 Open Access Article
Open Access ArticleDimethylaminoethyl methacrylate copolymer-siRNA nanoparticles for silencing a therapeutically relevant gene in macrophages†
Ratnesh
Jain
*ab,
Prajakta
Dandekar
*bc,
Brigitta
Loretz
c,
Marcus
Koch
d and
Claus-Michael
Lehr
c
aDepartment of Chemical Engineering, Institute of Chemical Technology, NP Marg, Matunga, Mumbai 400019, India. E-mail: rd.jain@ictmumbai.edu.in; Fax: +91 22 3361 1020; Tel: +91 22 3361 2029
bDepartment of Pharmaceutical Sciences and Technology, Institute of Chemical Technology, NP Marg, Matunga, Mumbai 400019, India. E-mail: pd.jain@ictmumbai.edu.in; Fax: +91 22 3361 1020; Tel: +91 22 3361 2221
cDepartment of Drug Delivery (DDEL), Helmholtz-Institute for Pharmaceutical Research Saarland (HIPS), Campus A4 1, Saarland University, Saarbrücken, 66123 Germany. E-mail: lehr@uni-saarland.de; Fax: +49 681 302 4677; Tel: +49 681 302 2039
dInnovative Electron Microscopy, INM – Leibniz Institute for New Materials, Service Group Physical Analysis, Campus D2 2, Saarland University, Saarbrücken, 66123 Germany. E-mail: marcus.koch@inm-gmbh.de; Fax: +49 681 9300 223; Tel: +49 681 9300 0
First published on 5th January 2015
Abstract
Therapeutic gene silencing using small-interfering RNA (siRNA) for treatment of bacterial infections has been neglected in comparison with cancer and viral infections. The aim of our investigation was to formulate siRNA-loaded nanoparticles, using an established cationic polymethacrylate polymer, to enhance the delivery of siRNA into the cytoplasm of macrophages that host many pathogenic bacterial species, including tuberculosis. Nanoparticles of cationic dimethylaminoethyl methacrylate copolymer (Eudragit® E 100) were successfully formulated and were found to efficiently bind the siRNA molecules (Cy3-siRNA, Bfl1/A1 siRNA). The efficiency of nanoparticles in overcoming cellular barriers to intracellular siRNA delivery and the precise pathway of endocytosis of nanoparticles were both confirmed using confocal microscopy. Through efficient siRNA release into the cytoplasm, the siRNA-loaded nanoparticles enabled a five-fold enhancement in the knockdown efficiency of the endogenous host gene Bfl1/A1, when the formulation was compared with free siRNA. Persistence of Bfl1/A1 is useful for phagolysosomal survival of tuberculosis bacteria in macrophages, and the nanoparticles offer a promising concept for exploitation as an anti-tuberculosis therapy.
Introduction
Extensive spread of pulmonary infections is a cause of global concern due to the progressive increase in the number of affected cases, particularly caused by the spread of acquired immune deficiency virus (AIDS) pandemic.1 The intracellular infectious pulmonary pathogens are taken up by the alveolar macrophages, wherein resistance to biocidal and biophylactic mechanisms enables their rampant multiplication, consequently resulting in severe respiratory infections.1–4 Amongst the various infectious microorganisms, Mycobacterium tuberculosis remains one of the world's most notorious pathogens, this organism being single-handedly responsible for millions of tuberculosis related mortalities, annually. Further, the emergence of multidrug-resistant M. tuberculosis underscores the need for new therapeutic approaches to combat this pathogen.5Amongst the numerous strategies being exploited worldwide to combat this pathogen, macrophage delivery is now considered as one of the most challenging and promising approaches.4,6 However, the success of this approach in delivering chemical or nucleotide based drugs to the intracellular bacteria is limited by inadequate specificity of the macrophages and their poor internalization of drug-carrier conjugates.6–9
Although numerous delivery carriers, such as liposomes and lipidic nanoparticles, have been previously explored for macrophage delivery,10,11 the use of polymeric nanoparticles has gained increasing interest due to their low toxicity in comparison with alternative approaches such as viral systems. Further, in comparison with lipidic nanocarriers, nanoparticles of cationic polymers are less immunogenic and impart more stability to the associated nucleic acids due to multivalent interactions with the latter. Other advantages of polymeric nanoparticles are their ability to participate in cellular interactions and undergo internalization, and the possibility of using optimized synthesis conditions to obtain the desired, reproducible physicochemical characteristics.12,13 Furthermore, polymeric nanoparticles are also receiving attention as vectors for therapeutically relevant nucleotides including antisense oligonucleotides (asODNs) and functional small interfering RNA (siRNA).14,15
Macrophages are interesting targets in M. tuberculosis therapy as they host the pathogens, demonstrate favorable uptake of NPs and last but not least, act as important mediators of immune response. Hence, in our investigation we aimed to exploit all these advantages of macrophages as potential tools to interfere with the host–pathogen interaction. However the challenge in targeting siRNA to macrophages was that although macrophages efficiently phagocytose particulate matter, they might prove problematic for delivery of functional nucleotides since they degrade phagocytosed molecules.
The gene of choice was Bfl1/A1, an anti-apoptotic member of the Bcl-2 family of proteins. It is an NF-κB (nuclear factor kappa B) dependent gene, which is up-regulated in response to various NF-κB pathway stimulators like TNF-α and IL-1β, CD40, phorbol ester, and lipopolysaccharide (LPS). Moreover, virulent mycobacteria have been shown to induce Bfl1/A1 in order to create a protective niche within the infected macrophages.16–18,21–23
In this investigation, we aimed to target anti-Bfl1/A1 siRNA to macrophages employing nanoparticles of the cationic polymethacrylate polymer Eudragit® E100 (Dimethylaminoethyl Methacrylate Copolymer; DMC). The cationic nanoparticles were hypothesized to overcome the problems associated with intracellular siRNA delivery, such as rapid degradation by nucleases, low intracellular uptake, and limited blood stability.19 The choice of polymer was supported by literature reports confirming the ability of Eudragit polymers to induce autophagy in macrophages, a primary defence mechanism of the immune system against infectious diseases like tuberculosis, thereby affecting host–bacterial interactions.12 Also, Eudragit polymers are reportedly more benign to the cells as compared to standard cationic transfection polymers like polyethylenimine (PEI) and poly(L-lysine) (PLL), which are known to possess both in vitro and in vivo toxicity.13 Our formulation was successfully able to deliver anti-Bfl1/A1 siRNA within the macrophage cell line and down-regulate this protein, considerably better than free siRNA. Success in these experiments indicates the potential of the nanoparticles for further studies in animal models to confirm their efficacy as a platform technology for gene silencing in intracellular infections.
