Open Access Article
Open Access ArticleCopper tolerance and virulence in bacteria
Erik
Ladomersky
ab and
Michael J.
Petris
*abc
aDepartment of Biochemistry, University of Missouri, Columbia, MO 65211, USA. E-mail: petrism@missouri.edu; Fax: +1-573-884-2537; Tel: +1-573-882-9685
bThe Christopher S. Bond Life Sciences Center, University of Missouri, Columbia, MO 65211, USA
cDepartment of Nutrition and Exercise Physiology, University of Missouri, Columbia, MO 65211, USA
First published on 23rd January 2015
Abstract
Copper (Cu) is an essential trace element for all aerobic organisms. It functions as a cofactor in enzymes that catalyze a wide variety of redox reactions due to its ability to cycle between two oxidation states, Cu(I) and Cu(II). This same redox property of copper has the potential to cause toxicity if copper homeostasis is not maintained. Studies suggest that the toxic properties of copper are harnessed by the innate immune system of the host to kill bacteria. To counter such defenses, bacteria rely on copper tolerance genes for virulence within the host. These discoveries suggest bacterial copper intoxication is a component of host nutritional immunity, thus expanding our knowledge of the roles of copper in biology. This review summarizes our current understanding of copper tolerance in bacteria, and the extent to which these pathways contribute to bacterial virulence within the host.
Introduction
Copper (Cu) has been used throughout much of human civilization as an antimicrobial agent. The earliest recorded medicinal use of copper can be traced to ancient Egyptian and Greek civilizations for the treatment of wounds and sterilization of water.1 Today, the antimicrobial properties of copper are utilized in many different materials. Between 2008 and 2011, the Environmental Protection Agency (EPA) registered more than 300 copper alloys as antimicrobial, underscoring the increasing application of copper-based materials in the manufacture of surfaces where the presence of microbes could lead to nosocomial infections.2 The realization that copper is used by the innate immune system is a relatively recent discovery. In contrast to other essential elements such as iron and manganese, which are withheld from the invading pathogen, host-derived copper appears to play a unique role in nutritional immunity by acting as a component of the antimicrobial arsenal produced by cells of the innate immune system. Several lines of evidence indicate that bacterial copper tolerance genes provide an important counter measure to this activity of the innate immune system. This review discusses our current understanding of bacterial copper tolerance pathways, and their contributions to pathogenesis within the host.The essentiality of copper
The inclusion of copper within the repertoire of elements essential for life is thought to have first arisen within ancient photosynthetic cyanobacteria following the release of oxygen into the atmosphere. The ensuing decrease in the bioavailability of iron due to its oxidative precipitation as insoluble Fe(III) hydroxides allowed for the incorporation of copper into energy capturing systems such as cytochrome oxidase.3 The essentiality of copper lies in its ability to undergo redox cycling between Cu(II), the oxidized cupric form, and Cu(I), the reduced cuprous form. As a soft Lewis acid, Cu(I) favors a tetrahedral coordination with soft bases such as hydrides, alkyl groups, phosphines, cysteinyl thiols and the thioether of methionine. Cu(II) is an intermediate Lewis acid that forms bonds with sulfates, nitrogen donors such as histidine, and oxygen donors such as glutamate and aspartate. The ability of copper to redox cycle between Cu(I) and Cu(II) endows copper-containing enzymes with redox potentials typically between +0.25 and +0.75 V,3 permitting the removal of electrons from diverse substrates such as catechols, superoxide, ascorbate and iron. Consequently, copper-dependent enzymes across kingdoms function in diverse processes including oxidative phosphorylation (cytochrome oxidase),4 iron homeostasis (ceruloplasmin, hephaestin),5 pigmentation (tyrosinase; laccase),6 superoxide dismutation (superoxide dismutases),7 and connective tissue formation (lysyl oxidases).8Mechanisms of bacterial copper toxicity
Despite the utility of copper as an enzymatic cofactor, its redox activity also creates a potential hazard to all life. Copper toxicity was probably an early and constant evolutionary pressure as suggested by the presence of homologous copper tolerance proteins within distantly related organisms such as bacteria and archaea.9 Several mechanisms have been ascribed to the toxic properties of copper. Under aerobic conditions, copper is proposed to catalyze the production of hydroxyl radicals via the Fenton and Haber–Weiss reactions:10| Cu(I) + H2O2 → Cu(II) + OH− + ˙OH |
| Cu(II) + ˙O2− → Cu(I) + O2 |
