Rational design of promiscuous binding modulators of p53 inducing E3(Ub)-ligases (Mdm2 and Pirh2) as anticancer agents: an in silico approach†
Abstract
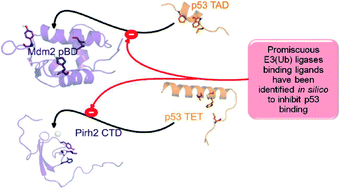
In this study, for the first time, we report on in silico based development of Mdm2 and Pirh2 promiscuous binding small molecule modulators. The methodology involves capturing the important Mdm2 and Pirh2 interacting residues of p53 TAD and TET, respectively from the literature; as well as from the protein–protein docking and molecular dynamics simulations applied in the present study. The knowledge of important residues that have been already delineated was used as the benchmark for the design of Mdm2 and Pirh2 focused ligand libraries, which were obtained by pharmacophore based screening of 3.9 million compounds deposited in MMsINC® database. This was followed by 2D fingerprint similarity based filtering of focused ligand libraries with respect to a known reference set of Mdm2 inhibitors, which further confined the chemical space to 608 molecules. These included 365 Mdm2-like small molecule mimetics of p53 TAD and 243 Mdm2-like small molecule mimetics of p53 TET. Docking iterations with respective targets and reverse docking resulted in twelve potential best fit molecules that showed favourable binding interactions with both Mdm2 pBD and Pirh2 CTD. The quality of the docking protocol was assessed using experimentally determined IC50 values of the known 213 Mdm2 inhibitors and their docking scores with a set of statistical measures, which showed good correlations.

 Please wait while we load your content...
Please wait while we load your content...