Results and discussion
Nanoparticle preparation and characterization
DMC belongs to a polymethacrylate class of polymers, which contain tertiary amine groups. Polymethacrylates have previously demonstrated the proton buffering ability similar to PEI, but with minimal cellular toxicity.23 As with other polymethacrylates, DMC contains tertiary amino groups, which were hypothesized to yield good transfection of nucleic acids.24 The importance of tertiary amino groups for imparting a good transfection capability has been already demonstrated in the case of branched PEI (polyethyleneimine) compared to linear PEI, through several transfection experiments in previous publications.25,26 However it is important to note that DMC, despite its similarity to PEI with regard to its tertiary amino groups, lacks the severe toxicity associated with the latter, which in turn enhances its applicability for transfecting mammalian cells.27 In addition, DMC is soluble up to pH 5.5 and swellable and permeable above pH 5.0. The polymer is protonated at this pH and is anticipated to provide a proton sponge effect to the endosome due to its cationic dimethylaminomethyl groups. Due to this effect, the siRNA loaded nanoparticles of Eudragit have been anticipated to enter the cytoplasm after breaking the endosomal compartment.28 Various polymethacrylates have already been evaluated for delivery of nucleotide based drugs;28 however, the application of commercial polymethacrylates for siRNA delivery has been attempted for the first time in this investigation, to the best of our knowledge.DMC (Fig. 1) exists in deprotonated form in organic solvents like acetone and ethyl acetate and also remains insoluble in water.29 Hence DMC nanoparticles were formulated by a solvent emulsification technique, after dissolving DMC in ethyl acetate and acetone. Further, the nanoparticles were optimized with respect to various parameters to provide the desired particle size, homogeneity and zeta potential for adequate complexation of nucleotides. The effect of various stabilizers like poloxamer 407, vitamin E-TPGS and PVA was evaluated; the results of which have been depicted in Fig. 1B and C. It was observed that ethyl acetate resulted in a lower particle size compared to acetone due to the partial solubility of ethyl acetate in water. It has been reported that solvents having partial solubility in water result in smaller nanoparticles than the solvents with high water solubility, due to the ability to provide low interfacial tension at the aqueous–organic interphase.30 Additionally, low interfacial tension also facilitates a narrow distribution profile of the nanoparticles, as reflected by their low polydispersity index. As seen in Fig. 1C, nanoparticle formulation containing DMC (ethyl acetate) in combination with PVA (100 μg mL−1) exhibited an optimum particle size (170 nm ± 1.65 nm), polydispersity index (0.09 ± 0.003) and zeta potential (34.8 ± 0.97 mV) and hence was chosen for further experiments involving complexation with nucleotide molecules. Various parameters like monodispersity, higher zeta potential, stability and lung delivery were considered before choosing the optimum nanoparticle formulation.
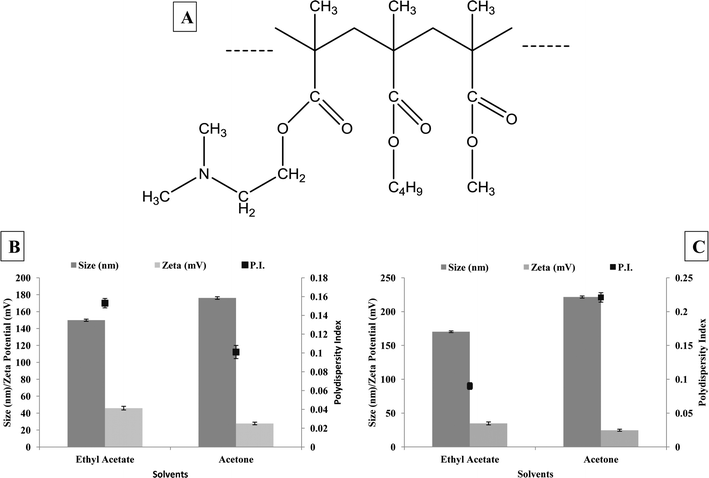 | ||
| Fig. 1 A. Structure of DMC; particle size, polydispersity index and zeta potential of nanoparticles with B. DMC: poloxamer : VTPGS (1 : 1 : 1) and C. DMC: PVA (1 : 1). | ||
Complexation of nanoparticles with nucleic acids
The optimized DMC nanoparticles demonstrated a positive zeta potential (34.8 ± 0.97 mV; Fig. 1), which allowed them to efficiently bind to the negatively charged nucleic acids. Preformed DMC nanoparticles were loaded with a combination of pUC 18-control plasmid DNA (pDNA) and siRNAs in a ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 w/w. The idea behind using preformed nanoparticles was to prevent the chemical, thermal or mechanical stresses associated with the process of nanoparticle formation from destroying the integrity and efficacy of the fragile siRNA molecules. The nucleotide combinations consisted of a cargo pDNA, non-functional in mammalian cells (pUC 18), and the respective siRNAs. The selection of the type of pDNA assured that any biological action of the nanoplexes could be attributed only to the particular siRNA molecule. pDNA was used to enable efficient loading of nanoparticles with siRNA due to its complex size and suitable molecular topography.31 siRNA molecules by themselves behave as stiff rods due to their very short length (21–23 bp). Since the minimum length of nucleotides required to efficiently complex with cationic carriers is typically reported to be about 80–120 bp, siRNA by themselves exhibit ample potential for poor loading and disordered complexing behavior, resulting in the formation of larger nanoparticles that may not be efficiently internalized.31
1 w/w. The idea behind using preformed nanoparticles was to prevent the chemical, thermal or mechanical stresses associated with the process of nanoparticle formation from destroying the integrity and efficacy of the fragile siRNA molecules. The nucleotide combinations consisted of a cargo pDNA, non-functional in mammalian cells (pUC 18), and the respective siRNAs. The selection of the type of pDNA assured that any biological action of the nanoplexes could be attributed only to the particular siRNA molecule. pDNA was used to enable efficient loading of nanoparticles with siRNA due to its complex size and suitable molecular topography.31 siRNA molecules by themselves behave as stiff rods due to their very short length (21–23 bp). Since the minimum length of nucleotides required to efficiently complex with cationic carriers is typically reported to be about 80–120 bp, siRNA by themselves exhibit ample potential for poor loading and disordered complexing behavior, resulting in the formation of larger nanoparticles that may not be efficiently internalized.31
It was hypothesized that such a combination of nucleotides would lower the amount of siRNA necessary to have the optimum binding to the nanoparticles. This is particularly important because high concentrations of siRNA have been reported to result in unwanted off-target effects.31 Alternatively, too low concentrations of siRNA when complexed with cationic nanoparticles with a high surface charge, as in the present investigation, would result in their excessively tight binding and thus hamper the siRNA release and efficacy. Previous reports have also demonstrated the ability of DMC nanoparticles to efficiently complex DNA, albeit without siRNA.32
Three different types of siRNAs, namely, Cy3 labeled control siRNA, anti-luciferase siRNA (ESI) and anti-Bfl1/A1 siRNA, were complexed with the DMC nanoparticles to demonstrate the cellular uptake and efficiency of the nanoplexes.