Because of the high standard reduction potential of the hydroxyl radical (>2 V),11 copper-catalyzed production of ˙OH is able to cause oxidative damage to most types of macromolecules.11,12 Because of its very short half-life (∼10−9 s), such damage would necessarily be diffusion-limited and thus restricted to macromolecules within the immediate vicinity of copper. Consistent with this notion, copper-mediated hydroxyl radical formation in Escherichia coli appears to be confined to the periplasmic regions where copper is most highly enriched.13 However, studies suggest that in the absence of oxygen, various non-Fenton based mechanisms are more important processes of copper toxicity.14 Such anoxic mechanisms involve the formation of adventitious Cu(I)–thiolate bonds, thus damaging enzymes that functionally depend on free cysteines or disulfide bonds.15 In E. coli, targets of anoxic copper toxicity include families of iron sulfur cluster proteins that are required for intermediary metabolism.14 In addition, excess copper is thought to lead to incorrect disulfide bond formation in the periplasm, as evidenced by studies showing that loss of the periplasmic disulfide bond isomerase, DsbC, which resolves incorrect disulfide bonds, renders E. coli more sensitive to copper toxicity.15
Mechanisms of microbial copper tolerance
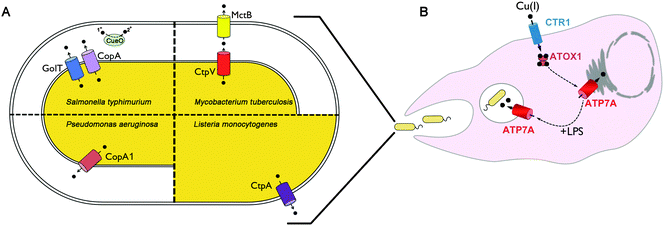
To avoid copper toxicity, all organisms have evolved copper handling machinery to maintain a cytoplasmic milieu that is devoid of free copper. This concept was initially based on studies of the E. coli copper-responsive transcription factor CueR.16 The finding that CueR induced the expression of copper tolerance genes at 10−21 M copper, which is many orders of magnitude lower than one free copper atom per cell, indicated that free unligated copper in the cytoplasm is not tolerated in bacteria. The principle mechanisms of copper tolerance in bacteria include: (1) transmembrane copper export, occurring from the cytoplasm into the periplasmic space or into the extracellular milieu; (2) copper sequestration by metallothioneins; and (3) oxidation of Cu(I) by multi-copper oxidases to generate the less toxic Cu(II) ion. Below is a general description of these mechanisms and their importance in bacterial virulence.While all bacteria appear to possess at least one copper exporting P1B-type ATPase, there is considerable diversity when it comes to alternative copper exporters among different bacteria. The E. coli CusABC complex is a large tripartite copper exporter found in the majority of gamma proteobacteria.24 Studies in E. coli demonstrate that the CusABC complex and its metallochaperone, CusF, mediates copper export across the inner and outer membranes via proton motive force,25,26 and is required for tolerance to moderately high copper concentrations, especially under anaerobic conditions.25 Interestingly, recent studies show that CusF acquires copper directly from the CopA suggesting it can function as a periplasmic target of this Cu(I)–ATPase.27 The Cus complex is comprised of an inner membrane proton–substrate carrier (CusA) and an outer membrane pore (CusC), which are connected by a linker protein, CusB in the periplasm.28–30 Recent studies suggest that copper-bound CusB facilitates cuprous ion delivery from the CusF metallochaperone to the CusABC complex.31,32 In the case of mycobacteria, copper export is dependent on the P1B-type ATPase, CtpV, located within the inner membrane,33 as well as MctB a pore-forming protein originally identified in the outer or inner membrane,34 although its precise role and function remain unknown. Mutation in either CtpV or MctB results in reduced copper tolerance due to hyperaccumulation of the metal.
There have been additional plasmid-encoded copper tolerance proteins identified in certain bacteria isolated from environments with extremely high copper concentrations. The pcoABCDRSE system of E. coli was initially discovered in a plasmid pRJ1004 within bacterial isolates from pigs fed a copper-supplemented diet.35 A homologous system, copABCDRS, was later found in plasmid pPT23D isolated from Pseudomonas syringae growing on tomato plants treated with copper-based fungicides.36,37 The pco and cop systems share four structural genes, pco/copABCD. PcoA and CopA are soluble periplasmic proteins with homology to multi-copper oxidases,38 and may function in a similar manner to the chromosomally encoded CueO multi-copper oxidase (see below). PcoB/CopB are located in the outer membrane with putative roles in copper translocation. CopC/PcoC are copper binding proteins located in the periplasm and may deliver copper to PcoD/CopD proteins located in the inner membrane.39,40 There is genetic evidence that PcoD may function in copper transport into the cytoplasm,41 however, this would appear to be at odds with its role in copper tolerance and remains to be demonstrated biochemically.
Recently, a novel mechanism of copper tolerance was identified in studies of yersiniabactin, a siderophore produced in Yersinia species and found in E. coli isolates from patients with urinary tract infection.43 Siderophores are small high-affinity iron chelating compounds secreted by microorganisms to scavenge iron within the host. Although required for iron acquisition, yersiniabactin is also capable of binding Cu(II); an interaction that confers copper tolerance by preventing the formation of the toxic Cu(I).43 In addition, recent studies using yersiniabactin-expressing E. coli demonstrated that the copper-bound siderophore possesses superoxide dismutase activity, which protects against the respiratory burst of macrophages.44 Thus, it would appear that yersiniabactin is a highly versatile siderophore capable of counteracting multiple host defenses including iron limitation, copper toxicity and the oxidative burst.
