Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Exploration of the HIF-1α/p300 interface using peptide and Adhiron phage display technologies†
Hannah F.
Kyle
abc,
Kate F.
Wickson
d,
Jonathan
Stott
d,
George M.
Burslem
ab,
Alexander L.
Breeze
ac,
Christian
Tiede
c,
Darren C.
Tomlinson
ac,
Stuart L.
Warriner
ab,
Adam
Nelson
ab,
Andrew J.
Wilson
ab and
Thomas A.
Edwards
*ac
aAstbury Centre for Structural Molecular Biology, University of Leeds, Woodhouse Lane, Leeds, LS2 9JT, UK. E-mail: t.a.edwards@leeds.ac.uk
bSchool of Chemistry, University of Leeds, Woodhouse Lane, Leeds, LS2 9JT, UK
cSchool of Molecular and Cellular Biology, Faculty of Biological Sciences, University of Leeds, Woodhouse Lane, Leeds, LS2 9JT, UK
dAstraZeneca Discovery Sciences, Structure & Biophysics, AstraZeneca Darwin Building, Cambridge, Science Park, Milton Road, Cambridge, CB4 0WG, UK
First published on 10th June 2015
Abstract
The HIF-1α/p300 protein–protein interaction plays a key role in tumor metabolism and thus represents a high value target for anticancer drug-development. Although several studies have identified inhibitor candidates using rationale design, more detailed understanding of the interaction and binding interface is necessary to inform development of superior inhibitors. In this work, we report a detailed biophysical analysis of the native interaction with both peptide and Adhiron phage display experiments to identify novel binding motifs and binding regions of the surface of p300 to inform future inhibitor design.
Introduction
An emerging opportunity for anticancer therapy is to identify ligands which affect metabolic and cellular processes enabling the survival and growth of tumours.1 Hypoxia is a hallmark of many solid tumours; the ability to adapt to hypoxic conditions is crucial to their growth and survival2,3 and occurs via a coordinated homeostatic response dominated by the transcription factor hypoxia-inducible factor 1 (HIF-1). Overexpression of HIF-1 in cancer patients has been associated with resistance to some therapies, increased risk of invasion and metastasis, and poor prognosis.4 Inhibition of the HIF-1 pathway may therefore have anticancer therapeutic utility.5HIF-1 is a heterodimer made up of two subunits: HIF-1α and HIF-1β (also known as the aryl hydrocarbon receptor nuclear translocator, ARNT). With oxygen at normal physiological concentrations, HIF-1α is rapidly degraded mostly via the von Hippel–Lindau pathway (an oxygen-dependent process).6 Under hypoxic conditions, however, the stability and thus transcriptional activity of HIF-1α increases. HIF-1α can then translocate to the nucleus, where it forms heterodimers with HIF-1β and recruits transcriptional coactivator proteins; one of which is p300.7,8 Transcriptional activation by the HIF-1α/p300 complex leads to the hypoxic response cascade, resulting in expression of multiple genes (e.g., VEGF) that mediate angiogenesis, various metabolic processes and cell proliferation and survival. In rapidly developing solid tumours, hypoxic conditions are generated; cancerous cells exploit this HIF-1 activated pathway to develop new vasculature which initiates the resupply of the tumour with oxygen.5
There has been considerable focus on inhibiting the hypoxic response, but this has been challenging because HIF’s function as a transcription factor is exerted predominantly through protein–protein interactions (PPIs), such as the HIF-1α/p300 interaction. Designing small molecule inhibitors of PPIs is challenging because the interfaces are generally large, and can lack well-defined pockets in comparison to enzyme active sites.9–12 Using mutational studies, Clackson and Wells demonstrated that a small subset of residues – termed “hot-spots” – tend to contribute much of the free energy of binding to many PPIs.13–16 By focusing only on the hot-spots and not the full interface, the challenge of designing small molecule PPI inhibitors becomes less daunting. Consequently identification and characterization of PPI hot-spots represents a precursor to effective small molecule design.17
