 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Characterisation of SEQ0694 (PrsA/PrtM) of Streptococcus equi as a functional peptidyl-prolyl isomerase affecting multiple secreted protein substrates†
Felicia
Ikolo
ab,
Meng
Zhang
a,
Dean J.
Harrington
c,
Carl
Robinson
d,
Andrew S.
Waller
d,
Iain C.
Sutcliffe
*a and
Gary W.
Black
a
aDepartment of Applied Sciences, Faculty of Health & Life Sciences, University of Northumbria at Newcastle, Newcastle upon Tyne, NE1 8ST, UK. E-mail: iain.sutcliffe@northumbria.ac.uk; Fax: +44 (0)191 227 3519; Tel: +44 (0)191 227 4071
bDepartment of Biochemistry, School of Medicine, St. George's University, True Blue, St. George's, Grenada
cDivision of Biomedical Science, School of Life Sciences, University of Bradford, West Yorkshire, BD7 1DP, UK
dCentre for Preventive Medicine, Animal Health Trust, Lanwades Park, Kentford, Newmarket, Suffolk CB8 7UU, UK
First published on 8th October 2015
Abstract
Peptidyl-prolyl isomerase (PPIase) lipoproteins have been shown to influence the virulence of a number of Gram-positive bacterial human and animal pathogens, most likely through facilitating the folding of cell envelope and secreted virulence factors. Here, we used a proteomic approach to demonstrate that the Streptococcus equi PPIase SEQ0694 alters the production of multiple secreted proteins, including at least two putative virulence factors (FNE and IdeE2). We demonstrate also that, despite some unusual sequence features, recombinant SEQ0694 and its central parvulin domain are functional PPIases. These data add to our knowledge of the mechanisms by which lipoprotein PPIases contribute to the virulence of streptococcal pathogens.
Introduction
In order to interact with their environments, bacteria translocate significant numbers of proteins across their plasma membranes, either for eventual release (secretion) or for localisation within the cell envelope.1–3 In pathogens, this ‘secretome’ plays a vital role in host–pathogen interactions and consequently the mechanisms of protein translocation are of much interest as ‘virulence-associated’ functions. Proteins exported by the Sec translocase emerge on the extracytoplasmic side of the plasma membrane as unfolded proteins and the subsequent correct folding of these proteins is therefore critical to their functioning. In ‘diderm’ bacteria (those with outer membranes), a variety of periplasmic chaperones are required to allow protein folding in the periplasm and/or translocation across or into the outer membrane.1,4 In monoderm Gram-positive bacteria, secreted proteins fold at the membrane-wall interface with the assistance of a range of accessory components of the Sec translocase.5 These include proteins belonging to the peptidyl-prolyl Isomerase (PPIase) family, which assist protein folding by catalysing cis–trans isomerisation of the peptide bond preceding proline residues.6,7 In many Gram-positive bacteria, these PPIases are N-terminally lipid-anchored lipoproteins, presumably because the localisation of a PPIase peripheral to the plasma membrane surface places it in an optimal position to engage with substrate proteins emerging from the Sec translocon.8Several lipoprotein PPIases have been shown to have significant roles in bacterial physiology, notably PrsA in Bacillus subtilis.9 Moreover, in some pathogens PPIases have been shown to affect virulence,7 including PrsA of Bacillus anthracis,10Enterococcus faecalis EF0685 and EF1534,11Listeria monocytogenes PrsA2,12,13Streptococcus pneumoniae SlrA and PpmA14 and Streptococcus pyogenes PrsA.15 Some of these PPIase belong to the cyclophilin subfamily (e.g. S. pneumoniae SlrA; E. faecalis EF1534) but many belong to the parvulin subfamily,16 including the members of PrsA family that appear to be ubiquitous in Firmicute genomes.
Streptococcus equi is the causative agent of the widespread equine disease Strangles.17,18 We have previously shown that the PrsA homologue of S. equi (UniProt: C0M9L5, originally denoted PrtM) plays a significant role in S. equi virulence, both in an air interface tissue culture model, a mouse model and, most significantly, in the equine host.19 PrtM is here referred to as SEQ0694, based on its annotation in the S. equi genome.17
To further investigate the role of SEQ0694 we have here characterised the recombinant protein as a functional PPIase and used a proteomic approach to demonstrate that SEQ0694 likely influences the folding and activity of multiple secreted proteins of S. equi, including at least two putative virulence factors.