While less information is available for other bacterial species, in general there is a positive correlation between copper sensitivity and virulence (Fig. 1A). Mutations in the P. aeruginosa copA1 gene (formerly cueA) renders this pathogen sensitive to high copper concentrations when grown in vitro, and significantly attenuates virulence as determined by bacterial colonization of the spleen in mice, and the number of bacteria required to kill mice.65 Similarly, mutation of the CtpA copper exporting P-type ATPase in Listeria monocytogenes significantly reduces liver colonization of in mice,66 and loss of the CopA copper exporting P-type ATPase of Streptococcus pneumonia reduces colonization of the lung and nasopharynx of infected mice compared to wild type strains.67 In S. typhimurium, loss of both the CopA and GolT copper exporting P-type ATPases causes copper hypersensitivity and reduces bacterial survival within cultured macrophages.63 However, such mutations did not affect tissue colonization using a systemic infection model in mice.63 In contrast, loss of the CueO multi-copper oxidase in S. typhimurium was found to reduce colonization of the lung and spleen following orogastric infection of mice, however, there was no effect on bacterial survival in cultured RAW264.7 macrophage cells.68 Taken as a whole, these studies indicate a strong, albeit imperfect, correlation between copper tolerance and virulence in the host.
Host-derived copper as a mechanism to control infections
Copper is important for both adaptive and innate immune function. Human infants with the copper deficiency disorder, Menkes disease, exhibit higher incidences of lung and bladder infection.69–72 Copper deficiency in animals has been shown to impair the production of antibody-producing cells,73 suppress the respiratory burst of neutrophils and macrophages,74,75 and limit the ability of the host to combat infection.76–81 It has been known for decades that infection or inflammation induces marked changes in copper homeostasis in the host. For example, copper concentrations in the serum are significantly elevated in response to inflammation.82–93 This is due, in part, to the increased production and secretion of ceruloplasmin, an acute phase response protein which contains approximately 85% of total serum copper.94 Copper accumulates at sites of inflammation and injury,95 including within granulomatous lung tissues of guinea pigs infected with M. tuberculosis.34 While the underlying mechanisms by which copper deficiency limits the ability to fight infection are not fully understood, a supply of copper to the phagocytic cells of the innate immune system appears to be of particular importance. Copper concentrations are known to accumulate within the phagolysosomal compartments of interferon-gamma activated peritoneal macrophages challenged with different mycobacteria species.96 Insight into the underlying molecular basis of these changes came from studies of RAW 267.4 macrophage cells in which activation with interferon-gamma or bacterial lipopolysaccharide (LPS) was found to stimulate copper uptake by increasing the expression of the copper importer, CTR197,98 (Fig. 1B). Moreover, these same inflammatory conditions triggered the increased expression and trafficking of the ATP7A copper pump from the Golgi complex to cytoplasmic vesicles and the phagolysosome, thus providing a possible mechanism for concentrating bactericidal copper within this compartment during infection97 (Fig. 1B). Consistent with this model, studies have demonstrated the ability of RAW264.7 cultured macrophages to kill E. coli was found to be dependent on the expression of ATP7A.97 It will be important to test the extent to which ATP7A is required for killing other bacterial pathogens and whether it plays a significant role in whole animal studies of infection.Concluding remarks
Whereas nutritional immunity is a term that has been historically applied to metals such as iron and manganese that are withheld from invading pathogens, the unique roles of copper in host immune defense now expand this concept to encompass nutrient intoxication. There are several outstanding questions to be addressed in the coming years: What is the mechanism by which host-derived copper kills bacteria? Why do certain mutations cause copper sensitivity in bacterial pathogens without affecting virulence? What is the role of copper-containing ceruloplasmin in host immune function and does this protein provide a means of mobilizing systemic copper to sites of infection? Can drugs that are designed to inhibit bacterial copper tolerance proteins, or enhance copper delivery to the pathogen,99,100 give rise to new classes of antibiotics? To what extent is host-derived copper effective against non-bacterial pathogens? Does the widespread use of copper as a dietary supplement in livestock contribute to the virulence of enteric pathogens that may enter the human food supply?101 The answers to such questions will require multidisciplinary approaches to unravel the genetic and physiological basis of copper handling pathways within both host and pathogen.Conflict of interest statement
The authors have no conflicts of interest to declare.Acknowledgements
The Petris lab is supported by funds from the NIH (DK093866). We apologize to any colleagues whose work could not be cited due to space limitations.References
- H. H. A. Dollwet and J. R. J. Sorenson, Historic uses of copper compounds in medicine, Trace Elem. Med., 1985, 2, 80–87 Search PubMed.
- A. L. Casey, D. Adams, T. J. Karpanen, P. A. Lambert, B. D. Cookson, P. Nightingale, L. Miruszenko, R. Shillam, P. Christian and T. S. Elliott, Role of copper in reducing hospital environment contamination, J. Hosp. Infect., 2010, 74, 72–77 CrossRef CAS PubMed.
- R. R. Crichton and J. L. Pierre, Old iron, young copper: from Mars to Venus, BioMetals, 2001, 14, 99–112 CrossRef CAS.
- P. A. Cobine, F. Pierrel and D. R. Winge, Copper trafficking to the mitochondrion and assembly of copper metalloenzymes, Biochim. Biophys. Acta, 2006, 1763, 759–772 CrossRef CAS PubMed.
- J. R. Prohaska, Impact of copper limitation on expression and function of multicopper oxidases (ferroxidases), Adv. Nutr., 2011, 2, 89–95 CrossRef CAS PubMed.