Several approaches to inhibit the HIF-1 hypoxic response pathway have been described, which can broadly be classified in to five categories:18 (1) HIF-1α mRNA expression,19 (2) HIF-1α protein translation,20–27 (3) HIF-1α protein degradation,28 (4) HIF-1α DNA binding29 and (5) HIF-1α transcriptional activity; which fall in to two main groups, disruptors of HIF-1 dimerization30,31 and disruption of binding of co-activator proteins.30,32–37 One approach for inhibition of the HIF-1 pathway, which falls into the category of transcriptional activity inhibition, is to target the interaction between HIF-1α and the co-activator protein p300.5 We and others have had some success in targeting this interaction using protein domain36 and α-helix mimetics35,37 as well as natural products,33 designed protein ligands (optimised from natural products)38 and small molecule arylsulfonamides.39,40
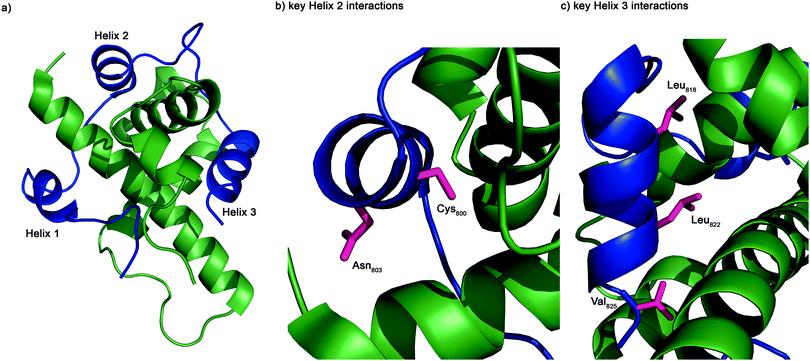
The structure of the complex of the CH1 domain of p300 bound to the C-TAD of HIF-1α was solved using multidimensional NMR methods (PDB IDs: 1L8C and 1L3E).7,8 The NMR structure (Fig. 1) suggests that the interaction is complex: when bound, HIF-1α C-TAD consists of three distinct helical regions and wraps around the p300 CH1 domain.8 A conventional small molecule41 could not cover the whole interacting interface; consequently a detailed understanding of the interface is necessary to inform inhibitor design. Mutational studies have proposed key binding residues of HIF-1α.42 Helices 2 and 3 of the HIF-1α C-TAD have been shown to be required for binding; helix 2 contains two key residues: Cys800 and Asn803,34,43,44 and helix 3 contains 3 key binding residues, Leu818, Leu822 and Val825.42 Additionally, other helix 3 residues have been suggested to be important in the interaction (Asp823 and Gln824).36,37 Beyond these mutational analyses, a number of studies provide contradictory conclusions as to the relative importance of various regions and residues on the HIF-1α C-TAD.36,37,42,45 The binding potency of sequences derived from HIF-1α C-TAD (HIF-1α776–826, HIF-1α786–826 HIF-1α788–8222 HIF-1α776–813) with p300 CH1 was compared using fluorescence polarization.45 From this experiment it was concluded that the C-terminus of HIF-1α C-TAD is important for binding, in agreement with the mutagenesis studies.7,42 However these data do not narrow down the interaction area to one of the individual helices and the interaction area is still a relatively large surface area; therefore a single cluster of residues (hot-spots) that could be targeted for disruption has yet to be identified and small molecule design is still challenging.
In this study we exploited two approaches to probe the HIF-1α binding surface on p300, to refine our understanding of the most productive regions to target using designed small-molecules: first, by analysis of the binding of shorter HIF-1α peptide fragments; and second, by phage display experiments. Binding analysis of fragments of the native peptide allow identification of the highest affinity region of the HIF-1α peptide, whereas phage display permits the unbiased exploration of the p300 CH1 protein surface to discover high affinity binders. Two phage display technologies were used:46 a peptide phage library (NEB47–49) and an Adhiron50 (commercially known as Affimers51,52) phage library. The location and binding mode of phage display derived ligands provides new information on suitable chemotypes for orthosteric small-molecule inhibitor development.
Results and discussion
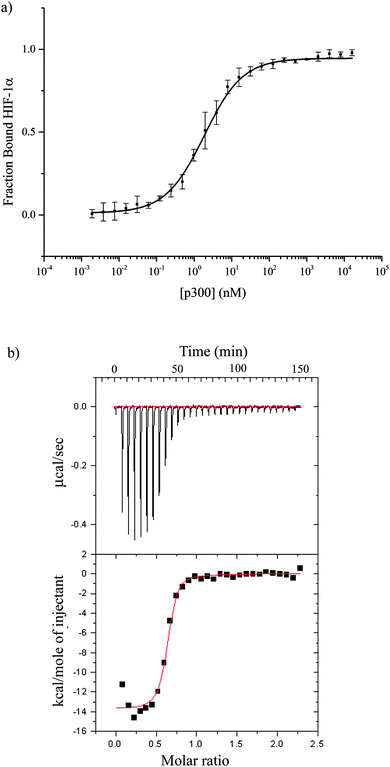
The CH1 domain of p300 (amino acids 330–420) was cloned and expressed as a GST fusion protein; p300 was subsequently cleaved from GST. The C-TAD of HIF-1α786–826 was purchased from ProteoGenix. The binding of p300 to HIF-1α786–826 was measured by fluorescence anisotropy (using FITC-HIF-1α786–826) and orthogonally by isothermal titration calorimetry. The Kd was determined to be 16.11 ± 0.06 nM by fluorescence anisotropy and 45 ± 10 nM by ITC (Fig. 2 and Table S1, ESI†).To investigate whether the binding interface could be restricted, smaller sections of the HIF-1α C-TAD were selected. The truncated peptides tested are summarised in Fig. 3. Whilst it is likely there will be contributions to binding along the length of the 41 amino acid HIF-1α peptide, each of these peptides was assessed for either direct binding to p300 CH1, or competitive inhibition of the HIF1α/p300 complex, to ascertain which regions are most important for binding.