The complexation efficiency was studied over various weight ratios of nanoparticles to nucleic acids (pUC 18 and anti-luciferase siRNA), ranging from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.01 to 1
0.01 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.2 (w/w). The results have been presented in Fig. 2A. As seen from the figure, complete binding of nucleotides was observed at a weight ratio of 1
0.2 (w/w). The results have been presented in Fig. 2A. As seen from the figure, complete binding of nucleotides was observed at a weight ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.05, whereas excess nucleotides at higher weight ratios were observed as fluorescence bands during gel electrophoresis. The weight ratio of 1
0.05, whereas excess nucleotides at higher weight ratios were observed as fluorescence bands during gel electrophoresis. The weight ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.05 was hence finalized for formulating the nanoparticles for further cellular evaluations. The particle size and polydispersity of the nanoparticles increased with the increase in nucleotide concentration. This was accompanied by a steady decrease in zeta potential due to the overall negative charge of the loaded nucleotides. The weight ratio of 1
0.05 was hence finalized for formulating the nanoparticles for further cellular evaluations. The particle size and polydispersity of the nanoparticles increased with the increase in nucleotide concentration. This was accompanied by a steady decrease in zeta potential due to the overall negative charge of the loaded nucleotides. The weight ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.05 resulted in the optimum particle size, distribution (183.2 ± 4.5 nm, 0.187 ± 0.008) and zeta potential (23.11 ± 1.7 mV). Higher weight ratios resulted in non-homogenous formulations, as indicated by the increased PI and substantial lowering of overall surface charge, and hence were not chosen for further studies.
0.05 resulted in the optimum particle size, distribution (183.2 ± 4.5 nm, 0.187 ± 0.008) and zeta potential (23.11 ± 1.7 mV). Higher weight ratios resulted in non-homogenous formulations, as indicated by the increased PI and substantial lowering of overall surface charge, and hence were not chosen for further studies.
Complexation with all the three siRNAs resulted in nanocomplexes with similar size, distribution and surface charge. The results have been depicted in Fig. 2B. However the overall surface charge of the nanoparticles was positive, in all cases, which was anticipated to improve their cellular uptake by interaction with negatively charged cell membranes.
Morphology
The particle size and morphology of the unloaded and nucleic acid loaded nanoparticles were observed to be similar after drying (Fig. 3A) and in the liquid film (ESI). ESEM is an important technique for visualizing the shape and surface texture of pDNA/siRNA loaded nanoparticles. ESEM is suitable for polymeric nanoparticles since it does not cause any physical damage to the sample (ref. 20, ESI). Fig. 3B depicts the ESEM image of nucleic acid loaded DMC nanoparticles. The surface of the nanoplexes exhibited a slight roughness, as compared to the unloaded nanoparticles (Fig. 3A), which may be attributed to the adsorption of pDNA-siRNA on the nanoparticle surface.Cytotoxicity studies
The safety of the nanoparticles for various cellular evaluations was established by the MTT assay.20,21 The results of the MTT assay have been depicted in Fig. 4. Nanoparticles equivalent to test concentrations of DMC were employed during the assay. The results indicate that the nanoparticles exhibited no significant toxicity even at the highest test concentration, equivalent to 200 μg mL−1 DMC. Furthermore at lower test concentrations, equivalent to 75–125 μg mL−1 DMC, only a 10% decrease in their mitochondrial activity was observed.From Fig. 4, it is evident that the cells exhibit almost complete viability when incubated with nanoparticles equivalent to 100 μg mL−1 DMC. This concentration of nanoparticles was hence chosen for further cellular evaluations. However, PEI, the positive control, tested at the same concentration was found to be significantly toxic to the cells. Thus the developed nanoparticles were non-toxic to the cells at the test concentrations, which indicated their potential for further cellular evaluations.
Intracellular trafficking
Fig. 5 depicts the results of FACS analysis. As can be clearly seen, the shift in the mean fluorescence intensity (MFI) peak of the cells treated with naked Cy3-siRNA was minor as compared to that of the cells treated with Cy3-siRNA loaded DMC nanoparticles, indicating that the cell associated fluorescence was higher in the latter case.
The above observations were further reinforced by percent MFI values. The percent MFI for DMC nanoparticles was 33.62 ± 4.6% as against 13.44 ± 0.2% for naked siRNA and 7.33 ± 0.063% for cell control. This may be attributed to the inability of the naked siRNA to be efficiently internalized into the cells due to its labile nature and unfavorable size and charge. Although the cell associated fluorescence was higher in the case of DMC nanoparticles, the internalization of particles could not be precisely confirmed by this analysis. To confirm this internalization and to understand the mechanism of internalization of nanoparticles, confocal microscopy was performed (Fig. 6). The images indicate evident internalization of Cy3-siRNA loaded nanoparticles (Fig. 6A and B), denoted using arrows and circles. To further confirm these observations, z-stacks of confocal images were analyzed (Fig. 6C and D). These images indicate the localization of nanoparticles between the cell membrane and nucleus. This enhanced cellular association and uptake may be attributed to electrostatic attractions, overall cationic charge of the nanoparticles and electronegative groups of the cell surfaces, in addition to hydrophobic association of the polymer with membrane structures as observed in earlier investigations.33 To further understand the uptake mechanism, the cell uptake experiments were conducted in the presence of inhibitors of various cell uptake pathways. Although various other types of Eudragit® polymers have been previously explored for ref. 34 and 35 nanoparticle formulations, this study is the first one to identify the precise endocytic pathway for DMC nanoparticles. The various inhibitors used during this experiment, along with the respective doses employed and the specific pathways inhibited by each, are shown in Table S1 in the ESI. The table also indicates the experimental protocol followed for each of these inhibitors with regard to their incubation time, with or without nanoparticles.