- C. A. Ramsden and P. A. Riley, Tyrosinase: the four oxidation states of the active site and their relevance to enzymatic activation, oxidation and inactivation, Bioorg. Med. Chem., 2014, 22, 2388–2395 CrossRef CAS PubMed.
- Y. Sheng, M. Chattopadhyay, J. Whitelegge and J. S. Valentine, SOD1 aggregation and ALS: role of metallation states and disulfide status, Curr. Top. Med. Chem., 2012, 12, 2560–2572 CrossRef CAS.
- J. Finney, H. J. Moon, T. Ronnebaum, M. Lantz and M. Mure, Human copper-dependent amine oxidases, Arch. Biochem. Biophys., 2014, 546, 19–32 CrossRef CAS PubMed.
- M. Solioz, H. K. Abicht, M. Mermod and S. Mancini, Response of gram-positive bacteria to copper stress, JBIC, J. Biol. Inorg. Chem., 2010, 15, 3–14 CrossRef CAS PubMed.
- S. I. Liochev and I. Fridovich, The Haber–Weiss cycle – 70 years later: an alternative view, Redox Rep., 2002, 7, 55–57 CrossRef CAS PubMed ; author reply 59–60.
- W. Freinbichler, M. A. Colivicchi, C. Stefanini, L. Bianchi, C. Ballini, B. Misini, P. Weinberger, W. Linert, D. Vareslija, K. F. Tipton and L. Della Corte, Highly reactive oxygen species: detection, formation, and possible functions, Cell. Mol. Life Sci., 2011, 68, 2067–2079 CrossRef CAS PubMed.
- Y. Yoshida, S. Furuta and E. Niki, Effects of metal chelating agents on the oxidation of lipids induced by copper and iron, Biochim. Biophys. Acta, 1993, 1210, 81–88 CrossRef CAS.
- L. Macomber, C. Rensing and J. A. Imlay, Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli, J. Bacteriol., 2007, 189, 1616–1626 CrossRef CAS PubMed.
- L. Macomber and J. A. Imlay, The iron–sulfur clusters of dehydratases are primary intracellular targets of copper toxicity, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 8344–8349 CrossRef CAS PubMed.
- A. Hiniker, J. F. Collet and J. C. Bardwell, Copper stress causes an in vivo requirement for the Escherichia coli disulfide isomerase DsbC, J. Biol. Chem., 2005, 280, 33785–33791 CrossRef CAS PubMed.
- A. Changela, K. Chen, Y. Xue, J. Holschen, C. E. Outten, T. V. O'Halloran and A. Mondragon, Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR, Science, 2003, 301, 1383–1387 CrossRef CAS PubMed.
- J. M. Arguello, E. Eren and M. Gonzalez-Guerrero, The structure and function of heavy metal transport P1B-ATPases, BioMetals, 2007, 20, 233–248 CrossRef PubMed.
- M. Andersson, D. Mattle, O. Sitsel, T. Klymchuk, A. M. Nielsen, L. B. Moller, S. H. White, P. Nissen and P. Gourdon, Copper-transporting P-type ATPases use a unique ion-release pathway, Nat. Struct. Mol. Biol., 2014, 21, 43–48 CAS.
- A. C. Rosenzweig and J. M. Arguello, Toward a molecular understanding of metal transport by P(1B)-type ATPases, Curr. Top. Membr., 2012, 69, 113–136 CAS.
- T. Padilla-Benavides, C. J. McCann and J. M. Arguello, The mechanism of Cu+ transport ATPases: interaction with Cu+ chaperones and the role of transient metal-binding sites, J. Biol. Chem., 2013, 288, 69–78 CrossRef CAS PubMed.
- Y. Fu, H. C. Tsui, K. E. Bruce, L. T. Sham, K. A. Higgins, J. P. Lisher, K. M. Kazmierczak, M. J. Maroney, C. E. Dann 3rd, M. E. Winkler and D. P. Giedroc, A new structural paradigm in copper resistance in Streptococcus pneumoniae, Nat. Chem. Biol., 2013, 9, 177–183 CrossRef CAS PubMed.
- O. Preisig, R. Zufferey and H. Hennecke, The Bradyrhizobium japonicum fixGHIS genes are required for the formation of the high-affinity cbb3-type cytochrome oxidase, Arch. Microbiol., 1996, 165, 297–305 CrossRef CAS.
- D. Osman, C. J. Patterson, K. Bailey, K. Fisher, N. J. Robinson, S. E. Rigby and J. S. Cavet, The copper supply pathway to a Salmonella Cu,Zn-superoxide dismutase (SodCII) involves P(1B)-type ATPase copper efflux and periplasmic CueP, Mol. Microbiol., 2013, 87, 466–477 CrossRef CAS PubMed.
- G. Hernandez-Montes, J. M. Arguello and B. Valderrama, Evolution and diversity of periplasmic proteins involved in copper homeostasis in gamma proteobacteria, BMC Microbiol., 2012, 12, 249 CrossRef CAS PubMed.
- F. W. Outten, D. L. Huffman, J. A. Hale and T. V. O'Halloran, The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli, J. Biol. Chem., 2001, 276, 30670–30677 CrossRef CAS PubMed.