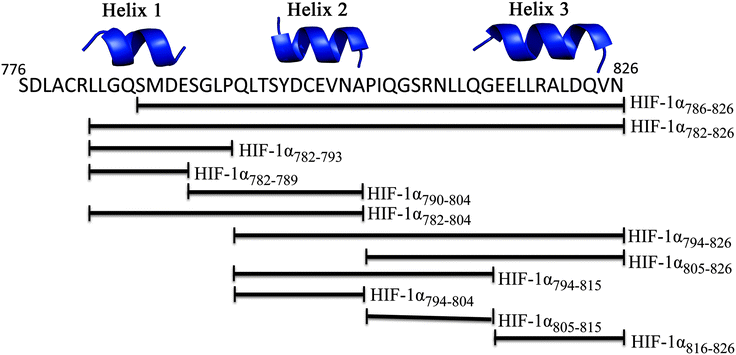 | ||
| Fig. 3 Schematic of peptide fragments to be investigated to highlight key binding regions of HIF-1α. | ||
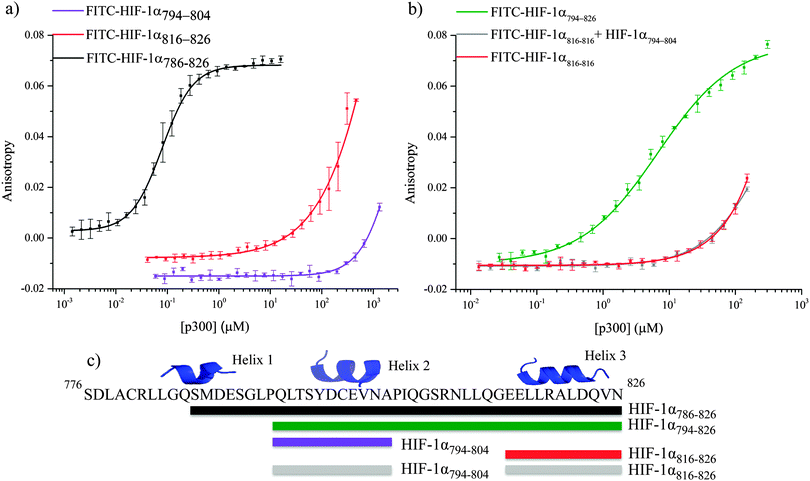
α-helices have been highlighted as major binding hot-spots of PPIs,53–55 so initial assessment centered on identification of the highest affinity helix. The affinity of helices 2 and 3 was investigated by fluorescence anisotropy using FITC-HIF-1α794–804 and FITC-HIF-1α816–826 (Fig. 4a). Both peptides showed weak binding to p300 (compared to 16 ± 0.06 nM for HIF-1α786–826): FITC-HIF-1α816–826Kd approximately 200 μM and FITC-HIF-1α794–804Kd > 1 mM. Notably, FITC-HIF-1α816–826 had a higher affinity than FITC-HIF-1α794–804, indicating that helix 3 has a higher affinity than helix 2; therefore helix 3 may be important for high affinity binding of the native complex.
To examine further the binding energy contributions of the individual helices to the interaction and the effect of linking, peptide FITC-HIF-1α794–826, comprising helices 2 and 3 with the intervening linker region, was also tested in the fluorescence anisotropy assay. A Kd of 6.74 ± 0.54 μM was measured, showing that linking helices 2 and 3 together results in an increase in affinity for p300 compared with either individual helix. Linking can increase binding affinity either through enhanced local concentration of each binding entity (avidity or chelate effect), or through co-operative interaction between them mediated through allosteric conformational change in the binding site. To distinguish between chelate and allosteric co-operativity, the fluorescence anisotropy assay was used to test the binding of FITC-HIF-1α816–826 in the presence of HIF-1α794–804 (unlabeled). This demonstrated that the presence of HIF-1α794–804 had no effect on the affinity of FITC-HIF-1α816–826, suggesting either an avidity (chelate) effect or a direct contribution from the linker, rather than allosteric co-operativity between helix 3 and helix 2 (Fig. 4b). The avidity effect is modest, however, as the affinity measured for FITC-HIF-1α794–826 implies less than additive contributions from the individual binding energies of helices 2 and 3. This is consistent with the linker sequence having a considerable degree of flexibility when not bound to p300.
The reduced affinity of the FITC-HIF-1α794–826 (Kd = 6.74 ± 0.54 μM), compared to FITC-HIF-1α786–826 (Kd = 16.11 ± 0.06 nM), indicates that the additional N-terminal residues have some role in binding, although this may be a consequence of enhanced stability of helix 2 in addition to any contribution from interaction of the N-terminal residues themselves. The importance of the N-terminal residues, which include helix 1, was explored by measuring the difference in inhibitor capabilities of fragments of HIF-1α to disrupt the HIF-1α786–826/p300 interaction.
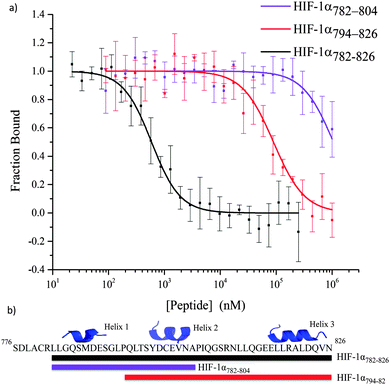
All of the peptides illustrated in Fig. 3 were tested in a fluorescence anisotropy competition assay (Table S2, ESI†); however only peptides that contained more than one helix were able to disrupt the HIF-1α786–826/p300 interaction (Fig. 5). HIF-1α782–826 inhibited the interaction with an IC50 = 0.59 ± 0.05 μM; this is superior to HIF-1α794–826 (IC50 = 89.26 ± 28 μM) and HIF-1α782–804 (IC50 > 1 mM). HIF-1α794–826 showed a greater capability to disrupt the complex than HIF-1α782–804 demonstrating the importance of the C-terminus of HIF-1α and reinforcing the importance of the helix 3 region.
