Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Elemental bioimaging of Cisplatin in Caenorhabditis elegans by LA-ICP-MS†
Barbara
Crone
a,
Michael
Aschner
b,
Tanja
Schwerdtle
c,
Uwe
Karst
a and
Julia
Bornhorst
*c
aUniversity of Münster, Institute of Inorganic and Analytical Chemistry, Corrensstraße 30, 48149 Münster, Germany
bDepartment of Molecular Pharmacology, Neuroscience, and Pediatrics, Albert Einstein College of Medicine, Bronx, NY, USA
cDepartment of Food Chemistry, Institute of Nutritional Science, University of Potsdam, Arthur-Scheunert-Allee 114-116, 14558 Nuthetal, Germany. E-mail: julia.bornhorst@uni-potsdam.de; Fax: +49-33200/88-5573; Tel: +49-33200/88-5262
First published on 14th May 2015
Abstract
cis-Diamminedichloroplatinum(II) (Cisplatin) is one of the most important and frequently used cytostatic drugs for the treatment of various solid tumors. Herein, a laser ablation-inductively coupled plasma-mass spectrometry (LA-ICP-MS) method incorporating a fast and simple sample preparation protocol was developed for the elemental mapping of Cisplatin in the model organism Caenorhabditis elegans (C. elegans). The method allows imaging of the spatially-resolved elemental distribution of platinum in the whole organism with respect to the anatomic structure in L4 stage worms at a lateral resolution of 5 μm. In addition, a dose- and time-dependent Cisplatin uptake was corroborated quantitatively by a total reflection X-ray fluorescence spectroscopy (TXRF) method, and the elemental mapping indicated that Cisplatin is located in the intestine and in the head of the worms. Better understanding of the distribution of Cisplatin in this well-established model organism will be instrumental in deciphering Cisplatin toxicity and pharmacokinetics. Since the cytostatic effect of Cisplatin is based on binding the DNA by forming intra- and interstrand crosslinks, the response of poly(ADP-ribose)metabolism enzyme 1 (pme-1) deletion mutants to Cisplatin was also examined. Loss of pme-1, which is the C. elegans ortholog of human poly(ADP-ribose) polymerase 1 (PARP-1) led to disturbed DNA damage response. With respect to survival and brood size, pme-1 deletion mutants were more sensitive to Cisplatin as compared to wildtype worms, while Cisplatin uptake was indistinguishable.
Introduction
Cancer is a leading cause of illness and death worldwide. Since its discovery in the late 1960s as a chemotherapy drug, cis-diamminedichloroplatinum(II) (Cisplatin) is one of the most commonly used anticancer drugs and is clinically proven to efficaciously combat various types of cancers, including carcinomas, germ cell tumors, lymphomas and sarcomas.1 It is generally considered that the DNA is the critical target for Cisplatin cytotoxicity by inhibiting DNA synthesis and damaging DNA, whereby different DNA adducts of Cisplatin, including intra- and interstrand cross-links, DNA–protein cross-links and DNA monoadducts are formed.2 Since Cisplatin can induce apoptosis, the signaling pathways that regulate apoptosis have significant impact on regulating cellular responsiveness.2,3 The success of Cisplatin therapy is compromised due to dose-limiting toxicity in the kidney, cochlea and peripheral nerves, with nephrotoxicity being the most well described.4The pharmacokinetics of Cisplatin has been studied in the past by determination of the total platinum concentration using atomic absorption spectroscopy (AAS), total reflection X-ray fluorescence (TXRF)5–7 or inductively coupled plasma-mass spectrometry (ICP-MS).8,9 However, none of these techniques allows for studies on Cisplatin distribution at the organ, cell and subcellular level. This question was addressed with the development of several novel analytical methods in the last several decades.10,11 For metal containing compounds, X-ray fluorescence and laser ablation-inductively coupled plasma-mass spectrometry (LA-ICP-MS) have become powerful bioimaging tools.12–14 LA-ICP-MS, in particular, offers an excellent modality for visualizing the elemental distribution in tissue samples. The method is fast and easy, highly sensitive and can be carried out with a high spatial resolution in the absence of complex and laborious sample preparation.15,16 Its broad field of applications include the examination of the distribution of the chemotherapy drug Cisplatin in tissues.17–24 As an example, Zoriy et al.17 as well as Moreno-Gordaliza et al.19 observed the distribution of platinum in kidneys to study Cisplatin-induced nephrotoxicity.