- S. Franke, G. Grass, C. Rensing and D. H. Nies, Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli, J. Bacteriol., 2003, 185, 3804–3812 CrossRef.
- T. Padilla-Benavides, A. M. George Thompson, M. M. McEvoy and J. M. Arguello, Mechanism of ATPase-mediated Cu+ export and delivery to periplasmic chaperones: the interaction of Escherichia coli CopA and CusF, J. Biol. Chem., 2014, 289, 20492–20501 CrossRef CAS PubMed.
- H. I. Zgurskaya and H. Nikaido, AcrA is a highly asymmetric protein capable of spanning the periplasm, J. Mol. Biol., 1999, 285, 409–420 CrossRef CAS PubMed.
- C. C. Su, F. Yang, F. Long, D. Reyon, M. D. Routh, D. W. Kuo, A. K. Mokhtari, J. D. Van Ornam, K. L. Rabe, J. A. Hoy, Y. J. Lee, K. R. Rajashankar and E. W. Yu, Crystal structure of the membrane fusion protein CusB from Escherichia coli, J. Mol. Biol., 2009, 393, 342–355 CrossRef CAS PubMed.
- E. H. Kim, D. H. Nies, M. M. McEvoy and C. Rensing, Switch or funnel: how RND-type transport systems control periplasmic metal homeostasis, J. Bacteriol., 2011, 193, 2381–2387 CrossRef CAS PubMed.
- T. D. Mealman, I. Bagai, P. Singh, D. R. Goodlett, C. Rensing, H. Zhou, V. H. Wysocki and M. M. McEvoy, Interactions between CusF and CusB identified by NMR spectroscopy and chemical cross-linking coupled to mass spectrometry, Biochemistry, 2011, 50, 2559–2566 CrossRef CAS PubMed.
- K. N. Chacon, T. D. Mealman, M. M. McEvoy and N. J. Blackburn, Tracking metal ions through a Cu/Ag efflux pump assigns the functional roles of the periplasmic proteins, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 15373–15378 CrossRef CAS PubMed.
- S. K. Ward, B. Abomoelak, E. A. Hoye, H. Steinberg and A. M. Talaat, CtpV: a putative copper exporter required for full virulence of Mycobacterium tuberculosis, Mol. Microbiol., 2010, 77, 1096–1110 CrossRef CAS PubMed.
- F. Wolschendorf, D. Ackart, T. B. Shrestha, L. Hascall-Dove, S. Nolan, G. Lamichhane, Y. Wang, S. H. Bossmann, R. J. Basaraba and M. Niederweis, Copper resistance is essential for virulence of Mycobacterium tuberculosis, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 1621–1626 CrossRef CAS PubMed.
- T. J. Tetaz and R. K. Luke, Plasmid-controlled resistance to copper in Escherichia coli, J. Bacteriol., 1983, 154, 1263–1268 CAS.
- C. L. Bender and D. A. Cooksey, Indigenous plasmids in Pseudomonas syringae pv. tomato: conjugative transfer and role in copper resistance, J. Bacteriol., 1986, 165, 534–541 CAS.
- J. S. Cha and D. A. Cooksey, Copper resistance in Pseudomonas syringae mediated by periplasmic and outer membrane proteins, Proc. Natl. Acad. Sci. U. S. A., 1991, 88, 8915–8919 CrossRef CAS.
- K. Y. Djoko, Z. Xiao and A. G. Wedd, Copper resistance in E. coli: the multicopper oxidase PcoA catalyzes oxidation of copper(I) in Cu(I)Cu(II)-PcoC, ChemBioChem, 2008, 9, 1579–1582 CrossRef CAS PubMed.
- F. Arnesano, L. Banci, I. Bertini, I. C. Felli, C. Luchinat and A. R. Thompsett, A strategy for the NMR characterization of type II copper(II) proteins: the case of the copper trafficking protein CopC from Pseudomonas Syringae, J. Am. Chem. Soc., 2003, 125, 7200–7208 CrossRef CAS PubMed.
- F. Arnesano, L. Banci, I. Bertini, S. Mangani and A. R. Thompsett, A redox switch in CopC: an intriguing copper trafficking protein that binds copper(I) and copper(II) at different sites, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 3814–3819 CrossRef CAS PubMed.
- J. S. Cha and D. A. Cooksey, Copper Hypersensitivity and Uptake in Pseudomonas syringae Containing Cloned Components of the Copper Resistance Operon, Appl. Environ. Microbiol., 1993, 59, 1671–1674 CAS.
- B. Gold, H. Deng, R. Bryk, D. Vargas, D. Eliezer, J. Roberts, X. Jiang and C. Nathan, Identification of a copper-binding metallothionein in pathogenic mycobacteria, Nat. Chem. Biol., 2008, 4, 609–616 CrossRef CAS PubMed.
- K. S. Chaturvedi, C. S. Hung, J. R. Crowley, A. E. Stapleton and J. P. Henderson, The siderophore yersiniabactin binds copper to protect pathogens during infection, Nat. Chem. Biol., 2012, 8, 731–736 CrossRef CAS PubMed.