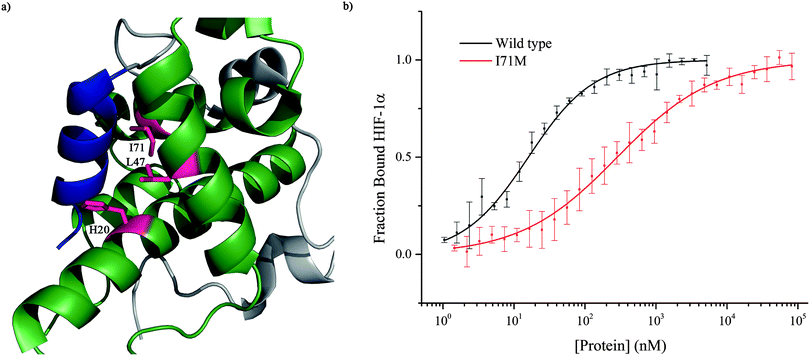
To further investigate the importance of helix 3, mutants of p300 (H20A, L47M, I71M, Fig. 6a) in the HIF-1α helix 3 binding pocket (as shown in the NMR structure7,8Fig. 1) were generated and their fold and stability confirmed by Circular Dichroism (CD) (Fig. S1, ESI†). H20A was selected, as the NMR structure shows this residue makes a contact to HIF-1α, and mutation to alanine would remove this contact. Both L47M and I71M were selected as we hypothesised they would introduce steric clashes. Each of the three mutants caused a reduction in the binding affinity of to FITC-HIF-1α786–826, with the greatest effect observed for I71M, which caused a 18-fold reduction in binding affinity (Table 1) confirming that helix 3 has a critical role in driving p300 binding.
| p300 | K d (nM) |
|---|---|
| Wild type | 16.11 ± 0.06 |
| H20A | 25 ± 0.35 |
| L47M | 29.55 ± 5.58 |
| I71M | 285 ± 5.06 |
Taken together, these data demonstrated that helix 3 appears to be the most important of the helical regions of HIF-1α in terms of affinity. However, the absence of a dominant high affinity fragment of the HIF-1α C-TAD sequence encouraged us to perform phage display studies to identify short but high potency peptide sequences that may serve as better starting points for small-molecule elaboration.
Phage display technologies can be used to present many different forms of binder; in this study 7mer and 12mer peptides libraries (NEB47–49) and Adhiron libraries50 were tested. For the peptide libraries, both the 7mer and 12mer libraries were chosen. The 7mer library should have full coverage (all amino acids represented in all positions47–49), however due to the length of the 7mer the generation of high affinity binding peptides was anticipated to be less likely. The 12mer library does not have full coverage, but the longer peptide may yield higher affinity peptide binders. As well as the two phage libraries, two different buffer conditions were used in the wash step: a high sodium chloride (1.5 M) and low sodium chloride (0.5 M) buffer (TBS + 0.1% tween) were used to eliminate any nonspecific electrostatic contributions. Elution using native HIF-1α786–826 (500 μM) was performed to ensure elution of specific binders. The experiment was also performed with a different protein (eIF4E); this parallel experiment acted as a selectivity control.
p300 was selectively N-terminally labelled with biotin using a biotin-depsipeptide and sortase A56 to enable immobilisation on to streptavidin plates. Three panning rounds were completed. Each round of panning led to enrichment of binders in the phage pool (Fig. S2, ESI† for the ELISA for each condition). The eluents for the unpanned libraries, round 1 and round 3 were sent for next generation sequencing using the Illumina platform.57 The top 5 clones from each condition and their frequency in the pool are summarized in the ESI† (Table S3). Three peptides were selected for synthesis: VHWDFRQWWQPS, (phage display derived peptide 1; PDDP1) SGVYKVAYDWQH (PDDP2) and ATNLFKS (PDDP3). The next generation sequencing showed an increase in enrichment of these peptides through the panning rounds. Initially the FITC-labelled peptides were tested for binding to p300 using a fluorescence anisotropy assay. The highest affinity peptide was PDDP1 with an affinity of 20.67 ± 3.17 μM. PDDP1 is predicted be 35% helical in isolation (as assessed by Agadir58), indicating that a helical bound conformation is plausible. Although this is a relatively low affinity for phage display-derived peptides, where low nM and pM binders are often generated,59,60 the phage display-derived peptide had a higher affinity for p300 than any of the HIF-1α C-TAD peptide fragments discussed above. PDDP1 showed selectivity for p300 over eIF4e (Fig. S3, ESI†).
