The use of metabolomics in the study of metals in biological systems
Oliver A. H.
Jones
*a,
Daniel A.
Dias
b,
Damien L.
Callahan
c,
Konstantinos A.
Kouremenos
d,
David J.
Beale
e and
Ute
Roessner
b
aSchool of Applied Sciences, RMIT University, GPO Box 2476, Melbourne, VIC 3001, Australia. E-mail: oliver.jones@rmit.edu.au; Fax: +61 (03)9925 3747; Tel: +61 (03)9925 2632
bMetabolomics Australia, School of Botany, The University of Melbourne, Parkville, VIC 3010, Australia
cCentre for Chemistry and Biotechnology, School of Life and Environmental Sciences, Deakin University, Burwood, Melbourne VIC 3125, Australia
dMetabolomics Australia, Bio21 Molecular Science and Biotechnology Institute, The University of Melbourne, 30 Flemington Road, VIC 3010, Australia
eCommonwealth Scientific and Industrial Research Organisation (CSIRO), Land and Water, 37 Graham Road, Highett, VIC 3190, Australia. Tel: +61 3 9252 6602
First published on 16th July 2014
Abstract
Metabolomics may be defined as the comprehensive quantitative and/or qualitative analysis of all metabolites present in a bio-fluid, cell, tissue, or organism. It is essentially the study of biochemical phenotypes (or metabotypes). Metabolic profiles are context dependent, and vary in response to a variety of factors including environment and environmental stimuli, health status, disease and a myriad of other factors; as such, metabolomics has been applied to a wide range of fields and has been increasingly utilised to the study of the roles played by metals in a range of biological systems as well as, encouragingly, in understanding the underlying biochemical mechanisms. The role of metals (and metalloids) in biological organisms is complex and the majority of studies in this area have been performed in plants but the fields of natural product chemistry, human health and even bacterial corrosion of water distribution systems have been investigated using this technique. In this review some of the novel approaches in which the metabolomics toolbox has been used to unravel the roles of metals and metalloids in a range of biological systems are discussed and suggestions made for future research.
1. Background/introduction
Metal ions are essential for the function of around one third of enzymes and are involved in a number of key biological processes, such as oxygen transport and electron transfer (oxidation–reduction) as well as structural co-factors such as zinc (Zn);1 indeed it has been estimated that as many as 1% of proteins in the human genome contain the zinc finger protein domain.2 While the lack of these essential metals may be detrimental to health they can also cause negative effects when present in excess concentrations. For example, iron (as Fe2+ and Fe3+) is a vital component of several biomolecules including haemoglobin, myoglobin, and cytochromes and is also an essential cofactor for non-heme enzymes such as ribonucleotide reductase (the limiting enzyme for DNA synthesis).3 However, excess Fe is toxic as it generates superoxide anions and hydroxyl radicals that react readily with many biomolecules, including proteins, lipids and nucleic acids.3 Elevated Fe (and copper, Cu) levels have also been implicated in heart disease.4In contrast, some metals such as lead (Pb), cadmium (Cd), mercury (Hg), as well as the metalloid arsenic (As) have only negative consequences to human and environmental health and present obvious concerns due to their persistence within ecosystems. Assessing the potential toxic effects and quantifying the risks associated with the exposure to these elements remains a major challenge for environmental scientists, risk assessors and regulators since, in many cases, the biological/biochemical mechanisms behind many observed toxic effects are not yet fully elucidated.
How then might one best investigate both the beneficial and detrimental activities of metals in biological systems? In recent years it has become apparent that metabolomics can provide novel insights in this area. Metabolomics (sometimes also known as metabonomics, or metabolic fingerprinting) is the analysis of endogenous and exogenous, low molecular mass metabolites within a cell, tissue, or biofluid of an organism, typically measured in a system which is responding to a change such as an external stress or disease.5 An organism's metabolome is its full complement of metabolites in the same way that its genome and proteome comprise its complete genetic and protein content, respectively. Whilst the concept of metabolic analysis is not new, one aspect of metabolomics, which distinguishes it from previous metabolically based studies, is the attempt to simultaneously detect and quantify as many metabolites as possible, in what is generally referred to as the ‘global approach’. Analytical techniques for metabolomics included nuclear magnetic resonance spectroscopy (NMR), gas and liquid chromatography mass spectrometry (GC and LC-MS) and where metals are concerned, inductively coupled plasma-mass spectroscopy (ICP-MS). The use of these techniques for metabolomics are not discussed here as they have been extensively reviewed elsewhere.6–8
The detection of biological indicators using metabolomics is not limited to systems in the natural state, as demonstrated by work undertaken on pipe biofilms by Beale et al. and Kouremenos et al.9–11 In these studies, samples isolated from biofilms known to cause microbial influenced corrosion (MIC) were exposed to Cu in consortia and as isolates. It was found that the biofilm metabolites of bacteria causing Cu pipe MIC comprised of a combination of organic acids, amino acids and lipids. These are common in biological metabolic processes, specifically those relating to soluble monomers and sulphur reducing bacteria substrates.9 In addition, the range of carboxylic acids released from the isolates (Bosea, Methylobacterium, Paenibacillus, Sphingomonas and Variovorax) suggest that the corrosion potential of these biofilms varies, which would account for localised pitting corrosion commonly observed in metallic pipes.10 Similarly, evidence has shown that the attachment of some bacterial cells to metal complexes does not take place randomly and that highly motile bacteria such as Leptospirillum ferrooxidans possess a chemo-sensory system that allows them to have the capacity to detect gradients of oxidizable substrates such as those extracted from ores.12
In this review we discuss recent studies using metabolomics to investigate the roles of metals and metal ions in a range of biological and environmental systems. We have purposely avoided discussing the assessment of metal toxicity via metabolomics since studies and reviews in this area are plentiful.13–16 However, this still leaves a wide range of areas to explore. The review begins with metabolomics studies on the roles of metals in plants as this is an area where the majority of research applying metabolomics in metal ion research has been carried out. We then discuss other important areas including natural products chemistry (as most natural products are derived from plants) then move to human health and disease (as improving health is a major use of natural products and compounds derived from them). The application and usefulness of metabolomics in determining the roles of metals is the unifying theme.