- K. S. Chaturvedi, C. S. Hung, D. E. Giblin, S. Urushidani, A. M. Austin, M. C. Dinauer and J. P. Henderson, Cupric yersiniabactin is a virulence-associated superoxide dismutase mimic, ACS Chem. Biol., 2014, 9, 551–561 CrossRef CAS PubMed.
- S. A. Roberts, A. Weichsel, G. Grass, K. Thakali, J. T. Hazzard, G. Tollin, C. Rensing and W. R. Montfort, Crystal structure and electron transfer kinetics of CueO, a multicopper oxidase required for copper homeostasis in Escherichia coli, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 2766–2771 CrossRef CAS PubMed.
- G. Grass and C. Rensing, CueO is a multi-copper oxidase that confers copper tolerance in Escherichia coli, Biochem. Biophys. Res. Commun., 2001, 286, 902–908 CrossRef CAS PubMed.
- J. L. Rowland and M. Niederweis, Resistance mechanisms of Mycobacterium tuberculosis against phagosomal copper overload, Tuberculosis, 2012, 92, 202–210 CrossRef CAS PubMed.
- G. Grass, K. Thakali, P. E. Klebba, D. Thieme, A. Muller, G. F. Wildner and C. Rensing, Linkage between catecholate siderophores and the multicopper oxidase CueO in Escherichia coli, J. Bacteriol., 2004, 186, 5826–5833 CrossRef CAS PubMed.
- F. W. Outten, C. E. Outten, J. Hale and T. V. O'Halloran, Transcriptional activation of an Escherichia coli copper efflux regulon by the chromosomal MerR homologue, cueR, J. Biol. Chem., 2000, 275, 31024–31029 CrossRef CAS PubMed.
- J. V. Stoyanov, J. L. Hobman and N. L. Brown, CueR (YbbI) of Escherichia coli is a MerR family regulator controlling expression of the copper exporter CopA, Mol. Microbiol., 2001, 39, 502–511 CrossRef CAS.
- J. S. Kim, M. H. Kim, M. H. Joe, S. S. Song, I. S. Lee and S. Y. Choi, The sctR of Salmonella enterica serova Typhimurium encoding a homologue of MerR protein is involved in the copper-responsive regulation of cuiD, FEMS Microbiol. Lett., 2002, 210, 99–103 CrossRef CAS PubMed.
- S. K. Checa, M. Espariz, M. E. Audero, P. E. Botta, S. V. Spinelli and F. C. Soncini, Bacterial sensing of and resistance to gold salts, Mol. Microbiol., 2007, 63, 1307–1318 CrossRef CAS PubMed.
- L. B. Pontel, M. E. Audero, M. Espariz, S. K. Checa and F. C. Soncini, GolS controls the response to gold by the hierarchical induction of Salmonella-specific genes that include a CBA efflux-coding operon, Mol. Microbiol., 2007, 66, 814–825 CrossRef CAS PubMed.
- G. P. Munson, D. L. Lam, F. W. Outten and T. V. O'Halloran, Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12, J. Bacteriol., 2000, 182, 5864–5871 CrossRef CAS.
- G. T. Smaldone and J. D. Helmann, CsoR regulates the copper efflux operon copZA in Bacillus subtilis, Microbiology, 2007, 153, 4123–4128 CrossRef CAS PubMed.
- T. Liu, A. Ramesh, Z. Ma, S. K. Ward, L. Zhang, G. N. George, A. M. Talaat, J. C. Sacchettini and D. P. Giedroc, CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator, Nat. Chem. Biol., 2007, 3, 60–68 CrossRef CAS PubMed.
- H. Teramoto, M. Inui and H. Yukawa, Corynebacterium glutamicum CsoR acts as a transcriptional repressor of two copper/zinc-inducible P(1B)-type ATPase operons, Biosci., Biotechnol., Biochem., 2012, 76, 1952–1958 CrossRef CAS PubMed.
- J. Baker, M. Sengupta, R. K. Jayaswal and J. A. Morrissey, The Staphylococcus aureus CsoR regulates both chromosomal and plasmid-encoded copper resistance mechanisms, Environ. Microbiol., 2011, 13, 2495–2507 CrossRef CAS PubMed.
- R. A. Festa, M. B. Jones, S. Butler-Wu, D. Sinsimer, R. Gerads, W. R. Bishai, S. N. Peterson and K. H. Darwin, A novel copper-responsive regulon in Mycobacterium tuberculosis, Mol. Microbiol., 2011, 79, 133–148 CrossRef CAS PubMed.
- X. Shi, R. A. Festa, T. R. Ioerger, S. Butler-Wu, J. C. Sacchettini, K. H. Darwin and M. I. Samanovic, The copper-responsive RicR regulon contributes to Mycobacterium tuberculosis virulence, mBio, 2014, 5, e00876 Search PubMed.
- J. E. Graham and J. E. Clark-Curtiss, Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS), Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 11554–11559 CrossRef CAS.
- D. M. Heithoff, C. P. Conner, P. C. Hanna, S. M. Julio, U. Hentschel and M. J. Mahan, Bacterial infection as assessed by in vivo gene expression, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 934–939 CrossRef CAS.
