. Truhlar, A new local density functional for main-group thermochemistry, transition metal bonding, thermochemical kinetics, and noncovalent interactions, J. Phys. Chem. A, 2006, 125, 194101 CrossRef PubMed.
- M. J. Frisch, et al., Gaussian 09, Revision A.02, Gaussian, Inc., Wallingford, CT, 2009 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: 1H NMR, 13C NMR and IR spectra of amines (3–6) (Fig. S1–S12), IR spectra of complexes (Fig. S13–S30), and mass spectra of complexes (Fig. S31–S48). CCDC 968863 [CuIILnapht2](ClO4), 968865 [CuIILH3](ClO4), 1002225 [CuIILOMe3](ClO4), 1002229 [CuIILH4](ClO4) and 1002230 [CuIILH5]2(ClO4)2. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c5nj01318f |
| This journal is © The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2015 |