Caenorhabditis elegans (C. elegans) represents an established model organism for biomedical research. Characteristics that have been contributed to its success include the genetic manipulability, the well-characterized genome, the ease of maintenance and the small body size.25,26 It takes three days for an embryo to reach adulthood and reproduce. A single hermaphrodite has the ability to produce approximately 300 offspring, which enables high-throughput analyses.27C. elegans is less complex than the mammalian system, while still sharing high genetic homology (60–80%).28 The model organism C. elegans has been previously used in a limited number of studies to identify interactions between Cisplatin and signaling pathways in vivo.29–31 Hypersensitivity to Cisplatin was noted in worms with mutations in asna-1, a metalloregulated ATPase gene promoting insulin secretion and membrane insertion of tail-anchored proteins, leading the authors to suggest that drugs targeting ASNA-1 may sensitize cancer cells to Cisplatin.30 In a follow up study, the same authors identified that depletion of the endoplasmic reticulum (ER) resident chaperone ENPL-1/GRP94 renders worms sensitive to Cisplatin.31 Since DNA repair mechanisms are known to occur in the genetically amenable nematode,32,33 deletion mutants have been used to identify various genes of the DNA repair pathways that are protective against Cisplatin toxicity. For example, translesion synthesis (TLS) (polh-1 and polk-1) polymerases which allow cells to complete DNA replication in the presence of damage, as well as xpf-1, which is involved in nucleotide excision repair and intrastrand crosslink repair, were pointed out as critical genes in protecting against platinum chemotherapy.34,35
Herein, a LA-ICP-MS method for Cisplatin bioimaging in the larval 4 (L4) stage wildtype (WT) worms was developed to localize Cisplatin distribution in C. elegans. Additionally, bioimaging was performed in poly(ADP-ribose)metabolism enzyme 1 (pme-1) deletion mutant worms following Cisplatin exposure. Loss of pme-1, which is the C. elegans ortholog of human poly(ADP-ribose) polymerase-1 (PARP-1), was posited to decrease genomic stability due to a disturbance of DNA repair pathways.36–38 Additionally, we assessed the response to Cisplatin treatment to determine appropriate dosing for the bioimaging studies as well as Cisplatin toxicity and its effect on the nematode's reproduction.
Experimental
Chemicals
All chemicals were of the highest purity available. Rhodium and cobalt ICP-standards (1000 μg mL−1) were obtained from SCP Science (Courtaboeuf, France). Nitric acid (67–69%, Optimat) for ICP-MS studies was purchased from Fisher Scientific (Loughborough, UK). Hydrochloric acid (30%, Suprapur) and nitric acid (65%, Suprapur) for ashing was obtained from Merck (Darmstadt, Germany).C. elegans strains and handling
Caenorhabditis elegans (C. elegans) strains were handled and maintained at 20 °C as previously described.26 The following strains were used in this study: N2, wildtype and the deletion mutant RB1042 (parp-1(ok988) I.) (Caenorhabditis Genetics Center, CGC). To obtain a synchronous population, worms were treated with an alkaline bleach solution (1% NaOCl and 0.25 M NaOH) as described.39 Synchronized L1 worms were placed on OP50-seeded NGM plates after hatching and experiments were performed using L4 stage nematodes.Cisplatin treatment in C. elegans and Cisplatin-induced lethality assay
Cisplatin solutions were prepared in 85 mM NaCl including 0.01% Tween shortly before each experiment. Treatment was performed using 500 (survival, brood size) or 1500 (Cisplatin uptake) L4 stage worms. L4 nematodes were exposed to Cisplatin (50, 100 μg mL−1) in siliconized tubes for 1 h or 2 h. Afterwards, worms were washed at least three times in 85 mM NaCl including 0.01% Tween. 30–50 worms were then placed on OP50-seeded NGM plates, pre-counted and the number of surviving worms was scored 24 h post-treatment.Reproductive profile assay
For brood size evaluations, worms were evaluated by counting the number of progenies from each individual worm day-by-day. Following Cisplatin treatment, as described above, individual worms were kept on OP50-seeded NGM plates and transferred every day to a new plate until day eight of adulthood. The number of progenies was scored every day.Cisplatin uptake in C. elegans