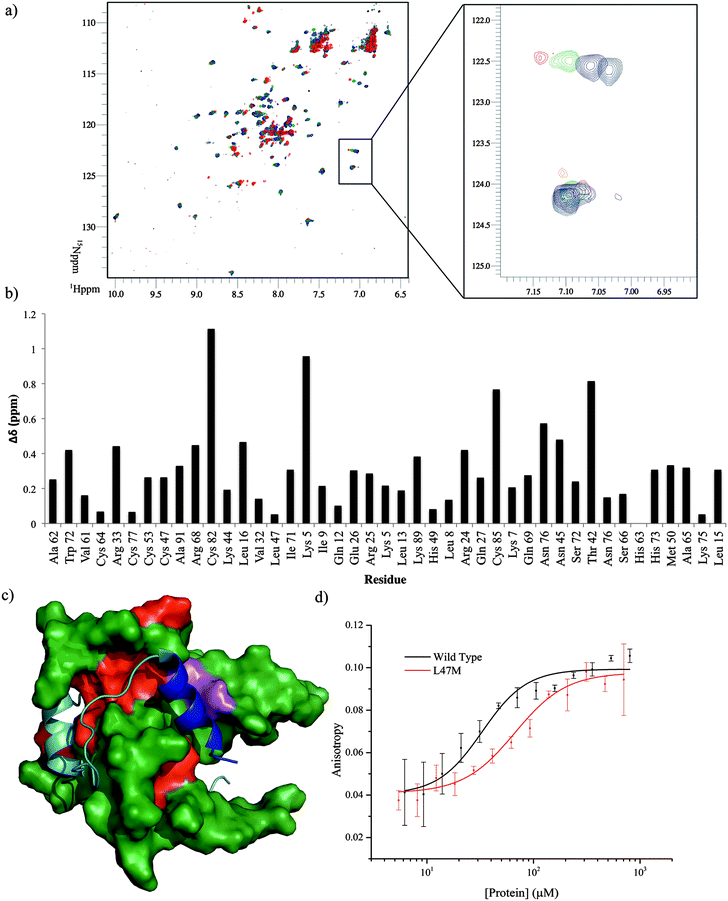
To locate the binding site of the phage display derived peptide on p300 15N–1H HSQC experiments were performed; the PDDP1 peptide was titrated (300–750 μM) into a solution of 15N-labelled p300 (230 μM). Peak shifts were observed confirming binding (Fig. 7a). The size of the shift for each peak (Fig. 7b) was measured and those with the largest shift were mapped on to p300 (assignments from BMRM: 626861). The location of the shifts indicates that PDDP1 may bind towards the top of the helix 3 binding pocket; the red area highlighted in Fig. 7c highlights a potential groove for interaction. In addition a shift was observed in the Trp indole peak; a change in the Trp environment has previously been used as an indicator of ligand binding to the helix 3 binding pocket.37
To further corroborate the binding of the phage display derived peptide in the helix 3 binding site, the binding of PDDP1 to both the wild type p300 and p300 with mutations in the helix 3 binding pocket (H20A, L47M, I71M) was investigated using fluorescence anisotropy (Table 2). The mutants L47M and I71M bound to PDDP1 with a statistically significant reduced affinity (p < 0.05) (Table 2). The reduction in binding affinity complements the NMR data, as the decrease in binding affinity of the mutant indicates that PDDP1 binds towards the top of the helix 3 binding pocket. The location of L47M and I71M is highlighted in Fig. 7c (magenta). This additionally highlights the HIF-1α helix 3 binding pocket as a key binding area.
| p300 | K d (μM) |
|---|---|
| *p < 0.05. | |
| Wild type | 20.67 ± 3.17 |
| H20A | 57.71 ± 7.87* |
| L47M | 57.76 ± 7.82* |
| I71M | 36.73 ± 11.0 |
The second phage display experiment used non-antibody binding proteins presented on the surface of the phage as opposed to short peptides; such scaffolds are designed to constrain and present variable peptide sequences for protein recognition.62 There are many different types of scaffold;63–69 the scaffold used in this study was the Adhiron scaffold (commercially named Affimer51,52). Adhirons are engineered non-antibody binding proteins which mimic the molecular recognition properties of antibodies but with improved properties (small, monomeric, thermostable, soluble and easy to express in E. coli and mammalian cells giving high protein yield).50 The Adhiron library has two randomised loops of 9 residues which are suitable for molecular recognition and are expected to adapt to form appropriate molecular contacts with a wide range of targets including protein pockets, protein surfaces, peptides and small molecules.50
Four panning rounds were carried out, and an ELISA was used to select those clones to be sequenced (Fig. S4, ESI†). Nine clones were sent for sequencing (5, 11, 12, 24, 34, 36, 37, 41 and 43; Fig. S4, ESI†) with four independent sequences present (Fig. S4 and Table S4, ESI†). Three Adhirons were taken forward: Ad24, Ad34 and Ad41 (note that the sequence of Ad41 is represented 6 times out of 9). These were initially tested using BLitz™ (ForteBio) to give an indication of affinity; BLitz™ is a dip and read system which enables the real time (kinetic) quantification of molecular interactions in solution.