Materials and methods
Bacterial strains and growth
S. equi strain 4047 (wild type) and its isogenic mutant strain (ΔprtM138–213) with a deletion of codons 138 to 213 in seq0694 (i.e. lacking the central domain of SEQ0694, ESI,† Fig. S1) are described in Hamilton et al.19S. equi strains were grown in Todd Hewitt media. Escherichia coli TOP10 and BL21 were grown in LB media.Production and purification of recombinant proteins
Genomic DNA from S. equi 4047 was isolated using a DNeasy extraction kit (Qiagen). To produce recombinant N-terminally His-tagged full-length SEQ0694 (rSEQ0694), the seq0694 ORF, minus the sequence encoding the signal peptide, was amplified from S. equi 4047 genomic DNA using the primer pair 5′ GATCGATC![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) TGTCAGTCTACAAATGACAATACAAGTG 3′ (forward primer, NdeI site underlined) and 5′ GATCGATC
TGTCAGTCTACAAATGACAATACAAGTG 3′ (forward primer, NdeI site underlined) and 5′ GATCGATC![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ATATTTTTCTGACTTAGATTTAGAAGATTGAC 3′ (reverse primer, XhoI site underlined) and KOD Hot Start polymerase (Merck Chemicals) according to the manufacturer's instructions. The amplified ORF was cloned into pET28a (Merck Chemicals) using NdeI-XhoI and expressed in E. coli BL21(DE3) grown at 37 °C with shaking at 200 rpm, to an absorbance of 0.6 at 600 nm, in LB medium supplemented with 100 μg mL−1 kanamycin. Induction was performed by the addition of isopropyl-1-thio-β-D-galactopyranoside to a concentration of 240 μg mL−1, followed by further incubation for 18 h at 30 °C with shaking at 100 rpm. rSEQ0694 was purified according to the method of Malik et al.,20 except that the purified protein was concentrated and the buffer exchanged into 18.2 MΩ cm−1 water using 10 kDa cut-off centrifugal concentrator units (Viva Science) The identity of rSEQ0694 was confirmed by peptide mass fingerprinting of trypsinized bands excised from Coomassie blue-stained SDS-polyacrylamide gels (see below).
ATATTTTTCTGACTTAGATTTAGAAGATTGAC 3′ (reverse primer, XhoI site underlined) and KOD Hot Start polymerase (Merck Chemicals) according to the manufacturer's instructions. The amplified ORF was cloned into pET28a (Merck Chemicals) using NdeI-XhoI and expressed in E. coli BL21(DE3) grown at 37 °C with shaking at 200 rpm, to an absorbance of 0.6 at 600 nm, in LB medium supplemented with 100 μg mL−1 kanamycin. Induction was performed by the addition of isopropyl-1-thio-β-D-galactopyranoside to a concentration of 240 μg mL−1, followed by further incubation for 18 h at 30 °C with shaking at 100 rpm. rSEQ0694 was purified according to the method of Malik et al.,20 except that the purified protein was concentrated and the buffer exchanged into 18.2 MΩ cm−1 water using 10 kDa cut-off centrifugal concentrator units (Viva Science) The identity of rSEQ0694 was confirmed by peptide mass fingerprinting of trypsinized bands excised from Coomassie blue-stained SDS-polyacrylamide gels (see below).
In addition to rSEQ0694, the section of the seq0694 ORF encoding the predicted parvulin domain of SEQ0694 (amino acids 148–242, ESI,† Fig. S1; rSEQ0694parv) was amplified using primer pair TGCCATAG![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ACTACTCAGGTCACTACTCTAGACAATG (forward, NdeI site underlined) and TGCCATAG
ACTACTCAGGTCACTACTCTAGACAATG (forward, NdeI site underlined) and TGCCATAG![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) TTAGGCTTTTTTGGTTACCTTAACA (reverse, XhoI site underlined), cloned, expressed and the protein purified as described above, except that 5 kDa, 6 mL cut-off concentrator units (Viva Science) were used.
TTAGGCTTTTTTGGTTACCTTAACA (reverse, XhoI site underlined), cloned, expressed and the protein purified as described above, except that 5 kDa, 6 mL cut-off concentrator units (Viva Science) were used.
The concentration of both purified proteins was determined using the Bradford Assay.
Protease-coupled peptidylprolyl isomerase (PPIase) assay
The standard protease-coupled PPIase assay12,21 was employed using three peptide substrates having a consensus sequence Suc-Ala-X-Pro-Phe-pNA (Suc, succinyl; X = alanine, lysine or phenylalanine; pNa, paranitroaniline). Assays were performed by mixing 10 μL of purified rSEQ0694 (60 mg mL−1) or rSEQ0694parv (40 mg mL−1) (diluted in 20 mM HEPES, pH 7.4; 140 mM NaCl; 10% v/v glycerol), cyclophilin (positive control) or diluent alone (negative control) with 480 μL of buffer (20 mM HEPES, pH 7.4; 140 mM NaCl; 1 mM DTT) and allowing the mixture to equilibrate on ice for 5 min. 10 μL of ice-cold chymotrypsin (20 mg mL−1 in 0.001 M HCl; 0.002 M CaCl2) was pipetted into a cuvette in a spectrophotometer (Spectronic Unicam Heλios-α, Thermos Electron Corporation), zeroed at 390 nm. The 490 μL ice-cold assay mixture was quickly added to and mixed with the chymotrypsin, followed by 500 μL tetrapeptide substrate in ice-cold 20 mM HEPES, pH 7.4, 140 mM NaCl, 1 mM DTT, so as to give a final concentration of 37.5–75 μM peptide, and mixed quickly by pipetting. The final chymotrypsin concentration in the reaction mixture was 0.2 mg mL−1. The rate of the reaction (cis–trans isomerization) was measured by following colour formation (absorbance at 390 nm) resulting from pNA release from the trans form of the tetrapeptide substrate by chymotrypsin, for a maximum of 6 min. Spectrophotometric readings were recorded automatically via Vision 32 (Unicam Ltd) software.Reported kinetic data are given as the mean value of triplicate measurements for every condition. To ascertain if these data reflected true Michaelis–Menten kinetics, a Lineweaver–Burk plot was constructed and used to determine value of Km (calculated by reciprocalising the X intercept in the Line-weaver-Burk plot). The specificity constant (M s) was determined by dividing Kcat by Kmα.