2. The role of metals in plants
Plants require certain concentrations of numerous elements (in various forms) to grow and reproduce. These elements are distinguished in macro-elements such as sodium (Na), potassium (K) and calcium (Ca) and micro-elements such as Cu, Fe, Zn, magnesium (Mg) and manganese (Mn). Thus, many metals, particularly the transition metals, are essential in biological processes in plants. Transition metal ions are characterised by their different chemical properties, which determine their redox potential, co-ordination geometry, charge and thermodynamic and kinetic properties of ligand exchange. These factors determine their specific function(s) and make them excellent cofactors for many metalloproteins.17 Metalloproteins are heavily involved in the photosynthetic electron transport chain and indeed, plant chloroplast activity and function is highly dependent on these molecules as in many cases electron transfer is mediated through the finely tuned redox potential of the metal centre by the bound protein. This means plants have a high demand for transition metals as part of their nutritional requirements.Both metal deficiency and metal excess strongly affect photosynthetic activity and thus any plant requires a finely tuned metal ion homeostasis network that strictly regulates metal uptake, chelation, trafficking and storage for survival, growth and reproduction. Small molecule metabolites are important compounds for chelation and carriage of metal ions. A pertinent example of the role of metabolites is in the uptake, transport and storage of Fe; the most common transition metal in living organisms. Fe is predominantly present in the form of Fe3+, however the Fe2+ form is physiologically more important for plants. Although Fe exists in high quantities in soils, it is usually highly insoluble or co-ordinated to other organic compounds and thus the availability to plants is usually very low. As a result, Fe deficiency (chlorosis) is a common problem in agriculture and leads to reduced photosynthetic activity and carbon fixation.18
One strategy to improve Fe nutrition in plants is to enhance the existing uptake mechanisms plants use to solubilise Fe from highly insoluble Fe(III) oxide hydrates, which they can absorb more easily than Fe ions. There are three chemical strategies that plants have evolved to do this: (i) release of protons (H+) from the root surface to enhance dissociation of Fe(III) oxides, (ii) reduction of Fe3+ to Fe2+, and (iii) chelation.19 Chelation takes advantage of Fe's propensity to form complexes through co-ordinative bonds and is based on the secretion of small molecules into the rhizosphere. Metal ions then form soluble co-ordination complexes allowing uptake into the plant through specialised transport systems.20 An example is the mugineic acid (MA) family of phytosiderophores (PS) utilised by graminaceous plants.14,15
Phytosiderophores are organic compounds with low molecular masses, which have the ability to bind a variety of metals and have a wide range of chemical structures and specific properties. Other examples of metabolites important for transition metal metabolism are porphyrins (e.g. Mg2+ binding chlorophyll), citrate or histidine (chelation with Zn2+),21 malate and citrate (detoxification of aluminum, Al2+/3+),22 phenolics (complexation with Fe2+/3+ for oxidative stress defence),23 and tannins (chelation of Zn2+, Fe2+/3+ and Cu2+).24
2.1. Metabolomics in the study of the role and function of elements in plants
The ability to analyse changes in metabolite levels in plants in response to imbalances of transition ion availability and impairments of uptake, transport and storage is important to gain a better insight into these processes and pave the path for approaches to overcome shortages or excesses of metals. In the past decade immense efforts have been made in the development of high-throughput and untargeted methodologies to analyse as many metabolites as possible in a given sample. Metabolomics has not yet been widely used to investigate plant responses to elemental or nutritional imbalances but this is slowly changing.Rellán-Álvarez et al.25 determined the metabolic changes induced by Fe deficiency in leaves and xylem sap. There were decreases in amino acids, N-related metabolites and carbohydrates and increases in tricarboxylic acid (TCA) cycle metabolites in xylem sap following Fe deficiency whereas in the leaves an increase in amino acids and N-related metabolites, carbohydrates and TCA cycle metabolites was observed. A clear correlation between Fe status of the plant and certain amino acids and TCA cycle intermediates were determined suggesting that in low photosynthesis, C-starved Fe-deficient plants anaplerotic reactions involving amino acids can be crucial for short-term survival.
Similar results were observed upon Fe deficiency in pea leaves by Kabir et al.26 The aim of this study was to identify mechanisms of Fe deficiency tolerance by comparison of tolerant and intolerant varieties. In the tolerant variety, Fe deficiency induced significant increases in N-, S- and TCA cycle metabolites, which was not detected in the intolerant variety. Increased glutathione and S-metabolites indicated protection of pea plants from Fe-deficiency induced oxidative stress. Additionally, an increase in citrate and malate in the leaves of the tolerant variety suggested long-distance transport of Fe is promoted by better xylem unloading mechanisms.
Metabolomics has been applied in mechanistic studies of Fe chelation in order to improve mineral nutrition to improve uptake in rice, which is one of the largest staple crops in the world. However, after milling it is very low in Fe, and Fe deficiency is one of the biggest problems in countries that obtain their caloric intake mainly through rice. Transgenic rice lines with overexpression of enzymes for nicotianamine synthesis have recently been demonstrated to have substantially increased Fe levels.27 Future work in this area will need to investigate how the inserted transgenes have altered metabolite levels, specifically Fe chelating compounds, in order to transport more Fe into the grain as well as if there are any unintended effects of transgene expression.