Cisplatin content was quantified after ashing Cisplatin exposed L4 stage worms by total reflection X-ray fluorescence (TXRF). A number of 1500 L4 stage nematodes were exposed to 50 or 100 μg mL−1 Cisplatin in siliconized tubes for 1 h or 2 h. Worms were then pelleted by centrifugation at 1600 rpm for 3 min, washed five times in 85 mM NaCl including 0.01% Tween and re-suspended in 0.5 mL 85 mM NaCl including 0.01% Tween. After sonification, an aliquot was taken for protein quantification using the bicinchoninic acid (BCA) assay-kit (Thermo Scientific). Thereafter, the suspension was mixed again, evaporated and incubated with ashing mixture (65% HNO3/30% H2O2 (1/1) (both from Merck)) at 95 °C until dryness. After dilution of the ash in 0.5 mL HNO3 (2%, (Fisher Scientific)), an internal standard solution of 500 μg mL−1 cobalt (ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was added. The analyte solution was transferred on quartz glass discs (Bruker AXS, Microanalysis GmbH, Berlin, Germany), previously cleaned in 1 mol L−1 nitric acid, coated with silicon oil for better wettability and controlled for platinum (Pt) contamination. The subsequent measurements were performed using a S2 PICOFOX system (Bruker AXS) with a low power X-ray tube with a molybdenum anode at 750 μA and 50 kV and an energy-dispersive, Peltier-cooled silicon drift-detector (SDD, XFlash). The signal integration time was 1000 s. Data evaluation was performed using the software Spectra Picofox Version 7.2.5.0 (Bruker AXS). The limit of detection (LOD) was calculated to 3.4 μg L−1 according to the approach of Klockenkämper.40 The limit of quantification (LOQ) was 11.3 μg L−1.
1) was added. The analyte solution was transferred on quartz glass discs (Bruker AXS, Microanalysis GmbH, Berlin, Germany), previously cleaned in 1 mol L−1 nitric acid, coated with silicon oil for better wettability and controlled for platinum (Pt) contamination. The subsequent measurements were performed using a S2 PICOFOX system (Bruker AXS) with a low power X-ray tube with a molybdenum anode at 750 μA and 50 kV and an energy-dispersive, Peltier-cooled silicon drift-detector (SDD, XFlash). The signal integration time was 1000 s. Data evaluation was performed using the software Spectra Picofox Version 7.2.5.0 (Bruker AXS). The limit of detection (LOD) was calculated to 3.4 μg L−1 according to the approach of Klockenkämper.40 The limit of quantification (LOQ) was 11.3 μg L−1.
Bioimaging of Cisplatin uptake in C. elegans
L4 stage worms (∼500) were exposed to Cisplatin as described above. Subsequently, worms were pelleted by centrifugation at 1600 rpm for 2 min and washed at least twice with 85 mM NaCl including 0.01% Tween. For methanol fixation, worms were re-suspended in ice-cold methanol (−20 °C, 2 h) and the fixed nematodes were dropped onto microscopic slides (Superfrost, Thermo Fisher Scientific). For plain bouin's fixation, worms were re-suspended in plain bouin's fix solution (75 mL saturated picric acid, 25 mL 37% formaldehyde solution and 5 mL glacial acetic acid) and vigorously shaken for 30 min. After one cycle of freeze–thawing in liquid nitrogen, the tube was shaken for additional 30 min. Subsequently, worms were washed five times with borate triton solution (25 mM borate buffer, 0.5% Triton X-100), and were dropped onto microscopic slides.Prior to analysis, a light-microscopic image was recorded from each worm with a microscope (BZ 9000, Keyence Deutschland GmbH, Neu-Isenburg, Germany) using the bright-field option and CFI 60-optics (Nikon, Chiyoda, Tokyo, Japan).
Bioimaging experiments were performed using a laser ablation system incorporating a frequency quintupled Nd:YAG laser (LSX-213, CETAC Technologies, Omaha, NE, USA) with a wavelength of 213 nm, coupled to a quadrupole-based inductively coupled plasma-mass spectrometer (ICP-MS) (iCAP Qc, Thermo Fisher, Bremen, Germany). The ablation was performed in a multiline scan, whereby the worms were ablated line by line using parameters listed in Table 1.