Effect of chymotrypsin on rSEQ0694 and rSEQ0694parv recombinant proteins
To determine if chymotrypsin had any significant effect on the recombinant proteins, 10 μL purified rSEQ0694 (60 mg mL−1) or rSEQ0694parv (40 mg mL−1) was incubated with chymotrypsin (10 μL, 20 mg mL−1) in 880 μL assay buffer (20 mM HEPES, pH 7.4; 140 mM NaCl; 1 mM DTT) for 20 s, 2 min and 5 min at 0 °C. The reaction was stopped by the addition of 100 μL 10 mM PMSF and subsequent incubation for 5 min at 0 °C. Incubations containing rSEQ0694 or rSEQ0694parv incubated with PMSF-inactivated chymotrypsin, chymotrypsin with PMSF, chymotrypsin alone, recombinant proteins with PMSF and recombinant proteins alone served as controls. The reactions were analyzed by SDS-PAGE.Proteomic & bioinformatic methods
To compare protein expression in S. equi 4047 and ΔprtM138–213, the strains were grown to mid-log phase in Todd Hewitt broth, harvested by centrifugation and total cell proteins prepared as described previously.22,23 After removal of cells, supernatant proteins were precipitated with 100% (w/v) trichloroacetic acid, washed three times with ice-cold acetone and processed as for total-cell proteins. Two dimensional electrophoresis (2DE) and protein spot identification following trypsinolysis and mass spectrometry were performed as described previously.22,23 Only proteins identified with ≥2 peptide matches and Mascot total scores ≥50 were included.Protein sequence alignments were performed using Clustal Omega24 (http://www.ebi.ac.uk/Tools/msa/clustalo/).
Physiological tests
Survival of S. equi strains in saline solutions was tested by resuspending early stationary phase cells in 0, 0.9%, 14.7% or 29.4% NaCl w/v essentially as described by Reffuveille et al.25 Cell suspensions were sampled after 24 and 48 h by serial dilution to 10−3 in the same medium and then plated on Todd Hewitt agar plates for enumeration of surviving colony forming units. Antibiotic sensitivity testing was performed by the standard disc diffusion method using discs containing ampicillin (10 μg per disc), penicillin G (6 μg per disc), streptomycin (500 μg per disc), norflaxacin (5 μg per disc) and vancomycin (30 μg per disc). Zones of inhibition were measured after 48 h incubation.Results and discussion
rSEQ0694 encodes a functional PPIase
Our earlier study of S. equi SEQ069419 confirmed that this lipoprotein is needed for full virulence but did not directly address its function. Bioinformatic analyses indicated that SEQ0694 exhibits significant pairwise homologies to members of the PrsA/parvulin family of PPIases. In Firmicutes, these proteins typically contain a central parvulin domain, flanked by N- and C-terminal domains with likely additional chaperone functions or roles in substrate recruitment,13,26,27 although these flanking domains show limited sequence homology (ESI,† Fig. S1). Notably, the parvulin domains of streptococcal and lactococcal PrsA/PrtM family members have been noted to lack key conserved residues13,28 (see below) and both L. lactis PpmA and S. pneumoniae PpmA apparently lack PPIase activity,14,29 although it is notable that these proteins can complement some, but not all phenotypes, of a L. monocytogenes prsA2 mutant.30To confirm in vitro PPIase activity of SEQ0694, we produced full-length SEQ0694 as a recombinant protein, rSEQ0694 (ESI,† Fig. S2), for assay using a standard protease-coupled PPIase assay in which the rate of cis to trans isomerisation of a tetrapeptide substrate is measured through selective and colourigenic chymotrypsin hydrolysis of the trans isomer.12,21 In addition we produced the central parvulin domain of SEQ0694 as a recombinant protein, rSEQ0694parv. Both recombinant proteins were assayed against three tetrapeptide substrates varying in the amino acid preceding the critical proline residue. Whereas no activity could be detected using tetrapeptide substrates containing lysine–proline or alanine–proline bonds (data not shown), both rSEQ0694 and rSEQ0694parv were found to exhibit PPIase activity using Suc-Ala-Phe-Pro-Phe-pNA as substrate (Fig. 1). However, both recombinant proteins exhibited notably lower activities than the calf thymus cyclophilin used as a positive control.
Recombinant protein stability to chymotrypsin under the assay conditions was assessed. Significant cleavage of rSEQ0694parv by chymotrypsin was observed (ESI,† Fig. S3), whereas rSEQ0694 remained relatively stable for up to 5 min. This meant that although rSEQ0694parv showed an apparently faster rate of reaction compared with rSEQ0694 (Fig. 1), enzyme kinetics could only be determined for the latter (Fig. 2). A KcatKm−1 of 5.84 × 106/M s for rSEQ0694 was calculated from triplicate PPIase assays, suggesting that rSEQ0694 is a moderately active PPIase compared to other members of the parvulin family, with a similar activity to E. coli PpiC (Table 1). This activity was somewhat surprising as sequence alignments indicate that several amino acids considered functionally significant in parvulins31–35 are not conserved in rSEQ0694 (Fig. 3). However, a candidate Asp (D187) which might fulfil the role of the critical conserved Asp/Cys could be identified in rSEQ0694 (Fig. 3). Although a role of this Asp/Cys as a catalytic nucleophile is not yet fully resolved,36 its conservation in rSEQ0694 is likely to be significant. Moreover, the conserved residues in bacterial PrsA proteins identified by Jakob et al.26 are well conserved in SEQ0694 (ESI,† Fig. S1).