The responses of rice to Zn deficiency (and bicarbonate excess) have also been assessed by metabolomics.28 The results showed a reduction in monosaccharide levels and increased concentrations of phenolics and nitrogen rich metabolites in the roots.28 Zn stress resulted in lower levels of the TCA cycle intermediates, notably succinate and the aromatic amino acid tyrosine. Genotypic differentiation revealed constitutively higher levels of D-gluconate, 2-oxoglutarate and two unidentified compounds in the Zn-efficient plant compared to the Zn-inefficient cultivar, suggesting a possible role for these metabolites in overcoming oxidative stress or improving metal re-distribution.
A recent addition to the omics family is ‘ionomics’; defined as “the high-throughput analysis of mineral nutrients and the trace element composition of an organism.”29 It has been extensively used in the plant sciences for screening mutant collections, forward and reverse genetic approaches and the investigation of mechanisms of elemental or ion uptake, transport, compartmentalisation and exclusion.30 Elemental profiling is exceedingly helpful when investigating a plant's response to salinity, nutrient deficiencies and toxicities or hyper-accumulation. The simultaneous analysis of the complete elemental profile can be achieved using inductively coupled plasma mass spectroscopy or optical emission spectroscopy coupled with mass spectrometry (ICP-MS and OES-MS, respectively) with appropriate sample preparation methods, these techniques allow for the analysis of 12–20 elements and ions simultaneously.30
2.2. Metal hyperaccumulation in plants
Interestingly, there is a disparate group of plants that can accumulate metal ions to concentrations which would be deadly to other organisms. These plants are collectively known as hyperaccumulators. Approximately 500 taxa have been reported to hyperaccumulate a range of metals and metalloids, and are found on metal rich soils throughout the world.31 The most striking example comes from the New Caledonian tree Pycnandra acuminata. The latex of this species is often a blue colour as it typically contains 6–7% Ni2+, fresh mass, or 26% dry mass; the highest concentration of Ni found in any living organism.32The accumulation of metal ions by hyperaccumulators is an active process and there are a number of theories behind this trait such as protection from herbivores and fungal pathogens.33 However, this latter theory is not completely convincing since herbivory has been observed in field specimens with leaf Ni concentrations exceeding 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 mg kg−1 (see Fig. 1).
000 mg kg−1 (see Fig. 1).
 | ||
Fig. 1 Panel A, photograph of the New Caledonian nickel hyperaccumulating tree P. acuminata. It has been estimated that one tree can contain approximately 40 kg of nickel (Ni). Panel B, Psychotria douarrei showing evidence of herbivory despite the fact that these leaves contain around 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 mg kg−1 of Ni (dry mass).34 300 mg kg−1 of Ni (dry mass).34 | ||
Some of the earliest small molecule analyses of hyperaccumulators were carried out in the 1970s and early 1980s, and showed a correlation between organic acids and Ni content in some hyperaccumulating plants.35,36 However, the metabolic traits employed by hyperaccumulators to resist stress from high metal-ion concentrations are still poorly understood. Small organic acids are present in high concentrations in the cell vacuoles of photosynthetic tissues and numerous studies have implied that these acids play a role in hyperaccumulation.34–44 Several physiological parameters in the metal ion transport system must also be involved, including, modification of the membrane structure and function, tissue water content and global changes to gene expression, protein, lipid and metabolite profiles.45 Genomic studies have shown that a feature of hyperaccumulation is the constitutive expression of a range of both metal-chelator biosynthetic and metal transporter genes.46 For example high expression of the metal pump HMA4 (Heavy metal ATPase 4) is observed in Arabidopsis halleri (a Zn and Cd hyperaccumulator).47
It is postulated that some plants detoxify metals by pumping the ions into vacuoles for storage. Organic acids, however, have relatively low association constants for metal ions at physiological pH and are not selective and therefore seem unlikely to be involved in chelating these inert, metal ion complexes in the cell vacuole. Labile metal–organic acid complexes also do not explain how plants resist protein damage and oxidative stress as the metal ions would not be sequestered in inert complexes under physiological conditions. Similarly, Alpine pennygrass (Noccaea caerulescens) has been shown to accumulate more than 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 mg Zn kg−1 leaf dry mass.48 This equates to 0.306 mol of Zn kg−1 leaf dry mass. A 1
000 mg Zn kg−1 leaf dry mass.48 This equates to 0.306 mol of Zn kg−1 leaf dry mass. A 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Zn–citrate complex would therefore account for 8% of the total leaf mass or 13% in a 1
1 Zn–citrate complex would therefore account for 8% of the total leaf mass or 13% in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 metal
2 metal![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ligand complex. Once could speculate that these values would be even higher if localised to a vacuole, therefore there may not be a specific ligand involved in storage. The role of organic acids may actually be in micronutrient transport rather than storage.
ligand complex. Once could speculate that these values would be even higher if localised to a vacuole, therefore there may not be a specific ligand involved in storage. The role of organic acids may actually be in micronutrient transport rather than storage.
Evidence for histidine as a metal transport ligand from roots to shoots has been shown in the Ni hyperaccumulator species Alyssum, N. caerulescens and Noccaea goesingense.49–51 Increased levels of histidine may therefore enhance the translocation of Ni from the roots to the shoots. Antioxidants appear to play a role in resistance to high Ni concentrations Noccaea hyperaccumulator species since higher concentrations of glutathione, cysteine and O-acetyl serine are present in Noccaea hyperaccumulators when compared to non Noccaea hyperaccumulators.52 Further evidence supporting the role of glutathione was observed in a transgenic Arabidopsis where increases in glutathione increased the resistance to Ni toxicity related to oxidative stress.52
Hyperaccumulators offer important models for studying the molecular mechanisms behind metal transport and detoxification in plants. There are also a number of key practical uses of these plants such as phytoremediation and phytomining (using plants to extract metals ions from soils) and bio-fortification for the improvement of micronutrient poor crops such as rice. However, there are many challenges faced when attempting to characterise the metal ion co-ordination chemistry with standard metabolomic approaches, which are primarily based on NMR spectroscopy and GC and LC. The typical tissue homogenisation and solvent extraction steps change the solution equilibria so the complexes detected may not reflect those present in vivo. Fractionation of metal complexes also presents a number of issues as the metal complex dissociation kinetics must be considered. Small metal complexes such as organic acids will dissociate under acidic conditions or through interaction with chromatograph column stationary phases. Additionally, co-elution of metal ions with other small molecules in the ionisation source can form complexes in the gas phase, producing false positives.