| Laser energy 8 J cm−2 (60% of the maximum energy). | |
|---|---|
| Repetition frequency | 20 Hz |
| Spot diameter | 5 μm |
| Scan rate | 5 μm s−1 |
| He flow | 800 mL min−1 |
| Additional Ar flow | 400 mL min−1 |
The dry aerosol produced was transported to the ICP, using He as carrier gas which is mixed with Ar downstream the ablation cell to improve transport efficiency. The PFA nebulizer and the cyclonic spray chamber between LA and ICP-MS instrument allowed introduction of wet aerosol into the ICP system. This improved the plasma stability and enabled to continuously deliver Rh as internal standard (10 ng L−1) to compensate for drift effects. For maximum sensitivity and to minimize possible interferences of e.g. [40Ar154Gd], [40Ar155Gd], the measurement was performed in kinetic energy discrimination mode (KED) of the ICP-MS instrument with helium (He) as cell gas. The important parameters for the ICP-MS system are shown in Table 2. The isotopes 194Pt, 195Pt and 103Rh were detected with dwell times of 0.45 s, 0.45 s and 0.1 s, respectively. The recorded ablation profiles were converted into 2D distribution images and were evaluated with ImageJ (National Institute of Health, Bethesda, MD, USA).
| Power | 1550 W |
| Nebulizer gas flow | 0.6 L min−1 |
| Auxiliary gas flow | 0.8 L min−1 |
| Cool gas flow | 14 L min−1 |
| Sampling depth | 5 mm |
| Measurement mode | KED |
| Cell gas flow (He, 99.999%) | 4.6 mL min−1 |
Statistics
Cisplatin dose–response curves and all histograms were generated using GraphPad Prism (GraphPad Software Inc.). Two-way analysis of variance (ANOVAs) were performed on the DA and SRT data and the basal slowing data, followed by Dunnett's multiple comparison post hoc test.Results
Sample preparation
In order to develop a method for Cisplatin bioimaging in L4 stage nematodes, a fast and easy sample preparation protocol was needed. Accordingly, cryosections as well as different fixation protocols were applied. While cryosections of 20 μm would guarantee a uniform thickness of the worms, it was a technical challenge to reproducibly section the worm along its longitudinal axis. Additionally, due to the high energy, which was needed to ablate the L4 stage worms because of their large diameter, it was necessary to develop a preparation protocol to assure that the worms did not detach from the slides. Therefore, L4 stage worms were washed after Cisplatin incubation, pelleted by centrifugation and a fixation with ice-cold methanol or plain bouin's solution was carried out. Subsequently, the fixed nematodes were dropped onto microscopic slides and air-dried in order to obtain single, separated worms. Since no differences of both fixation protocols could be observed (Fig. 1) and due to the fast and simple extraction protocol, the methanol fixation was the method of choice.Bioimaging of L4 stage nematodes
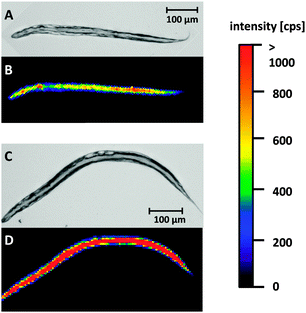
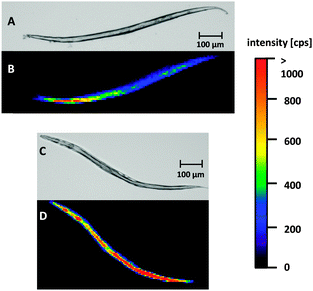
The developed LA-ICP-MS method and the optimized sample preparation were applied to evaluate Cisplatin uptake in WT worms and pme-1 deletion mutants following 50 μg mL−1 or 100 μg mL−1 Cisplatin exposure for 1 h or 2 h. Prior to the LA-ICP-MS, a microscopic image was recorded to correlate the elemental distribution with the anatomical structure of the worm.Fig. 2A and B are illustrating a WT L4 stage worm incubated with 100 μg mL−1 for 1 h. Both, the microscopic image (A) and the elemental image (B) demonstrate that the head of the worm is in the upper part and the tail in the lower part of the image. The elemental mapping of the platinum distribution shows that Cisplatin is predominantly located in the area of the intestine. Moreover, an accumulation in the head of the nematode could be observed. Overall, signal intensities were in the range of 500 cps to 1000 cps. A longer incubation time (2 h) led to an increased platinum intensity (Fig. 2C and D). Similar results for time- and dose-dependency could be detected incubating 50 μg mL−1 Cisplatin with an overall reduced signal intensity due to the lower incubation concentration (ESI†). Cisplatin exposure of the pme-1 deletion mutants resulted also in a Cisplatin uptake, which was indistinguishable from the WT worms at the respective dose and exposure time (Fig. 3).