The highest affinity Adhiron was Ad34 with an estimated Kd of 89 nM (Table 3). Therefore this Adhiron was taken forward to confirm binding by SPR, where an affinity of 157 nM (Chi2 0.114) was measured (Fig. S5, ESI†). The affinity of the phage display-derived Adhiron, Ad34, for p300 is higher than the phage display-derived peptides and the shorter native peptide sequences and is in the same range as HIF-1α786–826, which has an affinity of 16.11 ± 0.06 nM by fluorescence anisotropy and 45 ± 10 nM by ITC. However, Ad34 might be anticipated to bind at a more localised site than HIF-1α786–826. Each of the Adhirons was assessed for their ability to inhibit the HIF-1α/p300 interaction using the fluorescence anisotropy competition assay. All 3 Adhirons had an IC50 of 1–5 μM (Table 3, Fig. S6, ESI†). This is significantly better than fragments of native HIF-1α, or the phage peptides described above (which were unable to disrupt the interaction). However, the binding site of the Adhiron on p300 is not yet defined, and unfortunately excessive peak broadening in HSQC spectra on addition of Adhirons precludes interpretation of the resulting NMR shifts. These Adhiron sequences do not appear in a selection of Adhirons raised against a variety of other PPI targets, suggesting selectivity.
| Adhiron | K d (nM) | IC50 (μM) |
|---|---|---|
| Ad41 | 105 | 1.98 ± 0.32 |
| Ad34 | 89 | 4.78 ± 2.12 |
| Ad24 | 140 | 2.96 ± 0.46 |
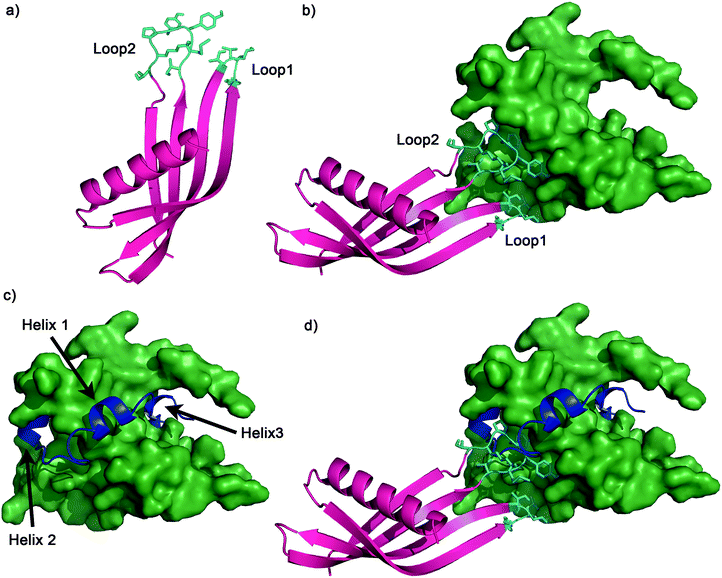
To further characterise Ad34 it was crystallised from 0.8 M di-sodium succinate pH 7; crystals were then supplemented with 20% glycerol for cryo-cooling and X-ray diffraction data collection. Data were phased to 2.8 Å (Fig. 8a).‡ Crystallographic data is summarised Table S5 (ESI†).
The binding mode of the Adhiron was then investigated by in silico docking. Ad34 was docked against the NMR structure of p300 (PDB 1L8C)8 using HADDOCK (High Ambiguity Driven protein–protein Docking).70 The docking results for the highest scored and most frequently predicted complex suggested that both variable loops are involved in binding, with both of the loops buried in hydrophobic areas of p300 (Fig. 8). Neither loop docked in the HIF-1α helix 3 region, suggesting that Ad34 may bind to a different region from the highest affinity phage display derived peptide (PDDP1) and the highest affinity native helix (HIF-1α816–826) (Fig. 8). The modelling suggests the two Adhiron loops bind in different crevices on p300; these pocket-like structures are in mutual proximity to each over. Given the proximity of the two sites identified, fragment-based approaches may be appropriate to link small molecules that target each site, although further experimental work is required to validate the binding site.
Conclusion
The purpose of this study was to understand the details of what drives the interaction between p300 and HIF-1α. Which may yield important information for inhibitor discovery and development. Characterisation of the native binding interface has demonstrated that the HIF-1α helix 3 region has the highest affinity for p300; we did not identify a short sequence of the native HIF-1α C-TAD peptide which has high affinity for p300 CH1 in isolation. We, and others, have targeted the helix 3 region using α-helix35,37 and protein domain mimetics36 allowing identification of HIF1α/p300 inhibitors with low micromolar affinity. Peptide phage display generated a 12mer peptide that had a higher affinity for p300 than any native HIF-1α fragment of the same length, providing a new template for small molecule development. Phage display generated competitive Adhirons of around 100 nM affinity which is in the same range as the binding affinity of the HIF-1α786–826 to p300. If binding occurs as predicted, mediated through the two 9mer loops, the Adhiron exploits a smaller binding interface than the HIF-1α786–826 peptide. The Adhirons themselves inhibit the HIF-1α/p300 interaction with a low micromolar IC50; therefore, mimicking the pharmacophore of the loops in a small molecule has the potential to generate novel high affinity inhibitors.Adhirons provide a simple and non-biased way to produce binders with high affinity and selectivity to blockade protein–protein interactions. Adhirons can be used as tool reagents for assay development work, as model interactions that can be used to direct pharmacophore development, and also have great potential as therapeutics in their own right.
The combination of a detailed biophysical analysis of the interaction between HIF-1α and p300, along with two orthogonal phage display techniques, has provided detailed information that can be used to inform design of highly potent compounds to disrupt an extremely challenging but pharmaceutically important protein–protein interaction.