Alternative means of separation such as size exclusion and capillary electrophoresis can overcome some of these issues. However, metabolomic analyses have been carried out on bulk tissue samples (e.g. whole roots or leaves). This results in the determination of “average” metabolite profiles across different tissue and cell types and therefore potentially overriding specific metabolic responses within a particular cell type. It is often difficult or even impossible to separate and isolate tissue types or single cell types from plant tissues without detailed taxonomic and dissection skills. High resolution direct imaging techniques such as mass spectrometry imaging may help address these problems.
2.3. Imaging approaches to increase our understanding of metal metabolism in plants
To be able to image the distribution of elemental and biochemical compositions has been an interest for a long time and new technologies are now paving the way in the plant sciences. Laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS) can be used to produce three-dimensional images of spatial element distributions in thin biological tissue sections (see Fig. 2). This technique has not been used extensively in the plant sciences but many potential applications become obvious, particularly when working in the field of metal metabolism.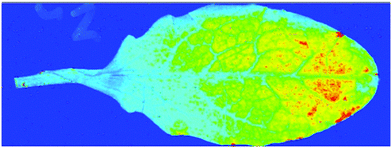 | ||
| Fig. 2 Phosphorous distribution in a leaf of Noccaea caerulescens obtained by LA-ICP-MS. Blue indicates low concentration, red indicates high concentration. | ||
More recently a promising new technology is now being adapted to determine the spatial distribution of metabolites. Matrix Assisted Laser Desorption Ionization (MALDI) is a well-known technique in peptide mass fingerprinting approaches and is used to raster across thin tissue slices and ionize compounds of interest (metabolites, lipids, or peptides – following proteolytic digestion of proteins) prior to analysis in the mass spectrometer.53 This technique allows for the determination of tissue type specific differences of target distributions and has been well developed for lipids due to their ease of ionization. Recently, efforts have been made to optimize this approach for the analysis of small molecule distributions with the addition of Fourier Transform Ion Cyclotron Resonance Mass Spectrometry (FT-ICR-MS) to the MALDI ionisation process.54,55
These two exciting new technologies will open many doors for plant researchers allowing for the co-localisation of metabolites/lipids and elements (including transition metals) across tissue sections (see Fig. 3). For example, it would be interesting to determine the localization of salinity defence mechanisms in a plant root through co-localisation of effects of salt surrounding the root (analysis of Na+, K+ and Ca2+) with alterations in the metabolite profile for selected osmoprotectants. Another application of both elemental (e.g. for Fe) and metabolite imaging (e.g. phytosiderophores) may provide indications as to which parts of the plant play major roles in Fe uptake, transport and storage. There is also scope to explore the role and potential benefits of metal and metalloid containing natural products from plants.
3. Metal containing natural products and their biological potential in human health
Natural products (NPs) can be broadly defined as any chemical substance produced by a living organism. However it is commonly used to refer to a chemical substance or substances found in nature that has distinctive pharmacological effects and indeed the main role in human society for products from natural sources is to improve health in one form or another.56 NPs and their derivatives often have potent physiological activities and therefore play important roles as both frontline treatments for many diseases and as the inspiration for chemically synthesised therapeutics.NPs are also an essential source of successful drug leads and can show a surprising amount of incorporation of metalloids and semi-metals; for example before the advent of penicillin arsenic compounds, were used extensively to treat syphilis. A more modern example is the recent discovery of Boron (B) containing NPs which have shown to posses antibiotic activity (Fig. 4).
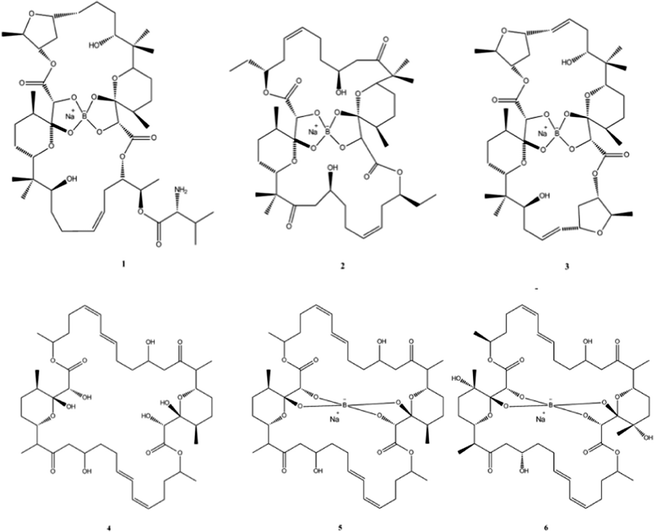 | ||
| Fig. 4 Chemical structures of boromycin (1), borophycin (2), aplasmomycin (3), tartrolon A (4), tartrolon B (5) and tartrolon C (6). | ||
The polyketide antibiotic boromycin, was identified in the 1960s from the bacteria Streptomyces antibioticus collected from soil in the Ivory Coast, Africa and contains significant amounts of boron. Boromycin inhibited the growth of Gram-positive bacteria and also strongly inhibited the replication of HIV.57,58 A structurally related analogue, borophycin isolated from a marine strain of the blue-green alga (Nostoc linckia) similarly showed antibacterial activity as well as appreciable cytotoxicity against human tumour cell lines.59 Okazaki et al., also reported the isolation of the boron containing antibiotic aplasmomycin which was effective in controlling Plasmodium berghei.60 The authors described that the chelation with other metals did not directly correspond with antibacterial activity and cation selectivity decreased in the order Rb > K > Cs = Na > Li, with no affinity towards divalent cations.61 Similarly, Irschik and colleagues isolated the antibiotics tartrolon A and tartrolon B from a German soil sample62 while Lewer et al. isolated an analogue, tartrolon C and demonstrated its potential as an insecticide.63
Another good example of NPs containing metalloids/semi-metals are those which include Selenium (Se). Usually found in plants, they are well documented in the literature and can be divided into the following three subgroups: selenite-accumulators (e.g. broccoli and cucumber), selenomethionine accumulators (e.g. grains such as wheat) and Se-methyl selenomethionine accumulators (e.g. garlic and onion).64 Selenium based-metabolomics was carried out by Arnaudguilhem et al.65 who detected 64 metabolites including 21 previously unreported compounds, 9 of which were subsequently identified using high resolution mass spectrometry. The data indicated the existence of 5 highly reactive precursors of selenium metabolites and shed new light not only on the selenium metabolomic pathway but also on the ability of metabolomics to make a contribution to our understanding of metal metabolism and associated biochemical pathways.