- D. Osman, K. J. Waldron, H. Denton, C. M. Taylor, A. J. Grant, P. Mastroeni, N. J. Robinson and J. S. Cavet, Copper homeostasis in Salmonella is atypical and copper-CueP is a major periplasmic metal complex, J. Biol. Chem., 2010, 285, 25259–25268 CrossRef CAS PubMed.
- S. K. Ward, B. Abomoelak, E. A. Hoye, H. Steinberg and A. M. Talaat, CtpV: a putative copper exporter required for full virulence of Mycobacterium tuberculosis, Mol. Microbiol., 2010, 77, 1096–1110 CrossRef CAS PubMed.
- W. R. Schwan, P. Warrener, E. Keunz, C. K. Stover and K. R. Folger, Mutations in the cueA gene encoding a copper homeostasis P-type ATPase reduce the pathogenicity of Pseudomonas aeruginosa in mice, Int. J. Med. Microbiol., 2005, 295, 237–242 CrossRef CAS PubMed.
- M. S. Francis and C. J. Thomas, Mutants in the CtpA copper transporting P-type ATPase reduce virulence of Listeria monocytogenes, Microb. Pathog., 1997, 22, 67–78 CrossRef CAS PubMed.
- S. Shafeeq, H. Yesilkaya, T. G. Kloosterman, G. Narayanan, M. Wandel, P. W. Andrew, O. P. Kuipers and J. A. Morrissey, The cop operon is required for copper homeostasis and contributes to virulence in Streptococcus pneumoniae, Mol. Microbiol., 2011, 81, 1255–1270 CrossRef CAS PubMed.
- M. E. Achard, J. J. Tree, J. A. Holden, K. R. Simpfendorfer, O. L. Wijburg, R. A. Strugnell, M. A. Schembri, M. J. Sweet, M. P. Jennings and A. G. McEwan, The multi-copper-ion oxidase CueO of Salmonella enterica serovar Typhimurium is required for systemic virulence, Infect. Immun., 2010, 78, 2312–2319 CrossRef CAS PubMed.
- J. Kreuder, A. Otten, H. Fuder, Z. Tumer, T. Tonnesen, N. Horn and D. Dralle, Clinical and biochemical consequences of copper-histidine therapy in Menkes disease, Eur. J. Pediatr., 1993, 152, 828–832 CrossRef CAS.
- T. R. Gunn, S. Macfarlane and L. I. Phillips, Difficulties in the neonatal diagnosis of Menkes' kinky hair syndrome–trichopoliodystrophy, Clin. Pediatr., 1984, 23, 514–516 CrossRef CAS PubMed.
- H. Uno and S. Arya, Neuronal and vascular disorders of the brain and spinal cord in Menkes kinky hair disease, Am. J. Med. Genet. Suppl., 1987, 3, 367–377 CrossRef CAS.
- F. Agertt, A. C. Crippa, P. J. Lorenzoni, R. H. Scola, I. Bruck, L. Paola, C. E. Silvado and L. C. Werneck, Menkes' disease: case report, Arq. Neuropsiquiatr., 2007, 65, 157–160 CrossRef PubMed.
- J. R. Prohaska and O. A. Lukasewycz, Copper deficiency suppresses the immune response of mice, Science, 1981, 213, 559–561 CAS.
- U. Babu and M. L. Failla, Respiratory burst and candidacidal activity of peritoneal macrophages are impaired in copper-deficient rats, J. Nutr., 1990, 120, 1692–1699 CAS.
- U. Babu and M. L. Failla, Copper status and function of neutrophils are reversibly depressed in marginally and severely copper-deficient rats, J. Nutr., 1990, 120, 1700–1709 CAS.
- D. G. Jones and N. F. Suttle, Some effects of copper deficiency on leucocyte function in sheep and cattle, Res. Vet. Sci., 1981, 31, 151–156 CAS.
- R. Boyne and J. R. Arthur, Effects of selenium and copper deficiency on neutrophil function in cattle, J. Comp. Pathol., 1981, 91, 271–276 CrossRef CAS.
- D. G. Jones and N. F. Suttle, The effect of copper deficiency on the resistance of mice to infection with Pasteurella haemolytica, J. Comp. Pathol., 1983, 93, 143–149 CrossRef CAS.
- A. Crocker, C. Lee, G. Aboko-Cole and C. Durham, Interaction of nutrition and infection: effect of copper deficiency on resistance to Trypanosoma lewisi, J. Natl. Med. Assoc., 1992, 84, 697–706 CAS.
- P. M. Newberne, C. E. Hunt and V. R. Young, The role of diet and the reticuloendothelial system in the response of rats to Salmonella typhilmurium infection, Br. J. Exp. Pathol., 1968, 49, 448–457 CAS.
- A. D. Smith, S. Botero and O. A. Levander, Copper deficiency increases the virulence of amyocarditic and myocarditic strains of coxsackievirus B3 in mice, J. Nutr., 2008, 138, 849–855 CrossRef CAS PubMed.
- D. E. Auer, J. C. Ng, H. L. Thompson, S. Inglis and A. A. Seawright, Acute phase response in horses: changes in plasma cation concentrations after localised tissue injury, Vet. Rec., 1989, 124, 235–239 CAS.
- A. Morimoto, Y. Sakata, T. Watanabe and N. Murakami, Characteristics of fever and acute-phase response induced in rabbits by IL-1 and TNF, Am. J. Physiol., 1989, 256, R35–R41 CAS.