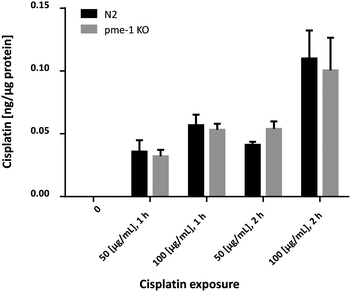
In order to corroborate the bioimaging data, the Cisplatin content in WT worms and pme-1 deletion mutants was determined by TXRF. Overall, the analyzed strains (Fig. 4) showed a significant dose-dependent increase. Additionally, Cisplatin uptake in the pme-1 deletion mutants was indistinguishable from WT worms for each of the respective doses and duration of Cisplatin exposure, corroborating the LA-ICP-MS data.
Cisplatin induced lethality and reproductive profile
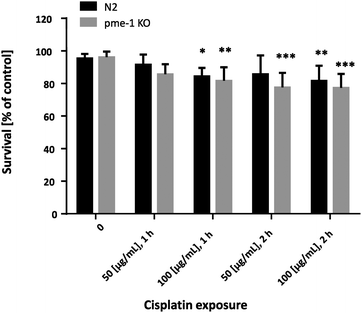
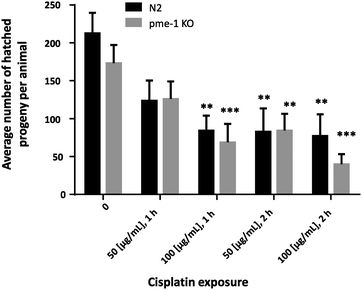
In order to determine appropriate Cisplatin concentrations for dosing in the bioimaging study and to determine Cisplatin sensitivity, survival curves following Cisplatin exposure were assessed. Therefore, WT as well as pme-1 deletion mutants were incubated with 50 μg mL−1 or 100 μg mL−1 1 h or 2 h and counted 24 h later. WT worms exhibited sensitivity to Cisplatin (100 μg mL−1 1 h or 2 h treatments). The genetic deletion of pme-1 increased mortality compared to WT worms following 2 h Cisplatin treatment. Cisplatin treatment at 100 μg mL−1 (1 h), 50 μg mL−1 (2 h) or 100 μg mL−1 (2 h) resulted in a significant decrease in surviving pme-1 deletion mutants (Fig. 5).To determine whether the Cisplatin doses used within the bioimaging study caused lasting effects on C. elegans reproduction, brood size was examined following Cisplatin incubation (Fig. 6). Overall, the analyzed strains showed a significantly decreased brood size following 100 μg mL−1 (1 h), 50 μg mL−1 (2 h) or 100 μg mL−1 (2 h) Cisplatin incubation.
Discussion
The LA-ICP-MS method provides information on the spatial distribution of Cisplatin in the model organism C. elegans in order to obtain more detailed assessment on the pharmacokinetics of Cisplatin and to achieve better understanding of complex biological processes at the tissue level. To the best of our knowledge, there are only limited data available regarding analytical bioimaging of various metals in C. elegans and the respective sample preparation in the worms. In expensive and time-consuming microbeam synchrotron radiation X-ray fluorescence studies, Mn, Fe and Zn distribution was imaged in C. elegans after worm preparation with liquid N2-chilled liquid propane and lyophilization.12 While the analysis of soft tissue by LA-ICP-MS is generally carried out using dewaxed sections19,41 or cryosections,42,43 the adaption of these techniques to C. elegans is quite complex due to its small size, different positions and torsion. Therefore, Mn measurements by LA-ICP-MS were carried out in L1 stage worms which have been air-dried on the microscopic slide following Mn treatment.13 However, within this study, older and thus bigger worms were used and air-drying resulted in an altered worm structure. Therefore, chemically-based tube-fixation preserving tissue morphology44 was used as a fast and simple preparation technique and the distribution of Cisplatin is not affected by metabolic functions of the organism. This sample preparation in combination with the advantages of C. elegans, including its quick life-cycle would allow for comparably high throughput analyses. While in previous studies, Cisplatin had been imaged in various tissues, this is the first study to image a whole organism by LA-ICP-MS.17–24 Additionally, the highly sensitive method allows the mapping of platinum with a lateral resolution of 5 μm. The Cisplatin uptake in WT worms was observed to be dose- and time-dependent, which was confirmed quantitatively by TXRF measurements. The elemental mapping of the distribution by LA-ICP-MS also suggests that Cisplatin is predominantly located in the area of the intestine, which is largely responsible for food digestion and assimilation. Additionally, the elemental mapping suggests that Cisplatin is also located in the head of the worms. Cisplatin-induced toxicity to the peripheral nervous system is a frequent adverse effect, leading to dose-limiting in Cisplatin chemotherapy.45 Whether Cisplatin leads to neurodegeneration in C. elegans has yet to be determined.The genetically amenable model system C. elegans can be used to identify additional genes, which