A extensive review by Tripathi et al.66 provided a comprehensive systems toxicology based evaluation of As exposure (arsenomics) in plants. The metabolomic analysis showed that thiol peptides such as glutathione and PCs play a central role in As detoxification, as well as various antioxidant defence system responses against As induced oxidative stress. However, the observed variation in metabolite profiling in different plant species signifies that plants modulate their metabolome to respond against As stress.
The use of such metallomes is, to date, a mostly unexplored frontier of omics based research with only a few metals and related metabolites being comprehensively assed by omic techniques. Nevertheless, metallomics and metabolomics have great potential to improve our understanding of metal related functional biochemistry.67
4. Metals, oxidative stress and human disease
Recently, ICP-MS was used in a metabolomic study of human cerebrospinal fluid (CSF) where it enabled the quantitative analysis of 33 metal ions.68 The authors were able to show that CSF is a reasonably rich reservoir of trace metals (33 different elements were detected) that ICP-MS can be effectively used to identify applied to study them. Since metals are known to play an important role in a wide number of diseases like Alzheimer's disease, Parkinson's disease and Wilson's disease, the measurement of trace levels of metals in human tissues, biofluids and even individual cells is of increasing interest. The techniques described earlier for plant science could therefore, potentially benefit studies of metals in human disease and health conditions, particular those involving the brain. There is a growing body of literature to suggest that metals may be involved in the progression of neurodegenerative diseases; these disorders constitute a set of pathological conditions originating from the slow, systematic and currently irreversible cell loss within the various regions of the brain and/or the spinal cord.69 The outcomes of neurodegeneration are very broad and diverse, ranging from the problems with movements to dementia, depending on the brain region affected.The presence of metal ions can catalyse the production of free radicals. These highly reactive free radicals can affect neurodegeneration as they can cause oxidative deterioration of DNA and proteins.70 Free radical production can also interfere with signal transduction pathways, deregulation of cell proliferation through the activation of transcription factors, the control of cell cycle progression and apoptosis.70 Reactive nitrogen intermediates (RNI) and reactive oxygen intermediates (ROI) are also produced under oxidative stress conditions.71
A link between Parkinson's disease (PD) and oxidative stress in brain tissue has been suggested, most likely due to a breakdown in the regulation of dopamine. It has also been demonstrated that the A53T alpha-synuclein mutation increases iron-dependent aggregation and toxicity by interfering with the vesicular storage of dopamine, which leads to this compound building up in the cytoplasm as well as the creation of reactive oxygen species (ROS).72 Some cell cultures overexpressing alpha-synuclein also show up to a four-fold increase in vulnerability to toxicity induced by Fe but why this should be so is not yet clear.73 Neurological effects associated with As toxicity have also been confirmed in animal studies, where changes in neurotransmitter levels such as dopamine, norepinephrine and 5-hydroxytryptamine were found in rats exposed to drinking water containing sodium arsenite.74
Interruption of brain iron homeostasis is also a known component in the early neuropathological events associated with Alzheimer's disease (AD).70 Fe has been shown to stimulate the formation of free radicals, the oxidation of proteins and peroxidation of lipids, as well as to decrease levels of cytochrome C oxidase.75 Many AD studies have been aimed at Amyloid-Beta protein (AB) – and AB toxicity is in turn linked with the occurrence of redox metals – mainly Cu and Fe.76 AB is also known to contain neurotoxic properties which stimulate the oxidation of ascorbate due to Cu.77 The breakdown products of lipid peroxidation, including increased levels of 4-hydroxy-2,3-nonenal, acrolein, F2-isoprostanes and malondialdehyde (MDA) have been found in AB heavy brains when compared to age-matched controls.78 For the interested reader, the role of systems biology in the study of large-scale cellular death and destruction, caused by poorly ligand Fe in Parkinson's, Huntington's, Alzheimer's, diseases and several other medical conditions was recently explored in detail by Kell et al.79
Wilson's disease (WD) is a severe disorder of Cu levels in the body and causes a wide spectrum of liver pathology and/or neurologic and psychiatric symptoms. It is caused by mutations in ATP7B, a gene encoding a copper-transporting ATPase with associated accumulation of Cu in tissues, especially in the liver. Although chelation therapy has been shown to be a useful treatment there is still a lot that is unknown about the disease and its progression. For example, there is no genotype–phenotype correlation and the treatment of neurologic symptoms is often only partially successful. WD was previously thought to act via oxidative damage to multiple tissues but a recent metabolomics based work has shed new light on the syndrome.