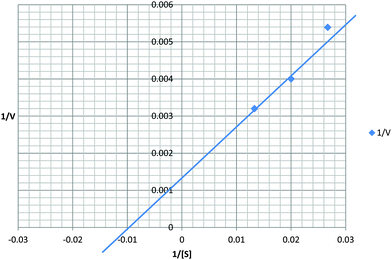 | ||
| Fig. 2 Kinetic analysis of rSEQ0694. The Kcat for rSEQ0694 was determined to be 583.75 s−1 and the Km 100 μM. Calculated Kcat/Km is 5.84 × 106 M−1 s−1. | ||
| Parvulin | Substratea | K cat/Km/M s | Ref. |
|---|---|---|---|
| a Data from protease-coupled assays where substrate is a colourigenic tetrapeptide Succ-Ala-X-Pro-Phe-pNA in which X is the amino acid indicated in the Table. b Estimation from Fig. 1 in Heikkinen et al.33. c Subsequently Weininger et al.49 have reported that PpiD is inactive as a PPIase using modified substrates in a protease-free assay. d Data from a protease-free assay using the tetrapeptide Succ-Ala-Ala-Pro-Phe-2,4-difluroanilide as substrate. | |||
| rSEQ0694 | Phe | 5.8 × 106 | This study |
| rSEQ0694 | Lys | Inactive | This study |
| rSEQ0694 | Ala | Inactive | This study |
| B. subtilis PrsA | Lys | 1.5 × 104 | 27 and 33 |
| B. subtilis PrsA | Ala | 0.6 × 104b | 33 |
| B. subtilis PrsA | Glu | 0.8 × 104b | 33 |
| S. aureus PrsA | Lys | 0.5 × 104b | 33 |
| S. aureus PrsA | Ala | 1.7 × 104b | 33 |
| S. aureus PrsA | Glu | 3.3 × 104 | 33 |
| E. coli PpiC (Par10) | Leu | 1.3 × 107 | 46 |
| E. coli PpiC (Par10) | Ser | 3.7 × 105 | 47 |
| E. coli PpiD (Par68) | Ala | 1.1 × 109c | 48 |
| E. coli PpiD (Par68) | Glu | 3.4 × 109c | 48 |
| E. coli PpiD (Par68) | Leu | 2.3 × 109c | 48 |
| Human Pin4 (Par14) | Arg | 3.9 × 103 | 46 |
| L. lactis PpmA | Ala | Inactived | 29 |
| S. pneumoniae PpmA | Ala, Phe, Gly, Val, Leu, Gln, Glu | Inactive | 14 |
 | ||
| Fig. 3 Sequence alignment of SEQ0694 with representative members of the parvulin family. Alignment produced with Clustal Omega. The signal peptide sequences of the Firmicutes proteins have been removed so that each sequence starts from the lipidated cysteine at the N-terminus of the mature protein. Key active site residues of the characterised parvulins are highlighted in yellow. For the longer bacterial sequences, the region aligning with the short E. coli PpiC sequence corresponds to the central parvulin domain. Realignment of the gapping in the central parvulin domain region in SEQ0694 could bring D187 into alignment with the critical D/C residue present in the characterised parvulins. The position of the region deleted in the S. equi mutant strain ΔprtM138–21319 is shown in bold. Abbreviations and UniProt accession codes for the sequences are: Bsu_PRSA (Q81U45); B. subtilis PrsA (P24327); Eco_PpiC, E. coli PpiC/Par10 (P0A9L5); Hsa_Par14, Homo sapiens Pin4 (Q9Y237); LMO_PrsA2, L. monocytogenes PrsA2 (Q71XE6); Sau_PrsA, Staphylococcus aureus PrsA (A6QI23); and SEQ0694, S. equi PrsA (C0M9L5). | ||
Proteomic analyses to identify putative SEQ0694 substrates
Having established that rSEQ0694 is a bona fide PPIase in vitro, we were interested to further explore the nature of its substrates. As SEQ0694 is a lipoprotein, we hypothesized that its substrates would be secreted proteins emerging from the Sec translocase, which need to fold rapidly en route to secretion. Misfolded proteins are typically turned over rapidly by extracytoplasmic proteases such as HtrA family members.37 Proteomic approaches have therefore been used to identify extracytoplasmic proteins for which folding is dependent on a lipoprotein PPIase.9,12,38,39 Thus we used proteomics to analyse differential protein expression in the proteomes of S. equi 4047 and an isogenic mutant, ΔprtM138–213, expressing a SEQ0694 N + C domains fusion protein lacking much of the central parvulin domain of SEQ069419 (Fig. 3). Note that as the seq0694 mutant strain was originally designated ΔprtM138–21319 for consistency we have retained this designation.Master 2D PAGE gels from 6 matched gel pairs (ESI,† Fig. S4) were analysed for differential protein expression and significant spots identified by mass spectrometry (Tables 2 and 3). Of the detectable total cell proteins, 12 differentially expressed proteins in 10 spots were identified (Table 2). The changes were primarily in cytoplasmic enzymes (e.g. enolase) which, because the proteins fold in the cytoplasm, may reflect general responses to stress due to lack of fully functional SEQ0694 (see below). Four of these proteins were also detected in the cell-free supernatant proteins (Table 3). In the cell-free supernatant proteomes, 13 proteins in 17 spots were found to be differentially expressed. As expected, the majority of these are proteins predicted to be either secreted or cell envelope localised and because of this could be plausible substrates for SEQ0694 (Table 3). As multiple proteins were found to be absent from the cell-free supernatant proteome of the mutant strain ΔprtM138–213, we hypothesise that SEQ0694 is likely to influence folding and secretion of multiple substrates rather than a specific substrate. Interestingly, two previously reported virulence factors of S. equi were notably absent from the cell-free supernatant proteome of the ΔprtM138–213 mutant: the truncated fibronectin-binding protein FNE40–42 and IgG endopeptidase IdeE2.43 FNE is noted to be misannotated as a pseudogene in the strain 4047 genome17 due to a misplaced start methionine. Our data therefore confirm the expression of FNE by strain 4047. SEQ0882, a putative DNase virulence factor homologous to S. pyogenes DNAse44 was also absent from the cell-free supernatant proteome of the ΔprtM138–213 mutant.