may modify the response to Cisplatin. Since the DNA is the critical target for Cisplatin toxicity and due to previous evidence that cells deficient in DNA repair are hypersensitive to Cisplatin,1 Cisplatin distribution and toxicity were analyzed in pme-1 deletion mutant worms. Pme-1 has been identified and characterized as structural and functional ortholog of mammalian PARP-1 in 2002 by Gagnon et al.46,47 PARP-1 is activated in response to DNA strand breaks and transfers ADP-ribosyl units from NAD+ onto themselves and other target proteins, thus producing protein-coupled ADP-ribose polymers of up to 200 units. Affected proteins are involved in many cellular processes including transcription, replication, telomere maintenance, genomic stability, chromatin organization and DNA repair.48 Thus, PARP-1 inhibition decreases the genomic stability due to a disturbance of DNA repair pathways.49 While in literature, Cisplatin resistance is associated with PARP hyperactivation,50 a recently published mechanistic study indicated that PARP inhibition selectively increases sensitivity to Cisplatin in lung cancer cells with low ERCC1, a key component in nucleotide excision repair, expression.51 This is in agreement with our study on C. elegans, showing increased sensitivity to Cisplatin in pme-1 deletion mutants compared to WT worms, in both the survival and brood size assays. However, the Cisplatin uptake as well as the elemental Cisplatin mapping in the pme-1 deletion mutants was indistinguishable from WT worms for each of the respective doses and Cisplatin exposure duration.
Conclusions
The newly developed laser ablation-inductively coupled plasma-mass spectrometry (LA-ICP-MS) method requires a fast and simple sample preparation and can be used rapidly and easily to visualize the Cisplatin distribution in the model organism C. elegans. A spatially-resolved elemental analysis with respect to the anatomic structure is possible. The data suggest that Cisplatin is taken up dose- and time-dependently and that it is predominantly located in the area of the intestine and in the head of the worms. Studies in the genetically tractable worm also established that deletion of pme-1 is associated with increased sensitivity to Cisplatin compared to WT worms.Taken together, spatially-resolved visualization of Cisplatin in a whole model organism opens up the possibility to correlate toxicological alterations with the amount of drug and might be helpful for a better understanding of Cisplatin pharmacokinetics and dose-efficiency studies of Cisplatin. Furthermore, the development of a quantification method might provide helpful additional information about concentration levels inside the worm.
Acknowledgements
We would like to acknowledge the Caenorhabditis Genetic Center (CGC), which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440), for providing the strains used in this manuscript. MA was supported in part by grants from the NIH, R01ES07331 and R01ES10563. Parts of this study were supported by the Cells in Motion Cluster of Excellence (CiM – EXC 1003), Münster, Germany (project FF-2013-17).References
- S. Dasari and P. B. Tchounwou, Eur. J. Pharmacol., 2014, 740, 364–378 CrossRef CAS PubMed.
- A. Basu and S. Krishnamurthy, J. Nucleic Acids, 2010, 2010, 201367 Search PubMed.
- R. H. Lee, J. M. Song, M. Y. Park, S. K. Kang, Y. K. Kim and J. S. Jung, Biochem. Pharmacol., 2001, 62, 1013–1023 CrossRef CAS.
- G. S. Oh, H. J. Kim, A. Shen, S. B. Lee, D. Khadka, A. Pandit and H. S. So, Electrolyte Blood Pressure, 2014, 12, 55–65 CrossRef PubMed.
- A. Meyer, S. Grotefend, A. Gross, H. Wätzig and I. Ott, J. Pharm. Biomed. Anal., 2012, 70, 713–717 CrossRef CAS PubMed.
- R. Fernández-Ruiz, Dev. Anal. Chem., 2014, 1, 1–14 Search PubMed.
- E. D. Greaves, L. M. Marćo Parra, A. Rojas and L. Sajo-Bohus, X-Ray Spectrom., 2000, 29, 349–353 CrossRef CAS.
- B. Gammelgaard, H. R. Hansen, S. Stürup and C. Moller, Expert Opin. Drug Metab. Toxicol., 2008, 4, 1187–1207 CrossRef CAS PubMed.
- B. Meermann and M. Sperling, Anal. Bioanal. Chem., 2012, 403, 1501–1522 CrossRef CAS PubMed.
- J. Pol, M. Strohalm, V. Havlicek and M. Volny, Histochem. Cell Biol., 2010, 134, 423–443 CrossRef CAS PubMed.