Using an animal model (the Atp7b−/− mouse), Huster et al.80 demonstrated that Cu-induced metabolic changes were inconsistent with widespread oxidative/free radical damage at least at the pre-symptomatic stages of the disease. The authors postulated that this was likely due to the fact that metallothionein levels are markedly higher in Atp7b−/− livers compared to wild type animals and this leads to the sequestration of cytosolic Cu. Instead, high Cu levels were found to selectively up-regulate a range of metabolites associated with the cell cycle and chromatin structure and down-regulate those related to lipid metabolism, particularly cholesterol biosynthesis.80 Reduced serum cholesterol levels (which diminished under therapy) were also found in human WD patients by Seessle et al.81 and a recent review by Burkhead et al.,82 which synthesised data from several omics based studies of the disease, also identified new metabolic pathways associated with WD disease progression, including lipid metabolism as well as cell cycle regulation and signalling and mRNA splicing.
The use of metabolomics and related systems biology and functional genomics methods in the study of WD shows the great potential of these techniques to increase the mechanistic analysis of human disorders of metal misbalance such as Menkes disease (Cu deficiency) and/or hemochromatosis (excess Fe levels). This is a potentially large areas of growth for future research since at present there are not many studies that make use of metabolomics in this area.
5. Conclusion
Although studies on metabolism have been performed for hundreds of years, metabolomics itself is one of the newest members of the omic family; and along with metallomics and ionomics, it shows great potential for the assessment of the roles metals play in biological systems and for investigating novel aspects of metal metabolism.83 Changes at the molecular level can also be linked to those at higher levels of organisation to create a greater holistic overview of the system. This in turn may enable biomarkers to be developed that allows for the rapid detection of effects at the biochemical level before changes become visible at the physiological level and may assist in interpreting how such physiological effects occur.Metabolomics can also be integrated with one or more of the other ‘omic’ sciences including genomics (genome sequencing and the annotation of function to genes), transcriptomics (gene expression at the transcription level) and proteomics (total protein expression) as part of both systems biology and functional genomics. The main goal being to develop a comprehensive and consistent knowledge of a biological system by investigating the behaviour of and interaction between its individual components.84
Additionally, the discovery of new information on how organisms maintain metal homeostasis and or deal with high levels of metal/metalloid ions in their systems could lead to advances in nutrition and health for humans as other organisms such as plants, bacteria and fungi. The utilisation of metal tolerant plants, fungi and bacteria could also be used to remediate contaminated sites while metabolic profiling could be employed to assess their efficiency. The integration of metallomics with metabolomics is thus ripe for exploration and discovery and while the majority of work to date is focused on plants; human health and wellbeing is likely be another major growth area in the near future.
Acknowledgements
The authors gratefully acknowledge colleagues from the Australia and New Zealand Metabolomics Network (ANZMN) and Proteomics and Metabolomics Victoria (PMV) for useful discussions on the manuscript.References
- J. J. R. F. Frausto da Silva, R. J. P. Williams, The Biological Chemistry of the Elements: The Inorganic Chemistry of Life, Oxford University Press, Oxford, 2001 Search PubMed.
- T. J. Lyons and D. J. Eide, in Biological Inorganic Chemistry: Structure and Reactivity, ed. I. Bertini, H. Gray, E. Stiefel and J. S. Valentine, University Science Books, Mill Valley, California, USA, 1st edn, 2007, ch. 5, pp. 57–77 Search PubMed.
- A.-S. Zhang and C. A. Enns, J. Biol. Chem., 2009, 284, 711–715 CrossRef CAS PubMed.
- N. Stadler, R. A. Lindner and M. J. Davies, Arterioscler., Thromb., Vasc. Biol., 2004, 24, 949–954 CrossRef CAS PubMed.
- O. A. H. Jones and V. L. Cheung, Comp. Med., 2007, 57, 436–442 CAS.
- R. Henry and T. Cassel, in The Handbook of Metabolomics, ed. T. W.-M. Fan, A. N. Lane and R. M. Higashi, Humana Press, 2012, ch. 5, pp. 99–125 Search PubMed.
- N. V. Reo, Drug Chem. Toxicol., 2002, 25, 375–382 CrossRef CAS PubMed.
- Z. Pan and D. Raftery, Anal. Bioanal. Chem., 2007, 387, 525–527 CrossRef CAS PubMed.
- D. J. Beale, M. S. Dunn, P. D. Morrison, N. A. Porter and D. R. Marlow, Corros. Sci., 2012, 55, 272–279 CrossRef CAS PubMed.
- D. J. Beale, P. D. Morrison, C. Key and E. A. Palombo, Water Sci. Technol., 2014, 69, 1–8 CrossRef CAS PubMed.
- K. A. Kouremenos, D. J. Beale, H. Antti and E. A. Palombo, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. DOI:10.1016/j.jchromb.2014.02.058.
- C. A. Jerez and M. Moo-Young, Comprehensive Biotechnology, Academic Press, Burlington, 2nd edn, 2011, pp. 717–729 Search PubMed.
- S. C. Booth, M. L. Workentine, A. M. Weljie and R. J. Turner, Metallomics, 2011, 3, 1142–1152 RSC.
- O. A. H. Jones, M. L. Maguire, J. L. Griffin, D. A. Dias, D. J. Spurgeon and C. Svendsen, Aust. J. Ecol., 2013, 38, 713–720 CrossRef PubMed.
- J. Bundy, M. Davey and M. Viant, Metabolomics, 2009, 5, 3–21 CrossRef CAS.
- M. A. Garcia-Sevillano, T. Garcia-Barrera and J. L. Gomez-Ariza, Metallomics, 2014, 6, 237–248 RSC.
- I. Yruela, Metallomics, 2013, 5, 1090–1109 RSC.
- P. B. Vose, J. Plant Nutr., 1982, 5, 233–249 CrossRef CAS.
- D. Staiger, Angew. Chem., Int. Ed., 2002, 41, 2259–2264 CrossRef CAS.
- V. Römheld, Plant Soil, 1991, 130, 127–134 CrossRef.
- W. E. Rauser, Cell Biochem. Biophys., 1999, 31, 19–48 CrossRef CAS PubMed.