Experimental
p300 protein purification
p300 CH1 was cloned into the pGex-6P-2 (GE Healthcare) plasmid and expressed as a GST fusion from which it was subsequently cleaved. GST-p300 containing BL21 Gold cells were grown to O.D600 0.6–0.8 and induced with 0.1 mM IPTG supplemented with 50 μM zinc sulphate and incubated for 18 hours at 18 °C. Cells were harvested and disrupted by sonication, then the soluble protein was separated from insoluble protein by centrifugation. GST-p300 was purified by affinity chromatography on glutathione beads (GE Healthcare); the GST was cleaved by PreScission protease and separated from p300 by size exclusion chromatography.p300 was biotin labeled by the chemoselective attachment of a biotin-depsipeptide to the N-terminal glycine of p300 catalysed by sortase A.56 The reaction consisted of 100 μM p300 (50 mM HEPES pH 7.5, 150 mM sodium phosphate and 5 mM calcium chloride), 300 μM biotin-depsipeptide and 20 μM sortase, the reaction was incubated at 37 °C with agitation for 3.5 hours. The biotin-p300 was separated from the unreacted biotin-depsipeptide by dialysis and from the sortase A by nickel affinity chromatography (the sortase A has a His tag). The biotinylatoin was confirmed by western blot and mass spectrometry.
15N labeled GST-p300 was purified after autoinduction with a glucose to lactose ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4; the full media recipe is outlined in Table S6 (ESI†). The cells were grown at 37 °C for four hours and then 20 °C for 40 hours. Cells were harvested and the protein purified in the same way as native p300.
4; the full media recipe is outlined in Table S6 (ESI†). The cells were grown at 37 °C for four hours and then 20 °C for 40 hours. Cells were harvested and the protein purified in the same way as native p300.
Mutants H20A, L47M and I71M were all made as per Q5 site- directed mutagenesis kit (NEB). They were expressed and purified in the same way as wild-type p300. CD was performed at 0.2 mg mL−1 in buffer: 40 mM sodium phosphate pH 7.5, 100 mM sodium chloride, 1 mM DTT and 5% glycerol.
eIF4E-SUMO expression by autoinduction and purification have been outlined previously.35
Peptides
All peptides outlined in Fig. 3 were purchased either FITC labeled of or unlabeled from PreoGenix (France), except HIF-1α794–804 and HIF-1α816–826 which were by standard Fmoc SPPS and purified by preparative HPLC. Characterisation outlined in ESI† Fig. S7–S10.Florescence anisotropy
| I = (2PG) + S | (1) |
| R = (S − PG)/I | (2) |
| Lb = (R − Rmin)/((λ(Rmax − R)) + R − Rmin) | (3) |
| y = ((Kd + x + [FL]) − √((Kd + x + [FL])2 − 4x[FL]))/2 | (4) |
• Cope with smaller files by not discarding partial blocks
• Allow for variable length adaptor sequences, and unequal adaptor sequences
• Allow for variable file tags (i.e. not just Illumina)
• Allow for variable length peptide libraries (tested with 7-mer and 12-mer)
Scripts were run over all ‘pure’ 7-mer and 12-mer libraries. Locus specific sequences rather than full primers were used and the quality control cut off was A.
| d[AB]/−dt = ka[A][B]·d[AB]/dt = kd[AB] |
| Kd = kd/ka |
Acknowledgements
We thank AstraZeneca and EPSRC for PhD studentships for G.M.B. and H.F.K. and the European Research Council (ERC-StG-240324) for support. We would also like to thank the Wellcome Trust (094232) for funding CD, ITC, X-ray generator and SPR facilities and the BBSRC (BB/L015056/1) for the crystallisation robotics and imaging facilities. We thank Arnout Kalverda and Gary Thompson for assistance with NMR experiments, Chi Trinh for assistance with crystallography, Iain Manfield for help with ITC and SPR and Daniel Williamson for help with biotin labeling of p300.Notes and references
- D. Hanahan and R. A. Weinberg, Cell, 2011, 144, 646–674 CrossRef CAS PubMed.
- P. Vaupel, O. Thews, D. K. Kelleher and M. Hoeckel, Adv. Exp. Med. Biol., 1998, 454, 591–602 CrossRef CAS.
- H. Zhong, A. M. De Marzo, E. Laughner, M. Lim, D. A. Hilton, D. Zagzag, P. Buechler, W. B. Isaacs, G. L. Semenza and J. W. Simons, Cancer Res., 1999, 59, 5830–5835 CAS.
- M. Hockel and P. Vaupel, Semin. Oncol., 2001, 28, 36–41 CrossRef CAS.
- I. K. Nordgren and A. Tavassoli, Chem. Soc. Rev., 2011, 40, 4307–4317 RSC.
- N. Vo and R. H. Goodman, J. Biol. Chem., 2001, 276, 13505–13508 CrossRef CAS PubMed.
- S. J. Freedman, Z. Y. Sun, F. Poy, A. L. Kung, D. M. Livingston, G. Wagner and M. J. Eck, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 5367–5372 CrossRef CAS PubMed.
- S. A. Dames, M. Martinez-Yamout, R. N. De Guzman, H. J. Dyson and P. E. Wright, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 5271–5276 CrossRef CAS PubMed.
- S. Surade and T. L. Blundell, Chem. Biol., 2012, 19, 42–50 CrossRef CAS PubMed.
- A. D. Thompson, A. Dugan, J. E. Gestwicki and A. K. Mapp, ACS Chem. Biol., 2012, 7, 1311–1320 CrossRef CAS PubMed.