| Spot #a | Protein identifiedb | Scorec | Matched peptidesd | % covere | Predicted functionf | Signal peptide |
|---|---|---|---|---|---|---|
| a Spot marked in ESI, Fig. S4. WT spots are upregulated or only detected in the wild type strain 4047, Prt spots were only detected in the ΔprtM138–213 mutant proteome. b As annotated in Holden et al.17 c Mascot score. d Number of non-redundant peptides identified for each protein. e Percent amino acid coverage of entire protein. f As determined from Uniprot annotation, BlastP and PFAM analysis. | ||||||
| WT2201 | SEQ0898 | 1229 | 17 | 54 | Enolase (PF00113,PF03952) | No |
| WT2201 | SEQ1657 | 117 | 3 | 8 | Cyclophilin PPIase (PF00160) | Lipoprotein |
| WT2201 | SEQ0210 | 91 | 2 | 26 | 10 kDa chaperonin GroES (PF00166) | No |
| WT3201 | SEQ1366 | 206 | 5 | 14 | Xaa-His dipeptidase (PF01546) | No |
| WT3601 | SEQ0434 | 158 | 3 | 14 | Mannose-6-phosphate isomerase (PF01238) | No |
| WT4001 | SEQ0408 | 318 | 6 | 68 | 30S ribosomal protein S6 (PF01250) | No |
| WT4204 | SEQ1025 | 188 | 3 | 25 | Asp23 domain protein (PF03780) | No |
| WT5302 | SEQ1354 | 184 | 3 | 23 | Purine nucleoside phosphorylase (PF01048) | No |
| WT5504 | SEQ0046 | 293 | 6 | 30 | Alcohol dehydrogenase (PF00107,PF08240) | No |
| WT6201 | SEQ1418 | 163 | 4 | 26 | Putative dTDP-4-keto-6-deoxyglucose-3,5-epimerase (PF00908) | No |
| WT6501 | SEQ1011 | 408 | 6 | 22 | 6-Phosphofructokinase (PF00365) | No |
| Prt9401 | SEQ1642 | 103 | 3 | 23 | Ribosome-recycling factor (PF01765) | No |
| Spot #a | Protein identifiedb | Scorec | Matched peptidesd | % covere | Predicted functionf | Signal peptide |
|---|---|---|---|---|---|---|
| a Spot marked in ESI, Fig. S4. b As annotated in Holden et al.17 c Mascot score. d Number of non-redundant peptides identified for each protein. e Percent amino acid coverage of entire protein. f As determined from Uniprot annotation, BlastP and PFAM analysis. | ||||||
| WT1002 | SEQ0210 | 174 | 4 | 57 | 10 kDa chaperonin GroES (PF00166) | No |
| WT1401 | SEQ1821 | 334 | 4 | 38 | PepSY (PF03413) protease inhibitor domain lipoprotein | Lipoprotein |
| WT1402 | SEQ1177 | 198 | 5 | 22 | Domain of Unknown Function (PF06207/DUF1002) | Present |
| WT2101 | SEQ1800 | 119 | 2 | 30 | Unknown function, no conserved domains. Restricted distribution within streptococci; spot position shifted compared to mutant Prt1103 | Present |
| WT2202 | SEQ1025 | 146 | 3 | 20 | Asp23 domain protein (PF03780) | No |
| WT2202 | FNE | 72 | 2 | 6 | Truncated fibronectin binding protein (PF08341) | Present |
| WT2401 | SEQ1177 | 526 | 8 | 36 | Domain of unknown function (PF06207/DUF1002) | Present |
| WT3301 | SEQ1657 | 409 | 6 | 35 | Cyclophilin type PPIase (PF00160) | Lipoprotein |
| WT7301 | SEQ0882 | 519 | 7 | 39 | DNA/RNA non-specific endonuclease | Present |
| WT7301 | FNE | 361 | 6 | 26 | Truncated fibronectin binding protein (PF08341) | Present |
| WT8401 | SEQ0938 | 331 | 6 | 19 | IdeE2 Mac family protein (PF09028) | Present |
| WT8501 | SEQ0938 | 204 | 4 | 11 | IdeE2 Mac family protein (PF09028) | Present |
| WT9202 | FNE | 221 | 5 | 13 | Truncated fibronectin binding protein (PF08341) | Present |
| WT9202 | SEQ0882 | 93 | 3 | 14 | DNA/RNA non-specific endonuclease | Present |
| WT9403 | SEQ0520 | 556 | 10 | 41 | Hydrolase/esterase (PF07859) | Present |
| Prt0301 | SEQ1171 | 165 | 4 | 25 | Sortase A (PF04203) | Signal anchor |
| Prt1103 | SEQ1800 | 133 | 3 | 36 | Unknown function, no conserved domains. Restricted distribution within streptococci; position shifted compared to mutant WT2101. | Present |
| Prt1202 | SEQ1919 | 221 | 3 | 6 | OppA olipopeptide binding lipoprotein (PF00496) | Lipoprotein |
| Prt2101 | SEQ0408 | 139 | 2 | 26 | 30S ribosomal protein S6 (PF01250) | No |
| Prt2301 | SEQ1919 | 86 | 3 | 6 | OppA olipopeptide binding lipoprotein (PF00496) | Lipoprotein |