- L. T. Yang, Y. P. Qi, H. X. Jiang and L. S. Chen, BioMed Res. Int., 2013, 1, 173682 Search PubMed.
- V. Chobot and F. Hadacek, Plant Signaling Behav., 2010, 5, 4–8 CrossRef CAS.
- M. Karamać, Int. J. Mol. Sci., 2009, 10, 5485–5497 CrossRef PubMed.
- R. Rellán-Álvarez, H. El-Jendoubi, G. Wohlgemuth, A. Abadía, O. Fiehn, J. Abadía and A. Alvarez-Fernández, Front. Plant Sci., 2011, 2, 66 Search PubMed.
- A. H. Kabir, N. G. Paltridge, U. Roessner and J. C. R. Stangoulis, Physiol. Plant., 2013, 147, 381–395 CrossRef CAS PubMed.
- A. A. Johnson, B. Kyriacou, D. L. Callahan, L. Carruthers, J. Stangoulis, E. Lombi and M. Tester, PLoS One, 2011, 6, e24476 CAS.
- M. T. Rose, T. J. Rose, J. Pariasca-Tanaka, T. Yoshihashi, H. Neuweger, A. Goesmann, M. Frei and M. Wissuwa, Planta, 2012, 236, 959–973 CrossRef CAS PubMed.
- D. E. Salt, I. Baxter and B. Lahner, Annu. Rev. Plant Biol., 2008, 59, 709–733 CrossRef CAS PubMed.
- I. Baxter, Curr. Opin. Plant Biol., 2009, 12, 381–386 CrossRef CAS PubMed.
- R. D. Reeves and A. J. M. Baker, in Phytoremediation of Toxic Metals, ed. I. Raskin and B. D. Ensley, J. Wiley, New York, 2000, pp. 193–229 Search PubMed.
- D. L. Callahan, U. Roessner, V. Dumontet, N. Perrier, A. G. Wedd, R. A. O'Hair, A. J. Baker and S. D. Kolev, Phytochemistry, 2008, 69, 240–251 CrossRef CAS PubMed.
- A. Kazemi-Dinan, S. Thomaschky, R. J. Stein, U. Krämer and C. Müller, New Phytol., 2014, 202, 628–639 CrossRef CAS PubMed.
- D. L. Callahan, U. Roessner, V. Dumontet, A. M. De Livera, A. Doronila, A. J. Baker and S. D. Kolev, Phytochemistry, 2012, 81, 80–89 CrossRef CAS PubMed.
- J. Lee, R. D. Reeves, R. R. Brooks and T. Jaffré, Phytochemistry, 1978, 17, 1033–1035 CrossRef CAS.
- W. J. Kersten, R. R. Brooks, R. D. Reeves and A. Jaffré, Phytochemistry, 1980, 19, 1963–1965 CrossRef CAS.
- U. Krämer, I. J. Pickering, R. C. Prince, I. Raskin and D. E. Salt, Plant Physiol., 2000, 122, 1343–1353 CrossRef PubMed.
- R.-L. Qiu, Y.-T. Tang, X.-H. Fang, R. L. Chaney, Y.-M. Li, J. S. Angle, W. Liu and X.-W. Zeng, Zhongshan Daxue Xuebao, Ziran Kexueban, 2004, 43, 144–149 CAS.
- G. de la Rosa, R. Peralta-Videa Jose, M. Montes, G. Parsons Jason, I. Cano-Aguilera and L. Gardea-Torresdey Jorge, Chemosphere, 2004, 55, 1159–1168 CrossRef CAS PubMed.
- X. Shan, H. Wang, S. Zhang, H. Zhou, Y. Zheng, H. Yu and B. Wen, Plant Sci., 2003, 165, 1343–1353 CrossRef CAS.
- G. de la Rosa, J. L. Gardea-Torresdey, J. R. Peralta-Videa, J. G. Parsons and M. Montes, Preprints of Extended Abstracts presented at the ACS National Meeting, American Chemical Society, Division of Environmental Chemistry, 2003, vol. 43, pp. 601–606.
- R. Boominathan and M. Doran Pauline, J. Biotechnol., 2003, 101, 131–146 CrossRef CAS.
- S. D. Bidwell, I. E. Woodrow, G. N. Batianoff and J. Sommer-Knudsen, Funct. Plant Biol., 2002, 29, 899–905 CrossRef CAS.
- A. R. Memon and M. Yatazawa, J. Plant Nutr., 1984, 7, 961–974 CrossRef CAS.
- U. Kramer, Annu. Rev. Plant Biol., 2010, 61, 517–534 CrossRef PubMed.
- M. Hanikenne and C. Nouet, Curr. Opin. Plant Biol., 2011, 14, 252–259 CrossRef CAS PubMed.
- M. Hanikenne, I. N. Talke, M. J. Haydon, C. Lanz, A. Nolte, P. Motte, J. Kroymann, D. Weigel and U. Kramer, Nature, 2008, 453, 391–395 CrossRef CAS PubMed.
- A. J. M. Baker, R. D. Reeves and A. S. M. Hajar, New Phytol., 1994, 127, 61–68 CrossRef CAS PubMed.
- D. L. Callahan, S. D. Kolev, R. A. J. O'Hair, D. E. Salt and A. J. M. Baker, New Phytol., 2007, 176, 836–848 CrossRef CAS PubMed.
- U. Kramer, J. D. Cotter-Howells, J. M. Charnock, A. J. M. Baker and J. A. C. Smith, Nature, 1996, 379, 635–638 CrossRef CAS.
- M. W. Persans, X. Yan, J. M. Patnoe, U. Kramer and D. E. Salt, Plant Physiol., 1999, 121, 1117–1126 CrossRef CAS PubMed.
- J. L. Freeman, M. W. Persans, K. Nieman, C. Albrecht, W. Peer, I. J. Pickering and D. E. Salt, Plant Cell, 2004, 16, 2176–2191 CrossRef CAS PubMed.