Clathrin dependent endocytosis has been reported to be one of the most common ‘entry ports’ for nanoparticulate carriers. Chlorpromazine (CHL), a cationic amphiphilic drug, is known to inhibit clathrin-coated cavity formation, by a reversible translocation of clathrin and its adapter proteins from the plasma membrane to intracellular vesicles.36 The confocal images of the cell uptake with this inhibitor have been depicted in Fig. 7A. It was observed that the cell uptake of nanoparticles was comparable to that observed in control cells with no inhibitors (images not shown). The possibility of involvement of a caveolae-dependent pathway was evaluated using nystatin (NYS), a known inhibitor of this pathway. Nystatin is also a preferred agent since it does not affect the clathrin-dependent internalization. Nystatin is a sterol-binding agent that disassembles caveolae and cholesterol in the membrane.37 The confocal images (Fig. 7B) indicate similar cell uptake to that observed in the case of CHL. The cell uptake, again, was comparable to the control cells. Further experiments were then conducted using 5-(N-ethyl-N-isopropyl) amiloride (AML), an inhibitor of macropinocytosis and phagocytosis. AML is an inhibitor of the Na+/H+ exchanger that specifically inhibits both these pathways.38 Confocal images (Fig. 7C) indicate that nanoparticle uptake was severely affected in the presence of AML and most of the nanoparticles remained outside the cells in the presence of this inhibitor as compared to the control cells. Qualitatively, CLSM images thus suggested micropinocytosis and phagocytosis as the possible uptake pathways for the nanoparticles. To further verify these results, quantification of nanoparticle uptake was conducted by a combined multiphoton pixel analysis method39 and manual counting.
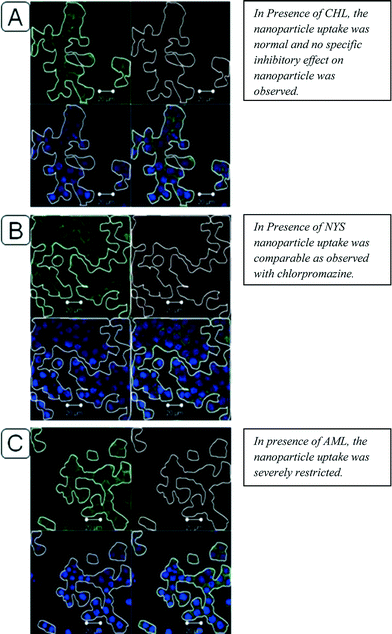 | ||
| Fig. 7 CLSM images of DMC nanoparticles loaded with Cy3-siRNA (red) in the presence of endocytic inhibitors. (Green: WGA stained Membrane. Blue: DAPI stained Nuclei. Red: Nanoparticles). | ||
The results support the observations shown by CLSM images and those of the manual method for quantification of nanoparticles. Also, our observations are in compliance with the earlier investigations with polymethacrylate (Eudragit) particles in various in vivo models, which speculate that in animal models these particles are taken up by macrophages via phagocytosis.40,41
In vitro gene silencing
The DMC nanoplexes (DMC nanoparticles loaded with anti-Luc siRNA) were evaluated for establishing the proof of biological efficacy of the formulated nanoparticles. These results have been discussed in depth in the ESI of this manuscript.
Bfl1/A1 gene silencing
Mycobacterium tuberculosis is an intracellular pathogen that primarily resides in host macrophages and utilizes the host cellular machinery for its survival. Although extensive research is under way to understand the ‘dependence factors’ of mycobacteria inside macrophages, knowledge and proof are rather limited. However, recent research has provided several new targets to reduce or diminish the bacterial growth inside the macrophages.45 One of the important survival pathways for these bacteria is to prevent apoptosis of the cell. Studies have suggested that apoptosis of infected macrophages is prevented by mycobacteria, especially due to the virulence associated with these organisms. It has been demonstrated that proteins of the Bcl-2 family (Bfl1/A1), cytokine tumour necrosis factor-α (TNF-α) and interleukin-10 (IL-10) modulate apoptosis of infected macrophages. Additionally, anti-apoptotic Bcl-2 proteins interact during mycobacterial infection and enhance their survival rate inside the macrophages.46–48 The role of Bfl1/A1, a member of the Bcl-2 family of proteins, has been comprehensively explored in the pathogenesis of tuberculosis and it has been demonstrated that inhibition of Bfl1/A1 can limit the mycobacterial growth and hence spread of infection.18 Thus in the present investigation DMC nanoparticles loaded with Bfl1/A1 siRNA were evaluated in the macrophages (RAW 264.7) for their ability to enhance knockdown of anti-Bfl1/A1 siRNA and thus down-regulate Bfl1/A1 expression. This was projected to have therapeutic significance for the treatment of tuberculosis.Expression of endogenous Bfl1/A1 was induced in RAW 264.7 cells using lipopolysaccharide from E. coli. Bfl1/A1 is an NF-κB (nuclear factor kappa B) dependent gene. Various studies have already demonstrated that LPS can stimulate NF-κB signaling and hence Bfl1/A1 expression in RAW 264.7 cells.16,17,49 The use of this model suggests an appropriate strategy to demonstrate anti-tubercular potential of the siRNA loaded polymeric nanoparticles without compromising on the safety issues associated with working with tuberculosis bacteria, in preliminary studies.
The stimulation of Bfl1/A1 expression in RAW 264.7 cells was confirmed by western blot analysis of the protein. Further, RAW 264.7 cells were treated with siRNA loaded DMC nanoparticles, naked anti-Bfl1/A1siRNA, control siRNA and anti-Bfl1/A1siRNA – jetPRIME™ transfection reagent combination. Bfl1/A1 protein and mRNA levels were analyzed using western blot and RT-PCR. The results were compared with the protein expression and mRNA levels in untreated cells and cells treated only with LPS. The results have been demonstrated in Fig. 8A and B, respectively.
Fig. 8A confirms the expression of Bfl1/A1 in Raw 264.7 cells when stimulated by LPS, as compared to its inherent expression in un-stimulated cells. Further, the siRNA-DMC nanoparticles were able to reduce the expression of Bfl1/A1 protein, a trend similar to that observed with luciferase gene (ESI). DMC nanoparticles loaded with 80 pmol of anti-Bfl1/A1 siRNA significantly reduced the protein expression as compared to the naked siRNA. These results were also reinforced by reduction in mRNA levels during RT-PCR studies (Fig. 8B). The protein and mRNA levels of Bfl1/A1 were not affected when the cells were treated with free anti-Bfl1/A1 siRNA or scrambled siRNA, either in free or complexed state (data not shown).
The reporter gene and functional gene silencing strongly prove the capability of DMC nanoparticles to act as effective siRNA carriers for macrophage delivery. This efficacy may be attributed to their ability to perform efficient intracellular trafficking. Commercial Eudragit polymers have been evaluated for their membrane disruption potential.50 It has been demonstrated that these polymers contain protonable amino nitrogen atoms, which makes them effective ‘proton sponges’. This effect is responsible for endosomal swelling and rupture, and provides successful escape of nanoparticles from endosomes, thus protecting the associated siRNA. The DMC polymer consists of a hydrophobic backbone due to esterification of all its carboxylic acid groups with methyl and butyl side groups. This hydrophobic backbone is responsible for the membrane-disrupting activities at acidic pH.50–55 At acidic pH of endosome, the polymer conformation changes from coil to globule due to protonation of carboxylic acid groups and polymer binding increases. The precise mechanism of disruption, however, is not well understood and it is believed that because of the electrostatic interaction between the endosomal membrane and tertiary amine groups of DMC, a positive charge is generated inside the hydrophobic layer of the membranes, which causes membrane disruption.51,52 This mechanism may be responsible for the endosomal escape of the siRNA-DMC nanoparticles and thus their enhanced biological functionality.