- R. McRae, P. Bagchi, S. Sumalekshmy and C. J. Fahrni, Chem. Rev., 2009, 109, 4780–4827 CrossRef CAS PubMed.
- G. McColl, S. A. James, S. Mayo, D. L. Howard, C. G. Ryan, R. Kirkham, G. F. Moorhead, D. Paterson, M. D. de Jonge and A. I. Bush, PLoS One, 2012, 7, e32685 CAS.
- S. Große Brinkhaus, J. Bornhorst, S. Chakraborty, C. A. Wehe, R. Niehaus, O. Reifschneider, M. Aschner and U. Karst, Metallomics, 2014, 6, 617–621 RSC.
- J. S. Becker, M. Zoriy, A. Matusch, B. Wu, D. Salber and C. Palm, Mass Spectrom. Rev., 2010, 29, 156–175 CAS.
- I. Konz, B. Fernandez, M. L. Fernandez, R. Pereiro and A. Sanz-Medel, Anal. Bioanal. Chem., 2012, 403, 2113–2125 CrossRef CAS PubMed.
- A. Matusch, C. Depboylu, C. Palm, B. Wu, G. U. Hoglinger, M. K. Schafer and J. S. Becker, J. Am. Soc. Mass Spectrom., 2010, 21, 161–171 CrossRef CAS PubMed.
- M. Zoriy, A. Matusch, T. Spruss and J. S. Becker, Int. J. Mass Spectrom., 2007, 260, 102–106 CrossRef CAS PubMed.
- O. Reifschneider, C. A. Wehe, I. Raj, J. Ehmcke, G. Ciarimboli, M. Sperling and U. Karst, Metallomics, 2013, 5, 1440–1447 RSC.
- E. Moreno-Gordaliza, C. Giesen, A. Lazaro, D. Esteban-Fernandez, B. Humanes, B. Canas, U. Panne, A. Tejedor, N. Jakubowski and M. M. Gomez-Gomez, Anal. Chem., 2011, 83, 7933–7940 CrossRef CAS PubMed.
- C. Herdering, C. A. Wehe, O. Reifschneider, I. Raj, G. Ciarimboli, K. Diebold, C. Becker, M. Sperling and U. Karst, Rapid Commun. Mass Spectrom., 2013, 27, 2588–2594 CrossRef CAS PubMed.
- J. Bianga, A. Bouslimani, N. Bec, F. Quenet, S. Mounicou, J. Szpunar, B. Bouyssiere, R. Lobinski and C. Larroque, Metallomics, 2014, 6, 1382–1386 RSC.
- A. E. Egger, S. Theiner, C. Kornauth, P. Heffeter, W. Berger, B. K. Keppler and C. G. Hartinger, Metallomics, 2014, 6, 1616–1625 RSC.
- A. E. Egger, C. Kornauth, W. Haslik, S. Hann, S. Theiner, G. Bayer, C. G. Hartinger, B. K. Keppler, U. Pluschnig and R. M. Mader, Metallomics, 2015, 7, 508–515 RSC.
- D. Pozebon, V. L. Dressler, A. Matusch and J. S. Becker, Int. J. Mass Spectrom., 2008, 272, 57–62 CrossRef CAS PubMed.
- M. C. Leung, P. L. Williams, A. Benedetto, C. Au, K. J. Helmcke, M. Aschner and J. N. Meyer, Toxicol. Sci., 2008, 106, 5–28 CrossRef CAS PubMed.
- S. Brenner, Genetics, 1974, 77, 71–94 CAS.
- C. G. Knight, M. N. Patel, R. B. Azevedo and A. M. Leroi, Evol. Dev., 2002, 4, 16–27 CrossRef.
- P. Chen, E. J. Martinez-Finley, J. Bornhorst, S. Chakraborty and M. Aschner, Front. Aging Neurosci., 2013, 5, 18 CAS.
- D. Bergamaschi, Y. Samuels, N. J. O'Neil, G. Trigiante, T. Crook, J. K. Hsieh, D. J. O'Connor, S. Zhong, I. Campargue, M. L. Tomlinson, P. E. Kuwabara and X. Lu, Nat. Genet., 2003, 33, 162–167 CrossRef CAS PubMed.
- O. Hemmingsson, G. Kao, M. Still and P. Naredi, Cancer Res., 2010, 70, 10321–10328 CrossRef CAS PubMed.