Cumulatively, these proteomic changes likely explain, at least in part, the attenuation of the ΔprtM138–213 mutant.19 However, as the ΔprtM138–213 mutant should still express a N + C domain fusion protein (lacking most of the parvulin domain), it may be that more dramatic proteome changes would be evident in an seq0694 null mutant, since a L. monocytogenes PrsA N + C construct partly complemented the proteome defect of a full prsA deletion12 and an N + C fusion construct of B. subtilis PrsA partially restored secretion of an AmyQ reporter protein (although it did not restore viability to PrsA-depleted cells27). In B. subtilis, the N and C domain is notable in driving dimerization of PrsA and, although lacking primary sequence homology, has structural similarity to other ‘foldases’ such as trigger factor.26 Without structural characterisation of the N + C fusion encoded by the S. equi ΔprtM138–213 mutant we cannot speculate whether this construct is likely to have a native-like conformation and functionality. However, it is notable that the sequence deletion removes not only the majority of the parvulin domain of SEQ0694 but also a conserved lysine of the Firmicutes PrsA protein N-domains. It is worth reemphasising that the partial deletion in the S. equi ΔprtM138–213 mutant is sufficient to cause significant attenuation of virulence in the natural host.19
It was interesting to note that SEQ1657, a cyclophilin PPIase lipoprotein (orthologous to S. pneumoniae SlrA14 and L. lactis PpiA29) was up-regulated in both the total cell and secreted proteins of the parental strain. Likewise, it was observed that the SEQ1171 sortase is up-regulated in the mutant strain, perhaps suggesting a need to remodel protein localisation within the mutant cell envelope.
As the proteomic data suggested a range of protein functions are likely to be perturbed in strain ΔprtM138–213, including stress responses, we performed several physiological tests. Although the mutant strain grows normally in nutrient rich broth, we observed pleiotropic changes including increased sensitivity to salt stress (ESI,† Table S1) and increased sensitivity to various antibiotics with diverse cellular targets (ESI,† Fig. S5). Increased sensitivity to salt stress has previously been observed in a prsA mutant of E. faecalis11 and a prsaA2 mutant of L. monocytogenes.30 A range of findings have been observed regarding antibiotic susceptibilities of other prsA mutants. Similar to our findings, a prsaA2 mutant of L. monocytogenes displayed increased sensitivity to bacitracin, penicillin and vancomycin but not gentamicin30 and a mutant in Staphylococcus aureus prsA showed increased sensitivity to vancomycin.45 However, a prsA mutant of E. faecalis was unaffected in its sensitivity to ampicillin and norflaxin,11 in contrast to our findings. Cumulatively, our data suggest a general perturbation in cell envelope function in the ΔprtM138–213 mutant, which likely reflects multiple changes in the extracytoplasmic proteome of the mutant (consistent with our proteomic data). This is conclusion is consistent with the pleiotropic effects of PrsA mutation in other Firmicutes.11,28,30,45
Conclusions
The data presented here confirm that rSEQ0694 is a moderately active PPIase, despite lacking conservation of several amino acids previously considered to be significant to the activity of other parvulin PPIases. This observation thus focusses attention on the conserved Asp/Cys identified as likely critical for catalysis. Furthermore, proteomic experiments confirm that loss of the lipoprotein PPIase activity in strain ΔprtM138–213 affects multiple cell envelope proteins, including virulence factors, and is likely to generate diverse phenotypic effects. As strain ΔprtM138–213 is attenuated,19 these findings further suggest that streptococcal PPIases, and PPIases generally,7 are interesting targets for novel therapeutic strategies. By analogy with other bacterial PPIases, it would also be of interest to determine whether the N- and C-terminal domains of SEQ0694 possess additional chaperone activities that contribute to post-translocational protein folding.Abbreviations
| pNa | Paranitroaniline |
| PPIase | Peptidyl-prolyl isomerase |
| rSEQ0694 | Recombinant N-terminally His-tagged mature SEQ0694 |
| rSEQ0694parv | Recombinant N-terminally His-tagged parvulin domain of SEQ0694 |
References
- R. E. Dalbey and A. Kuhn, FEMS Microbiol. Rev., 2012, 36, 1023–1045 CrossRef CAS PubMed.