Materials and methods
Materials
Eudragit® E100 or DMC (Mw approx. 47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1) was kindly gifted by Evonik, Germany. Ethyl acetate (Fluka Chemie GmbH, Buchs, Switzerland) and polyvinyl alcohol (PVA; Mowiol 4-88; Kuraray Specialities Europe GmbH, Frankfurt, Germany) were used as obtained from the suppliers. Bfl1/A1 siRNA, scrambled siRNA sequence and anti-Bfl1/A1 antibody were purchased from Santa Cruz Biotechnology Inc. (Germany). Total RNA isolation and RT-PCR kits were purchased from Qiagen (Germany).
000 g mol−1) was kindly gifted by Evonik, Germany. Ethyl acetate (Fluka Chemie GmbH, Buchs, Switzerland) and polyvinyl alcohol (PVA; Mowiol 4-88; Kuraray Specialities Europe GmbH, Frankfurt, Germany) were used as obtained from the suppliers. Bfl1/A1 siRNA, scrambled siRNA sequence and anti-Bfl1/A1 antibody were purchased from Santa Cruz Biotechnology Inc. (Germany). Total RNA isolation and RT-PCR kits were purchased from Qiagen (Germany).
Dimethyl sulfoxide (DMSO) for the MTT assay was purchased from Sigma Chemical Co. (St. Louis, MO, USA). Fluorescein wheat germ agglutinin (FITC-WGA) was obtained from Vector (Vector Laboratories, Inc., Burlingame, CA), and 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) was procured from Fluka Chemie GmbH (Buchs, Switzerland). All the other chemicals and solvents employed were of analytical grade. Highly purified water (Milli Q Plus system, Millipore, Bedford, MA, USA) was used whenever required.
The cellular uptake studies were conducted by employing Silencer® Cy™3 negative control siRNA (Ambion, Inc., TX, USA) for loading onto the nanoparticles. The knockdown of Bfl1/A1 gene, responsible for intra-macrophage survival of tuberculosis bacteria, was studied by complexing the nanoparticles with anti-Bfl1/A1 siRNA and scrambled siRNA (Santa Cruz Biotechnology, CA, USA). The concentrations were chosen as per the manufacturer's suggestions.
Cell culture
RAW 264.7 mouse macrophage line was obtained from American Type Culture Collection (ATCC cat. no. TIB-71, Manassas, VA). RAW 264.7 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS) (both from GIBCO, Karlsruhe, Germany). The cells were maintained at 37 °C, 85% relative humidity and 5% CO2.Nanoparticle preparation and siRNA loading
DMC nanoparticles were formulated using the solvent emulsion diffusion method20 with slight modifications. DMC nanoparticles were optimized using various stabilizers e.g. poloxamer 407, vitamin E-TPGS and PVA. These stabilizers were used in 1 mg mL−1 concentration in each formulation. Briefly, a solution of DMC in ethyl acetate or acetone (1.0 mg mL−1; 1 mL) was emulsified for 10 min with an aqueous solution of polyvinyl alcohol (PVA; 1 mg mL−1; 1 mL) at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 rpm, using a high-speed homogenizer (Ultra Turrax® Ika®, Staufen, Germany). Further, water (8 mL) was added into the mixture to promote diffusion of the organic phase into the aqueous phase. The resulting nanoparticles were stirred for 10 h to allow complete evaporation of the organic solvent. The final volume of the nanoparticle dispersion was adjusted to 10 mL with water, equivalent to a DMC content of 0.1 mg mL−1. pUC 18 DNA (pDNA; 1.2 nmol) was added into the nanoparticle formulation. Twenty microliters of siRNA (400 pmol) in nuclease-free water was added to 1 mL of DMC nanoparticles, while stirring, and incubated for 1 hour prior to further use.
500 rpm, using a high-speed homogenizer (Ultra Turrax® Ika®, Staufen, Germany). Further, water (8 mL) was added into the mixture to promote diffusion of the organic phase into the aqueous phase. The resulting nanoparticles were stirred for 10 h to allow complete evaporation of the organic solvent. The final volume of the nanoparticle dispersion was adjusted to 10 mL with water, equivalent to a DMC content of 0.1 mg mL−1. pUC 18 DNA (pDNA; 1.2 nmol) was added into the nanoparticle formulation. Twenty microliters of siRNA (400 pmol) in nuclease-free water was added to 1 mL of DMC nanoparticles, while stirring, and incubated for 1 hour prior to further use.
Photon correlation spectroscopy and zeta potential
The hydrodynamic size and surface charge of DMC nanoparticles (unloaded and loaded) were determined by photon correlation spectroscopy using a Zetasizer Nano ZS (Malvern Instruments, Malvern, UK). The experiments were conducted at 25 °C after diluting the formulation with water to guarantee that its light scattering intensity was within the instrument's sensitivity range. Measurements were conducted in triplicate at a wavelength of 633 nm and a backscattering angle of 173°.ESEM
The morphological characteristics of the DMC nanoparticles (unloaded and loaded) were examined using a high resolution environmental scanning electron microscope (ESEM, FEI Quanta 400 FEG). For ESEM, one drop of dilute nanoparticles was placed on a small piece of silicon wafer and dried in the SEM chamber. Images were recorded at room temperature with an accelerating voltage of 20 kV and a pressure p = 100 Pa water. Wet-STEM investigations were done with a 2 μl drop of dilute nanoparticle solution placed on a carbon coated copper TEM grid at an accelerating voltage of 30 kV and a pressure p = 800 Pa (T = 276 K).Complexation of nanoparticles with nucleic acids
Gel electrophoresis was performed to evaluate the complexation of siRNA with DMC nanoparticles. The details of this method have been provided in the ESI.Cell viability
To evaluate cytotoxicity of nanoparticles, the MTT assay was carried out using the standard protocol.21 We have used PEI as the positive control in the experiment. The experimental details of the MTT assay have been described in the ESI.Intracellular trafficking
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well. The nanoparticle suspension was diluted in a ratio of 1
000 cells per well. The nanoparticle suspension was diluted in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 with Krebs Ringer Buffer (KRB) (pH 7.4). The cell treatment procedure is indicated in the CLSM section. Analysis by flow cytometry was performed after 24 h with a FACS Calibur flow cytometer from Becton Dickinson (BD) Biosciences (Heidelberg, Germany) using the CellQuest™ Pro Version 4.02 (BD Biosciences) software program. For this purpose, the cells were washed twice with PBS and detached using a cell scrapper. Further, the cell suspension was centrifuged at 500 × g and the resulting pellet was re-suspended in cell medium (500 μL). The cell-associated fluorescence (Cy3) was elicited by excitation at 543 nm and measured after passing through a 560/615 nm band pass filter. The instrument was adjusted with untreated cells. During each run 10
4 with Krebs Ringer Buffer (KRB) (pH 7.4). The cell treatment procedure is indicated in the CLSM section. Analysis by flow cytometry was performed after 24 h with a FACS Calibur flow cytometer from Becton Dickinson (BD) Biosciences (Heidelberg, Germany) using the CellQuest™ Pro Version 4.02 (BD Biosciences) software program. For this purpose, the cells were washed twice with PBS and detached using a cell scrapper. Further, the cell suspension was centrifuged at 500 × g and the resulting pellet was re-suspended in cell medium (500 μL). The cell-associated fluorescence (Cy3) was elicited by excitation at 543 nm and measured after passing through a 560/615 nm band pass filter. The instrument was adjusted with untreated cells. During each run 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells were counted. The percentage of cells associated with DMC/siRNA (Cy3 labeled) nanoparticles was evaluated after gating and selection of a fluorescence threshold referred to as non-treated cells. The experiment was performed in triplicate.