- S. Kaspar, M. Peukert, A. Svatos, A. Matros and H. P. Mock, Proteomics, 2012, 11, 1840–1850 CrossRef PubMed.
- Y. J. Lee, D. C. Perdian, Z. Song, E. S. Yeung and B. J. Nikolau, Plant J., 2012, 70, 81–95 CrossRef CAS PubMed.
- M. Peukert, A. Matros, G. Lattanzio, S. Kaspar, J. Abadía and H. P. Mock, New Phytol., 2012, 193, 806–815 CrossRef CAS PubMed.
- B. B. Mishra and V. K. Tiwari, Eur. J. Med. Chem., 2011, 46, 4769–4807 CrossRef CAS PubMed.
- R. Hütter, W. Keller-Schierlein, F. Knüsel, V. Prelog, G. C. Rodgers, P. Suter, G. Vogel, W. Voser and H. Zähner, Helv. Chim. Acta, 1967, 50, 1533–1539 CrossRef PubMed.
- J. Kohno, T. Kawahata, T. Otake, M. Morimoto, H. Mori, N. Ueba, M. Nishio, A. Kinumaki, S. Komatsubara and K. Kawashima, Biosci., Biotechnol., Biochem., 1996, 60, 1036–1037 CrossRef CAS PubMed.
- T. Hemscheidt, M. P. Puglisi, L. K. Larsen, G. M. L. Patterson and R. E. Moore, J. Org. Chem., 1994, 59, 3467–3471 CrossRef CAS.
- T. Okazaki, T. Kitahara and Y. Okami, J. Antibiot., 1975, 28, 176–184 CrossRef CAS.
- T. J. Stout, J. Clardy, I. C. Pathirana and W. Fenical, Tetrahedron, 1991, 47, 3511–3520 CrossRef CAS.
- H. Irschik, D. Schummer, K. Gerth, G. Hofle and H. Reichenbach, J. Antibiot., 1995, 48, 26–30 CrossRef CAS.
- P. Lewer, E. L. Chapin, P. R. Graupner, J. R. Gilbert and C. Peacock, J. Nat. Prod., 2003, 66, 143–145 CrossRef CAS PubMed.
- P. D. Whanger, J. Am. Coll. Nutr., 2002, 21, 223–232 CrossRef CAS.
- C. Arnaudguilhem, K. Bierla, L. Ouerdane, H. Preud'homme, A. Yiannikouris and R. Lobinski, Anal. Chim. Acta, 2012, 757, 26–38 CrossRef CAS PubMed.
- R. D. Tripathi, P. Tripathi, S. Dwivedi, S. Dubey, S. Chatterjee, D. Chakrabarty and P. K. Trivedi, Front. Plant Physiol., 2012, 3, 275 CAS.
- J. Szpunar, Analyst, 2005, 130, 442–465 RSC.
- R. Mandal, A. Guo, K. Chaudhary, P. Liu, F. Yallou, E. Dong, F. Aziat and D. Wishart, Genome Med., 2012, 4, 1–11 CrossRef PubMed.
- L. Breydo and V. N. Uversky, Metallomics, 2011, 3, 1163–1180 RSC.
- K. J. Barnham and A. I. Bush, Curr. Opin. Chem. Biol., 2008, 12, 222–228 CrossRef CAS PubMed.
- C. Nathan, J. Clin. Invest., 2003, 111, 769–778 CrossRef CAS.
- J. Lotharius, S. Barg, P. Wiekop, C. Lundberg, H. K. Raymon and P. Brundin, J. Biol. Chem., 2002, 277, 38884–38894 CrossRef CAS PubMed.
- N. Ostrerova-Golts, L. Petrucelli, J. Hardy, J. M. Lee, M. Farer and B. Wolozin, J. Neurosci., 2000, 20, 6048–6054 CAS.
- G. M. Kannan, N. Tripathi, S. N. Dube, M. Gupta and S. J. Flora, J. Toxicol., Clin. Toxicol., 2001, 39, 675–682 CrossRef CAS PubMed.
- W. Droge, Physiol. Rev., 2002, 82, 47–95 CAS.
- M. P. Cuajungco, L. E. Goldstein, A. Nunomura, M. A. Smith, J. T. Lim, C. S. Atwood, X. Huang, Y. W. Farrag, G. Perry and A. I. Bush, J. Biol. Chem., 2000, 275, 19439–19442 CrossRef CAS PubMed.
- S. I. Dikalov, M. P. Vitek and R. P. Mason, Free Radical Biol. Med., 2004, 340–347 CrossRef CAS PubMed.
- S. Artl, U. Beisiegel and A. Kontush, Curr. Opin. Lipidol., 2002, 13, 289–294 CrossRef PubMed.
- D. B. Kell, Arch. Toxicol., 2010, 11, 825–889 CrossRef PubMed.
- D. Huster, T. D. Purnat, J. L. Burkhead, M. Ralle, O. Fiehn, F. Stuckert, N. E. Olson, D. Teupser and S. Lutsenko, J. Biol. Chem., 2007, 282, 8343–8355 CrossRef CAS PubMed.
- J. Seessle, A. Gohdes, D. Gotthardt, J. Pfeiffenberger, N. Eckert, W. Stremmel, U. Reuner and K. Weiss, Lipids Health Dis., 2011, 10, 1–6 CrossRef PubMed.
- J. L. Burkhead, L. W. Gray and S. Lutsenko, BioMetals, 2012, 24, 455–466 CrossRef PubMed.
- P. A. Muller and L. W. Klomp, Am. J. Clin. Nutr., 2008, 88, 821S–825S CAS.
- O. A. H. Jones and V. L. Cheung, Comp. Med., 2007, 57, 436–442 CAS.
| This journal is © The Royal Society of Chemistry 2015 |