- M. Desvaux, M. Hébraud, R. Talon and I. R. Henderson, Trends Microbiol., 2009, 17, 139–145 CrossRef CAS PubMed.
- R. Freudl, Res. Microbiol., 2013, 164, 664–674 CrossRef CAS PubMed.
- C. Goemans, K. Denoncin and J. F. Collet, Biochim. Biophys. Acta, 2014, 1843, 1517–1528 CrossRef CAS PubMed.
- M. Sarvas, C. R. Harwood, S. Bron and J. M. van Dijl, Biochim. Biophys. Acta, 2004, 1694, 311–327 CAS.
- J. Fanghänel and G. Fischer, Front. Biosci., 2004, 9, 3453–3478 CrossRef PubMed.
- C. M. Ünala and M. Steinert, Microbiol. Mol. Biol. Rev., 2014, 78, 544–571 CrossRef PubMed.
- M. I. Hutchings, T. Palmer, D. J. Harrington and I. C. Sutcliffe, Trends Microbiol., 2009, 17, 13–21 CrossRef CAS PubMed.
- H. L. Hyyryläinen, B. C. Marciniak, K. Dahncke, M. Pietiäinen, P. Courtin, M. Vitikainen, R. Seppala, A. Otto, D. Becher, M. P. Chapot-Chartier, O. P. Kuipers and V. P. Kontinen, Mol. Microbiol., 2010, 77, 108–127 CrossRef PubMed.
- R. C. Williams, M. L. Rees, M. F. Jacobs, Z. Pragai, J. E. Thwaite, L. W. Baillie, P. T. Emmerson and C. R. Harwood, J. Biol. Chem., 2003, 278, 18056–18062 CrossRef CAS PubMed.
- F. Reffuveille, N. Connil, M. Sanguinetti, B. Posteraro, S. Chevalier, Y. Auffray and A. Rince, Infect. Immun., 2012, 80, 1728–1735 CrossRef CAS PubMed.
- F. Alonzo, 3rd, B. Xayarath, J. C. Whisstock and N. E. Freitag, Mol. Microbiol., 2011, 80, 1530–1548 CrossRef PubMed.
- L. A. Cahoon and N. E. Freitag, Front. Cell. Infect. Microbiol., 2014, 4, 13 Search PubMed.
- P. W. Hermans, P. V. Adrian, C. Albert, S. Estevao, T. Hoogenboezem, I. H. Luijendijk, T. Kamphausen and S. Hammerschmidt, J. Biol. Chem., 2006, 281, 968–976 CrossRef CAS PubMed.
- Y. Ma, A. E. Bryant, D. B. Salmi, S. M. Hayes-Schroer, E. McIndoo, M. J. Aldape and D. L. Stevens, J. Bacteriol., 2006, 188, 7626–7634 CrossRef CAS PubMed.
- J. U. Rahfeld, K. P. Rücknagel, B. Schelbert, B. Ludwig, J. Hacker, K. Mann and G. Fischer, FEBS Lett., 1994, 352, 180–184 CrossRef CAS.
- M. T. Holden, Z. Heather, R. Paillot, K. F. Steward, K. Webb, F. Ainslie, T. Jourdan, N. C. Bason, N. E. Holroyd, K. Mungall, M. A. Quail, M. Sanders, M. Simmonds, D. Willey, K. Brooks, D. M. Aanensen, B. G. Spratt, K. A. Jolley, M. C. Maiden, M. Kehoe, N. Chanter, S. D. Bentley, C. Robinson, D. J. Maskell, J. Parkhill and A. S. Waller, PLoS Pathog., 2009, 5, e1000346 Search PubMed.
- A. S. Waller, R. Paillot and J. F. Timoney, J. Med. Microbiol., 2011, 60, 1231–1240 CrossRef CAS PubMed.
- A. Hamilton, C. Robinson, I. C. Sutcliffe, J. Slater, D. J. Maskell, N. Davis-Poynter, K. Smith, A. Waller and D. J. Harrington, Infect. Immun., 2006, 74, 6907–6919 CrossRef CAS PubMed.
- V. Malik, M. Zhang, L. G. Dover, J. S. Northen, A. Flinn, J. J. Perry and G. W. Black, Mol. BioSyst., 2013, 9, 2816–2822 RSC.
- J. Hani, B. Schelbert, A. Bernhardt, H. Domdey, G. Fischer, K. Wiebauer and J. U. Rahfeld, J. Biol. Chem., 1999, 274, 108–116 CrossRef CAS PubMed.
- Q. Yang, M. Zhang, D. J. Harrington, G. W. Black and I. C. Sutcliffe, Int. J. Med. Microbiol., 2010, 300, 331–337 CrossRef CAS PubMed.
- M. Zhang, F. M. McDonald, S. S. Sturrock, S. J. Charnock, I. Humphery-Smith and G. W. Black, Proteomics, 2007, 7, 1379–1390 CrossRef CAS PubMed.
- F. Sievers, A. Wilm, D. Dineen, T. J. Gibson, K. Karplus, W. Li, R. Lopez, H. McWilliam, M. Remmert, J. Soding, J. D. Thompson and D. G. Higgins, Mol. Syst. Biol., 2011, 7, 539 CrossRef PubMed.