000 cells were counted. The percentage of cells associated with DMC/siRNA (Cy3 labeled) nanoparticles was evaluated after gating and selection of a fluorescence threshold referred to as non-treated cells. The experiment was performed in triplicate.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well, 24 hours prior to the experiment. The cells were maintained employing the conditions described in section 2.2. For uptake experiments cells were treated with DMC/siRNA (Cy3 labeled) nanoparticles (100 μl). The nanoparticle suspension was diluted in a ratio of 1
000 cells per well, 24 hours prior to the experiment. The cells were maintained employing the conditions described in section 2.2. For uptake experiments cells were treated with DMC/siRNA (Cy3 labeled) nanoparticles (100 μl). The nanoparticle suspension was diluted in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 with Krebs Ringer Buffer (KRB) (pH 7.4). Cells were incubated with this medium containing nanoparticles for 4 h in a CO2 incubator. Afterwards, the incubation medium was replaced with normal cell culture medium without nanoparticles. Untreated cells, cells with unloaded DMC nanoparticles and plain siRNA (Cy3 labeled) were used as controls. After 24 h of incubation, the cells were fixed and stained to enable visualization of the nuclei (DAPI) and cell membranes (FITC-WGA). The staining procedures have been provided in the ESI.
4 with Krebs Ringer Buffer (KRB) (pH 7.4). Cells were incubated with this medium containing nanoparticles for 4 h in a CO2 incubator. Afterwards, the incubation medium was replaced with normal cell culture medium without nanoparticles. Untreated cells, cells with unloaded DMC nanoparticles and plain siRNA (Cy3 labeled) were used as controls. After 24 h of incubation, the cells were fixed and stained to enable visualization of the nuclei (DAPI) and cell membranes (FITC-WGA). The staining procedures have been provided in the ESI.
The imaging was performed using a CLSM (LSM 510; Zeiss, Jena, Germany) equipped with an argon/neon laser and a 63× water immersion objective. Zeiss LSM510 software was employed for all the measurements and analyses. The excitation wavelengths employed were 488 nm for FITC-WGA, 360 nm for DAPI and 543 nm for Cy3-siRNA containing nanoplexes and the fluorescence signals were collected at band pass filters of 500/530 nm for FITC-WGA, 390/465 nm for DAPI and 560/615 nm for the Cy3-siRNA containing nanoparticles, respectively.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well) were plated in 24-well plates as indicated in section 2.8.2. Stock solutions of chlorpromazine (CHL; C8138, Sigma-Aldrich, Taufkirchen, Germany) and 5-(N-ethyl-N-isopropyl) amiloride (EIPA; A3085, Sigma-Aldrich, Taufkirchen, Germany) in DMSO and nystatin (NYS; N3503, Sigma-Aldrich, Taufkirchen, Germany) in water, diluted with cell medium to the final concentrations of 10 μg mL−1 (CHL), 5 μg mL−1 (EIPA) and 10 μg mL−1 (NYS), were used for evaluation. The procedural details have been discussed in the ESI.
000 cells per well) were plated in 24-well plates as indicated in section 2.8.2. Stock solutions of chlorpromazine (CHL; C8138, Sigma-Aldrich, Taufkirchen, Germany) and 5-(N-ethyl-N-isopropyl) amiloride (EIPA; A3085, Sigma-Aldrich, Taufkirchen, Germany) in DMSO and nystatin (NYS; N3503, Sigma-Aldrich, Taufkirchen, Germany) in water, diluted with cell medium to the final concentrations of 10 μg mL−1 (CHL), 5 μg mL−1 (EIPA) and 10 μg mL−1 (NYS), were used for evaluation. The procedural details have been discussed in the ESI.
Therapeutic gene silencing: Bfl1-A1
After establishing the proof of efficacy of nanoparticles by down-regulating the transiently expressed luciferase gene (ESI) in RAW 264.7 cells, the nanoparticles were further evaluated for their efficiency to knockdown a gene relevant for pathogenesis of tuberculosis.RAW 264.7 cells were treated with LPS (10 μg mL−1) (derived from E. coli) to stimulate Bfl1/A1 expression. This particular LPS was chosen due to its close similarity to the LPS present on mycobacterial cell walls.22 LPS induced Bfl1/A1 expression was analyzed by western blot analysis. After treating with LPS overnight, the medium containing LPS was replaced with a medium containing nanoparticles, formulated with anti-Bfl1/A1 siRNA (Santa Cruz Biotechnology, CA, USA). Untreated cells, cells treated with uncomplexed siRNA, siRNA formulated with jetPRIME™ reagent and nanoparticles formulated with scrambled siRNA were maintained as controls. For all the samples, siRNA was used at a concentration of 80 pmol per well.
The knockdown efficiency of siRNA bound to nanoparticles was analyzed by western blot analysis and RT-PCR, and compared with that of the controls. Western blot analysis was conducted by the procedure described in the ESI. Mouse monoclonal IgG1 (A1 (B-3) antibody: SC-166943; Santa Cruz Biotechnology, CA, USA) at a dilution of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 in blocking buffer was employed as the primary antibody during the experiment.