- B. Natarajan, R. Gaur, O. Hemmingsson, G. Kao and P. Naredi, Worm, 2013, 2, e24059 CrossRef PubMed.
- H. Lans and W. Vermeulen, Mol. Biol. Int., 2011, 2011, 542795 Search PubMed.
- K. Asagoshi, W. Lehmann, E. K. Braithwaite, L. Santana-Santos, R. Prasad, J. H. Freedman, B. Van Houten and S. H. Wilson, Nucleic Acids Res., 2012, 40, 670–681 CrossRef CAS PubMed.
- B. Meier, S. L. Cooke, J. Weiss, A. P. Bailly, L. B. Alexandrov, J. Marshall, K. Raine, M. Maddison, E. Anderson, M. R. Stratton, A. Gartner and P. J. Campbell, Genome Res., 2014, 24, 1624–1636 CrossRef CAS PubMed.
- S. F. Roerink, W. Koole, L. C. Stapel, R. J. Romeijn and M. Tijsterman, PLoS Genet., 2012, 8, e1002800 CAS.
- T. M. Kauppinen, Neurochem. Int., 2007, 50, 954–958 CrossRef CAS PubMed.
- M. Javle and N. J. Curtin, Ther. Adv. Med. Oncol., 2011, 3, 257–267 CrossRef CAS PubMed.
- S. W. Yu, H. Wang, T. M. Dawson and V. L. Dawson, Neurobiol. Dis., 2003, 14, 303–317 CrossRef CAS PubMed.
- S. W. Caito, W. M. Valentine and M. Aschner, J. Neurochem., 2013, 127, 837–851 CrossRef CAS PubMed.
- R. Klockenkämper, Spectrosc. Int., 1990, 2, 26 Search PubMed.
- A. Matusch, A. Bauer and S. Becker, Int. J. Mass Spectrom., 2011, 307, 240–244 CrossRef CAS PubMed.
- J. S. Becker, J. Mass Spectrom., 2013, 48, 255–268 CrossRef PubMed.
- J. A. Pugh, A. G. Cox, C. W. McLeod, J. Bunch, M. J. Writer, S. L. Hart, A. Bienemann, E. White and J. Bell, Anal. Bioanal. Chem., 2012, 403, 1641–1649 CrossRef CAS PubMed.
- J. S. Duerr, WormBook, 2006, pp. 1–61 DOI:10.1895/wormbook.1.105.1.
- S. Amptoulach and N. Tsavaris, Chemother. Res. Pract., 2011, 2011, 843019 Search PubMed.
- S. N. Gagnon, M. O. Hengartner and S. Desnoyers, Biochem. J., 2002, 368, 263–271 CrossRef CAS PubMed.
- F. Dequen, S. N. Gagnon and S. Desnoyers, DNA Repair, 2005, 4, 814–825 CrossRef CAS PubMed.
- S. Beneke, Front. Genet., 2012, 3, 169 CAS.
- J. Bornhorst, S. Meyer, T. Weber, C. Boker, T. Marschall, A. Mangerich, S. Beneke, A. Burkle and T. Schwerdtle, Mol. Nutr. Food Res., 2013, 57, 1255–1269 CAS.
- J. Michels, I. Vitale, L. Galluzzi, J. Adam, K. A. Olaussen, O. Kepp, L. Senovilla, I. Talhaoui, J. Guegan, D. P. Enot, M. Talbot, A. Robin, P. Girard, C. Orear, D. Lissa, A. Q. Sukkurwala, P. Garcia, P. Behnam-Motlagh, K. Kohno, G. S. Wu, C. Brenner, P. Dessen, M. Saparbaev, J. C. Soria, M. Castedo and G. Kroemer, Cancer Res., 2013, 73, 2271–2280 CrossRef CAS PubMed.
- H. Cheng, Z. Zhang, A. Borczuk, C. A. Powell, A. S. Balajee, H. B. Lieberman and B. Halmos, Carcinogenesis, 2013, 34, 739–749 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1: light microscopic images and Pt images by LA-ICP-MS of WT worms and pme-1 deletion mutants of C. elegans L4 stage larvae, incubated with 50 μg mL−1 Cisplatin for 1 or 2 h; (A) WT worm after Cisplatin incubation for 1 h, (B) WT worm after Cisplatin incubation for 2 h, (C) pme-1 deletion mutant after Cisplatin incubation for 1 h, (D) pme-1 deletion mutant after Cisplatin incubation for 2 h. See DOI: 10.1039/c5mt00096c |
| This journal is © The Royal Society of Chemistry 2015 |