- F. Reffuveille, P. Serror, S. Chevalier, A. Budin-Verneuil, R. Ladjouzi, B. Bernay, Y. Auffray and A. Rince, Microbiology, 2012, 158, 816–825 CrossRef CAS PubMed.
- R. P. Jakob, J. R. Koch, B. M. Burmann, P. A. Schmidpeter, M. Hunkeler, S. Hiller, F. X. Schmid and T. Maier, J. Biol. Chem., 2015, 290, 3278–3292 CrossRef CAS PubMed.
- M. Vitikainen, I. Lappalainen, R. Seppala, H. Antelmann, H. Boer, S. Taira, H. Savilahti, M. Hecker, M. Vihinen, M. Sarvas and V. P. Kontinen, J. Biol. Chem., 2004, 279, 19302–19314 CrossRef CAS PubMed.
- S. Drouault, J. Anba, S. Bonneau, A. Bolotin, S. D. Ehrlich and P. Renault, Appl. Environ. Microbiol., 2002, 68, 3932–3942 CrossRef CAS.
- N. Trémillon, E. Morello, D. Llull, R. Mazmouz, J. J. Gratadoux, A. Guillot, M. P. Chapot-Chartier, L. Monlezun, V. Solé, H. Ginisty and I. Poquet, PLoS One, 2012, 7, e33516 Search PubMed.
- L. A. Cahoon and N. E. Freitag, Infect. Immun., 2015, 83, 4028–4041 CrossRef CAS PubMed.
- M. L. Bailey, B. H. Shilton, C. J. Brandl and D. W. Litchfield, Biochemistry, 2008, 47, 11481–11489 CrossRef CAS PubMed.
- C. D. Behrsin, M. L. Bailey, K. S. Bateman, K. S. Hamilton, L. M. Wahl, C. J. Brandl, B. H. Shilton and D. W. Litchfield, J. Mol. Biol., 2007, 365, 1143–1162 CrossRef CAS PubMed.
- O. Heikkinen, R. Seppala, H. Tossavainen, S. Heikkinen, H. Koskela, P. Permi and I. Kilpeläinen, BMC Struct. Biol., 2009, 9, 17 CrossRef PubMed.
- J. W. Mueller, N. M. Link, A. Matena, L. Hoppstock, A. Rüppel, P. Bayer and W. Blankenfeldt, J. Am. Chem. Soc., 2011, 133, 20096–20099 CrossRef CAS PubMed.
- H. Tossavainen, P. Permi, S. L. Purhonen, M. Sarvas, I. Kilpeläinen and R. Seppala, FEBS Lett., 2006, 580, 1822–1826 CrossRef CAS PubMed.
- A. Barman and D. Hamelberg, Biochemistry, 2014, 53, 3839–3850 CrossRef CAS PubMed.
- G. Hansen and R. Hilgenfeld, Cell. Mol. Life Sci., 2013, 70, 761–775 CrossRef CAS PubMed.
- F. Alonzo, 3rd and N. E. Freitag, Infect. Immun., 2010, 78, 4944–4957 CrossRef PubMed.
- L. Guo, T. Wu, W. Hu, X. He, S. Sharma, P. Webster, J. K. Gimzewski, X. Zhou, R. Lux and W. Shi, Mol. Oral Microbiol., 2013, 28, 154–165 CrossRef CAS PubMed.
- A. Lidén, A. Karlstrom, J. Lannergård, S. Kalamajski, B. Guss, K. Rubin and C. Rydén, Biochem. Biophys. Res. Commun., 2006, 340, 604–610 CrossRef PubMed.
- H. Lindmark, M. Nilsson and B. Guss, Infect. Immun., 2001, 69, 3159–3163 CrossRef CAS PubMed.
- M. Tiouajni, D. Durand, K. Blondeau, M. Graille, A. Urvoas, M. Valerio-Lepiniec, A. Guellouz, M. Aumont-Nicaise, P. Minard and H. van Tilbeurgh, FEBS J., 2014, 281, 5513–5531 CrossRef CAS PubMed.
- G. Hulting, M. Flock, L. Frykberg, J. Lannergård, J. I. Flock and B. Guss, FEMS Microbiol. Lett., 2009, 298, 44–50 CrossRef CAS PubMed.
- J. E. Korczynska, J. P. Turkenburg and E. J. Taylor, Nucleic Acids Res., 2012, 40, 928–938 CrossRef CAS PubMed.
- A. Jousselin, A. Renzoni, D. O. Andrey, A. Monod, D. P. Lew and W. L. Kelley, Antimicrob. Agents Chemother., 2012, 56, 3629–3640 CrossRef CAS PubMed.
- T. Uchida, F. Fujimori, T. Tradler, G. Fischer and J. U. Rahfeld, FEBS Lett., 1999, 446, 278–282 CrossRef CAS.
- R. Golbik, C. Yu, E. Weyher-Stingl, R. Huber, L. Moroder, N. Budisa and C. Schiene-Fischer, Biochemistry, 2005, 44, 16026–16034 CrossRef CAS PubMed.
- C. Dartigalongue and S. Raina, EMBO J., 1998, 17, 3968–3980 CrossRef CAS PubMed.
- U. Weininger, R. P. Jakob, M. Kovermann, J. Balbach and F. X. Schmid, Protein Sci., 2010, 19, 6–18 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5mb00543d |
| This journal is © The Royal Society of Chemistry 2015 |