200 in blocking buffer was employed as the primary antibody during the experiment.
RT-PCR was used to analyze the expression of Bfl1/A1 according to the previously described procedure, with slight modifications. The amplification cycle consisted of an initial denaturation of 5 min at 94 °C and 35 cycles of 94 °C for 1 min, 60 °C for 1 min and 72 °C for 1.5 min, a final amplification at 72 °C for 5 min, followed by a melt curve analysis (from 72 °C to 98 °C in 0.5 °C steps for 5 s).18 The primer sequences for Bfl1/A1 (Santa Cruz Biotechnology, CA, USA) were primer sense sequence 5′-TAC AGG CTG GCT CAG GAC TAT C-3′ and primer antisense sequence 5′-GGT ATC CAC ATC CGG GGC AAT-3′.
Statistics
The Student's ‘t’ test and ANOVA were applied to evaluate the data.Conclusion
The investigation proves the potential of DMC nanoparticles for efficient siRNA delivery into the macrophages which are host for the tuberculosis bacteria. The DMC nanoparticles effectively complex with various different siRNAs and protect them from degradation. Additionally, the nanoplexes are well internalized via phagocytosis/macropinocytosis and are believed to escape from the endosomes by DMC's membrane disruption properties at acidic pH. Further, the conformational change in the DMC polymer allows the nanoplexes to release the associated siRNA into the cytosol and provide enhanced knockdown of targeted genes. Down-regulation of Bfl1/A1 gene by DMC nanoparticles provides unique opportunity to further explore them in silencing genes specific to intracellular bacteria, especially Mycobacterium tuberculosis. The study suggests potential of the developed nanoparticles in RNAi therapy of bacterial diseases, a relatively nascent research arena.Acknowledgements
Dr. Ratnesh Jain is thankful to Alexander von Humboldt Fellowship, Germany. Dr. Prajakta Dandekar is thankful to ERS-Marie Curie Fellowship. The authors are thankful to Mr. Leon Mujis for technical help in confocal microscopy. We thank Professor Eduard Arzt for ongoing support to our work at INM.Notes and references
- S. Chono, T. Tanino, T. Seki and K. Morimoto, J. Controlled Release, 2008, 127, 50–58 CrossRef CAS PubMed.
- J. D. McKinney, K. Honer zu Bentrup, E. J. Munoz-Elias, A. Miczak, B. Chen, W. T. Chan, D. Swenson, J. C. Sacchettini, W. R. Jacobs Jr and D. G. Russell, Nature, 2000, 406, 735–738 CrossRef CAS PubMed.
- J. J. Ellner, M. J. Goldberger and D. M. Parenti, J. Infect. Dis., 1991, 163, 1326–1335 CrossRef CAS PubMed.
- T. R. Frieden, T. R. Sterling, S. S. Munsiff, C. J. Watt and C. Dye, Tuberculosis, 2003, 362, 887–899 Search PubMed.
- G. Harth, P. C. Zamecnik, J.-Y. Tang, D. Tabatadze and M. A. Horwitz, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 418–423 CrossRef CAS.
- W. Yeeprae, S. Kawakami, F. Yamashita and M. Hashida, J. Controlled Release, 2006, 114, 193–201 CrossRef CAS PubMed.
- D. L. Lefkowitz and S. S. Lefkowitz, Immunol. Cell Biol., 2001, 79, 502–506 CrossRef CAS PubMed.
- T. E. Wileman, M. R. Lennartz and P. D. Stahl, Identification of the macrophage mannose receptor as a 175-kDa membrane protein, Proc. Natl. Acad. Sci. U. S. A., 1986, 83, 2501–2505 CrossRef CAS.
- M. E. Taylor, J. T. Conary, M. R. Lennaria, P. D. Stahl and K. Drickamer, J. Biol. Chem., 1990, 265, 12156–12162 CAS.
- R. S. Haltiwanger and R. L. Hill, J. Biol. Chem., 1986, 261, 15696–15702 CAS.
- B. L. Largent, K. M. Walton, C. A. Hoppe, Y. C. Lee and R. L. Schnaar, J. Biol. Chem., 1984, 259, 1764–1769 CAS.
- H. Eidi, O. Joubert, C. Némos, S. Grandemange, B. Mograbi, B. Foliguet, J. Tournebize, P. Maincent, A. Le Faou, I. Aboukhamis and B. H. Rihn, Int. J. Pharm., 2012, 422, 495–503 CrossRef CAS PubMed.
- S. M. Moghimi, A. C. Hunter and J. C. Murray, Pharmacol. Rev., 2001, 53, 283–318 CAS.
- Y. L. Chiu, A. Ali, C. Y. Chu, H. Cao and T. M. Rana, Chem. Biol., 2004, 11, 1165–1175 CrossRef CAS PubMed.
- Y. Kakizawa, S. Furukawa and K. Kataoka, J. Controlled Release, 2004, 97, 345–356 CrossRef CAS PubMed.
- W. X. Zong, L. C. Edelstein, C. Chen, J. Bash and C. Gélinas, Genes Dev., 1999, 13, 382–387 CrossRef CAS.
- M. Fukui, R. Imamura, M. Umemura, T. Kawabe and T. Suda, J. Immunol., 2003, 171, 1868–1874 CrossRef CAS.
- R. Dhiman, M. Kathania, M. Raje and S. Majumdar, Biochim. Biophys. Acta, 2008, 1780, 733–742 CrossRef PubMed.
- R. H. Plasterk, Science, 2002, 29, 1263–1265 CrossRef PubMed.
- R. Jain, P. Dandekar, B. Loretz, A. Melero, T. Stauner, G. Wenz, M. Koch and C.-M. Lehr, Int. J. Pharm., 2011, 420, 147–155 CrossRef CAS PubMed.
- P. Dandekar, R. Jain, B. Loretz, T. Stauner, G. Wenz, M. Koch and C.-M. Lehr, Macromol. Biosci., 2012, 12, 184–194 CrossRef CAS PubMed.
- N. A. Begum, M. Kobayashi, Y. Moriwaki, M. Matsumoto, K. Toyoshima and T. Seya, Genomics, 2002, 80, 630–645 CrossRef CAS.
- A. Basarkar and J. Singh, Int. J. Nanomed., 2007, 2, 353–360 CAS.
- C. Zhu, S. Jung, G. Si, R. Cheng, F. Meng, X. Zhu, T. G. Park and Z. Zhong, J. Polym. Sci., Part A: Polym. Chem., 2010, 48, 2869–2877 CrossRef CAS.
- W. F. Lai, Expert Rev. Med. Devices, 2011, 8, 173–185 CrossRef CAS PubMed.