- E. Akcil, G. Yavuz and M. Kocak, Effects of inflammation and antiinflammatory treatment on serum trace elements concentrations, Biol. Trace Elem. Res., 2003, 93, 95–104 CrossRef CAS.
- R. Milanino, M. Marrella, R. Gasperini, M. Pasqualicchio and G. Velo, Copper and zinc body levels in inflammation: an overview of the data obtained from animal and human studies, Agents Actions, 1993, 39, 195–209 CrossRef CAS.
- W. F. Ward, A. Molteni, C. Ts'ao and H. Ischiropoulos, Serum copper concentration as an index of experimental lung injury, Adv. Exp. Med. Biol., 1989, 258, 287–302 CAS.
- I. Shah, L. M. Lewkow and U. Khilanani, Correlation of hypercupremia with other acute phase reactants in malignant lymphoma, Cancer, 1983, 51, 851–854 CrossRef CAS.
- A. Yilmaz, R. A. Sari, M. Gundogdu, N. Kose and E. Dag, Trace elements and some extracellular antioxidant proteins levels in serum of patients with systemic lupus erythematosus, Clin. Rheumatol., 2005, 24, 331–335 CrossRef PubMed.
- A. Koyanagi, D. Kuffo, L. Gresely, A. Shenkin and L. E. Cuevas, Relationships between serum concentrations of C-reactive protein and micronutrients, in patients with tuberculosis, Ann. Trop. Med. Parasitol., 2004, 98, 391–399 CrossRef CAS PubMed.
- U. Srinivas, J. H. Braconier, B. Jeppsson, M. Abdulla, B. Akesson and P. A. Ockerman, Trace element alterations in infectious diseases, Scand. J. Clin. Lab. Invest., 1988, 48, 495–500 CrossRef CAS.
- R. Hallgren, N. Feltelius and U. Lindh, Redistribution of minerals and trace elements in chronic inflammation–a study on isolated blood cells from patients with ankylosing spondylitis, J. Rheumatol., 1987, 14, 548–553 CAS.
- A. Conforti, L. Franco, R. Milanino, A. Totorizzo and G. P. Velo, Copper metabolism during acute inflammation: studies on liver and serum copper concentrations in normal and inflamed rats, Br. J. Pharmacol., 1983, 79, 45–52 CrossRef CAS PubMed.
- A. Seyrek, A. Kocyigit and O. Erel, Essential trace elements selenium, zinc, copper, and iron concentrations and their related acute-phase proteins in patients with vivax malaria, Biol. Trace Elem. Res., 2005, 106, 107–115 CrossRef CAS.
- M. Correale, A. Totaro, S. Abruzzese, M. Di Biase and N. D. Brunetti, Acute phase proteins in acute coronary syndrome: an up-to-date, Cardiovasc. Hematol. Agents Med. Chem., 2012, 10, 352–361 CrossRef CAS.
- S. J. Beveridge, I. R. Garrett, M. W. Whitehouse, B. Vernon-Roberts and P. M. Brooks, Biodistribution of 64Cu in inflamed rats following administration of two anti-inflammatory copper complexes, Agents Actions, 1985, 17, 104–111 CrossRef CAS.
- D. Wagner, J. Maser, B. Lai, Z. Cai, C. E. Barry 3rd, K. Honer Zu Bentrup, D. G. Russell and L. E. Bermudez, Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell's endosomal system, J. Immunol., 2005, 174, 1491–1500 CrossRef CAS.
- C. White, J. Lee, T. Kambe, K. Fritsche and M. J. Petris, A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity, J. Biol. Chem., 2009, 284, 33949–33956 CrossRef CAS PubMed.
- C. White, T. Kambe, Y. G. Fulcher, S. W. Sachdev, A. I. Bush, K. Fritsche, J. Lee, T. P. Quinn and M. J. Petris, Copper transport into the secretory pathway is regulated by oxygen in macrophages, J. Cell Sci., 2009, 122, 1315–1321 CrossRef CAS PubMed.
- R. A. Festa, M. E. Helsel, K. J. Franz and D. J. Thiele, Exploiting innate immune cell activation of a copper-dependent antimicrobial agent during infection, Chem. Biol., 2014, 21, 977–987 CrossRef CAS PubMed.
- A. Speer, T. B. Shrestha, S. H. Bossmann, R. J. Basaraba, G. J. Harber, S. M. Michalek, M. Niederweis, O. Kutsch and F. Wolschendorf, Copper-boosting compounds: a novel concept for antimycobacterial drug discovery, Antimicrob. Agents Chemother., 2013, 57, 1089–1091 CrossRef CAS PubMed.
- A. R. Freitas, T. M. Coque, C. Novais, A. M. Hammerum, C. H. Lester, M. J. Zervos, S. Donabedian, L. B. Jensen, M. V. Francia, F. Baquero and L. Peixe, Human and swine hosts share vancomycin-resistant Enterococcus faecium CC17 and CC5 and Enterococcus faecalis CC2 clonal clusters harboring Tn1546 on indistinguishable plasmids, J. Clin. Microbiol., 2011, 49, 925–931 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2015 |