- W. T. Godbey, K. K. Wu and A. G. Mikos, J. Controlled Release, 1999, 60, 149–160 CrossRef CAS.
- J. R. Flora, B. Baker, D. Wybenga, H. Zhu and C. M. Aelion, Chemosphere, 2008, 70, 1077–1084 CrossRef CAS PubMed.
- M. Gargouri, A. Sapin, S. Bouli, P. Becuwe, J. L. Merlin and P. Maincent, Cancer Res. Treat., 2009, 8, 433–444 CrossRef CAS PubMed.
- D. A. Quinteros, V. R. Rigo, A. F. Kairuz, M. E. Olivera, R. H. Manzo and D. A. Allemandi, Eur. J. Pharm. Sci., 2008, 33, 72–79 CrossRef CAS PubMed.
- K. C. Song, H. S. Lee, I. Y. Choung, K. I. Cho, Y. Ahn and E. J. Choi, Colloids Surf., A, 2006, 276, 162–167 CrossRef CAS PubMed.
- H. Rhinn, C. Largeau, P. Bigey, R. L. Kuen, M. Richard, D. Scherman and V. Escriou, Biochim. Biophys. Acta, 2009, 1790, 219–230 CrossRef CAS PubMed.
- E. Esposito, S. Sebben, R. Cortesi, E. Menegatti and C. Nastruzzi, Int. J. Pharm., 1999, 189, 29–41 CrossRef CAS.
- R. V. Alasino, V. Leonhard, I. D. Bianco and D. M. Beltramo, Colloids Surf., B, 2012, 91, 84–89 CrossRef CAS PubMed.
- P. Khachane, A. A. Date and M. S. Nagarsenker, J. Biomed. Nanotechnol., 2011, 7, 590–597 CrossRef CAS PubMed.
- P. Dandekar, R. Jain, C. Kumar, S. Subramanian, G. Samuel, M. Venkatesh and V. Patravale, J. Biomed. Nanotechnol., 2009, 5, 445–455 CrossRef PubMed.
- D. Vercauteren, R. E. Vandenbroucke, A. T. Jones, J. Rejman, J. Demeester, S. C. De Smedt, N. N. Sanders and K. Braeckmans, Mol. Ther., 2010, 18, 561–569 CrossRef CAS PubMed.
- X. D. Zhu, Y. Zhuang, J. J. Ben, L. L. Qian, H. P. Huang, H. Bai, J. H. Sha, Z. G. He and Q. Chen, J. Biol. Chem., 2011, 286, 8231–8239 CrossRef CAS PubMed.
- T. Fukushima, T. K. Waddell, S. Grinstein, G. G. Goss, J. Orlowski and G. P. Downey, J. Cell Biol., 1996, 132, 1037–1052 CrossRef CAS.
- H. I. Labouta, T. Kraus, L. K. El-Khordagui and M. Schneider, Int. J. Pharm., 2011, 413, 279–282 CrossRef CAS PubMed.
- D. S. Kohane, D. G. Anderson, C. Yu and R. Langer, Pharm. Res., 2010, 20, 1533–1538 CrossRef.
- W. N. Haining, D. G. Anderson, S. R. Little, M. S. von Bergwelt-Baildon, A. A. Cardoso, P. Alves, K. Kosmatopoulos, L. M. Nadler, R. Langer and D. S. Kohane, J. Immunol., 2004, 173, 2578–2585 CrossRef CAS.
- S. J. Reich, J. Fosnot, A. Kuroki, W. Tang, X. Yang, A. M. Maguire, J. Bennett and M. J. Tolentino, Mol. Vision, 2003, 9, 210–216 CAS.
- S. Hossain, A. Stanislaus, M. J. Chua, S. Tada, Y. Tagawa, E. H. Chowdhury and T. Akaike, J. Controlled Release, 2010, 147, 101–108 CrossRef CAS PubMed.
- S. Mao, M. Neu, O. Germershaus, O. Merkel, J. Sitterberg, U. Bakowsky and T. Kissel, Bioconjugate Chem., 2006, 17, 1209–1218 CrossRef CAS PubMed.
- G. Griffiths, B. Nyström, S. B. Sable and G. K. Khuller, Nat. Rev. Microbiol., 2010, 11, 827–834 CrossRef PubMed.
- M. F. Rodrigues, M. M. Barsante, C. C. Alves, M. A. Souza, A. P. Ferreira, G. P. Amarante-Mendes and H. C. Teixeira, Immunology, 2009, 128, e691–699 CrossRef PubMed.
- J. Zhang, R. Jiang, H. Takayama and Y. Tanak, Microbiol. Immunol., 2005, 49, 845–852 CrossRef CAS PubMed.
- V. A. Ríos-Barrera, V. Campos-Peña, D. Aguilar-León, L. R. Lascurain, M. A. Meraz-Ríos, J. Moreno, V. Figueroa-Granados and R. Hernández-Pando, Eur. J. Immunol., 2006, 36, 345–353 CrossRef PubMed.
- E. Jones, I. M. Adcock and B. Y. Ahmed, J. Inflammation, 2007, 23, 1–9 Search PubMed.
- R. V. Alasino, S. F. Ausar, I. D. Bianco, L. F. Castagna, M. Contigiani and D. M. Beltramo, Macromol. Biosci., 2005, 5, 207–213 CrossRef CAS PubMed.
- C. Kusonwiriyawong, P. van de Wetering, J. A. Hubbell, H. P. Merkle and E. Walter, Eur. J. Pharm. Biopharm., 2003, 56, 237–246 CrossRef CAS.
- S. M. Sparks, C. L. Waite, A. M. Harmon, L. M. Nusblat, C. M. Roth and K. E. Uhricha, Macromol. Biosci., 2011, 11, 1192–1200 CrossRef CAS PubMed.
- N. Iverson, N. Plourde, S. Sparks, J. Wang, E. Patel, P. Shah, D. Lewis, K. Zablocki, G. Nackman, K. Uhrich and P. Moghe, Biomaterials, 2011, 32, 8319–8327 CrossRef CAS PubMed.
- J. Wu, D. Yamanouchi, B. Liu and C.-C. Chu, J. Mater. Chem., 2012, 22, 18983–18991 RSC.
- A. Swami, M. Reagan, P. Basto, Y. Mishima, N. Kamaly, S. Glavey, S. Zhang, M. Moschetta, D. Seevaratnam, Y. Zhang, J. Liu, M. Memarzadeh, J. Wu, S. Manier, J. Shi, N. Bertrand, Z. Lu, K. Nagano, R. Baron, A. Sacco, A. Roccaro, O. Farokhzad and I. Ghobria, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 10287–10292 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4md00490f |
| This journal is © The Royal Society of Chemistry 2015 |