- S. Jones and J. M. Thornton, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 13–20 CrossRef CAS.
- L. Lo Conte, C. Chothia and J. Janin, J. Mol. Biol., 1999, 285, 2177–2198 CrossRef CAS.
- M. R. Arkin and J. A. Wells, Nat. Rev. Drug Discovery, 2004, 3, 301–317 CrossRef CAS PubMed.
- A. A. Bogan and K. S. Thorn, J. Mol. Biol., 1998, 280, 1–9 CrossRef CAS PubMed.
- T. Clackson and J. A. Wells, Science, 1995, 267, 383–386 CrossRef CAS.
- Z. Hu, B. Ma, H. Wolfson and R. Nussinov, Proteins, 2000, 39, 331–342 CrossRef CAS.
- W. Guo, J. A. Wisniewski and H. Ji, Bioorg. Med. Chem. Lett., 2014, 24, 2546–2554 CrossRef CAS PubMed.
- B. Onnis, A. Rapisarda and G. Melillo, J. Cell. Mol. Med., 2009, 13, 2780–2786 CrossRef CAS PubMed.
- L. M. Greenberger, I. D. Horak, D. Filpula, P. Sapra, M. Westergaard, H. F. Frydenlund, C. Albaek, H. Schroder and H. Orum, Mol. Cancer Ther., 2008, 7, 3598–3608 CrossRef CAS PubMed.
- A. Rapisarda, B. Uranchimeg, O. Sordet, Y. Pommier, R. H. Shoemaker and G. Melillo, Cancer Res., 2004, 64, 1475–1482 CrossRef CAS.
- E. Laughner, P. Taghavi, K. Chiles, P. C. Mahon and G. L. Semenza, Mol. Cell. Biol., 2001, 21, 3995–4004 CrossRef CAS PubMed.
- E. W. Newcomb, M. A. Ali, T. Schnee, L. Lan, Y. Lukyanov, M. Fowkes, D. C. Miller and D. Zagzag, Neuro-Oncology, 2005, 7, 225–235 CrossRef CAS PubMed.
- M. Mayerhofer, P. Valent, W. R. Sperr, J. D. Griffin and C. Sillaber, Blood, 2002, 100, 3767–3775 CrossRef CAS PubMed.
- P. K. Majumder, P. G. Febbo, R. Bikoff, R. Berger, Q. Xue, L. M. McMahon, J. Manola, J. Brugarolas, T. J. McDonnell, T. R. Golub, M. Loda, H. A. Lane and W. R. Sellers, Nat. Med., 2004, 10, 594–601 CrossRef CAS PubMed.
- G. V. Thomas, C. Tran, I. K. Mellinghoff, D. S. Welsbie, E. Chan, B. Fueger, J. Czernin and C. L. Sawyers, Nat. Med., 2006, 12, 122–127 CrossRef CAS PubMed.
- J. Y. Han, S. H. Oh, F. Morgillo, J. N. Myers, E. Kim, W. K. Hong and H. Y. Lee, J. Natl. Cancer Inst., 2005, 97, 1272–1286 CrossRef CAS PubMed.
- G. L. Wang, B. H. Jiang and G. L. Semenza, Biochem. Biophys. Res. Commun., 1995, 216, 669–675 CrossRef CAS PubMed.
- J. S. Isaacs, Y. J. Jung, E. G. Mimnaugh, A. Martinez, F. Cuttitta and L. M. Neckers, J. Biol. Chem., 2002, 277, 29936–29944 CrossRef CAS PubMed.
- D. Kong, E. J. Park, A. G. Stephen, M. Calvani, J. H. Cardellina, A. Monks, R. J. Fisher, R. H. Shoemaker and G. Melillo, Cancer Res., 2005, 65, 9047–9055 CrossRef CAS PubMed.
- E. Miranda, I. K. Nordgren, A. L. Male, C. E. Lawrence, F. Hoakwie, F. Cuda, W. Court, K. R. Fox, P. A. Townsend, G. K. Packham, S. A. Eccles and A. Tavassoli, J. Am. Chem. Soc., 2013, 135, 10418–10425 CrossRef CAS PubMed.
- S. R. Mooring, H. Jin, N. S. Devi, A. A. Jabbar, S. Kaluz, Y. Liu, E. G. Van Meir and B. Wang, J. Med. Chem., 2011, 54, 8471–8489 CrossRef CAS PubMed.
- D. H. Shin, Y. S. Chun, D. S. Lee, L. E. Huang and J. W. Park, Blood, 2008, 111, 3131–3136 CrossRef CAS PubMed.
- A. L. Kung, S. D. Zabludoff, D. S. France, S. J. Freedman, E. A. Tanner, A. Vieira, S. Cornell-Kennon, J. Lee, B. Wang, J. Wang, K. Memmert, H. U. Naegeli, F. Petersen, M. J. Eck, K. W. Bair, A. W. Wood and D. M. Livingston, Cancer Cell, 2004, 6, 33–43 CrossRef CAS PubMed.
- L. K. Henchey, S. Kushal, R. Dubey, R. N. Chapman, B. Z. Olenyuk and P. S. Arora, J. Am. Chem. Soc., 2010, 132, 941–943 CrossRef CAS PubMed.
- G. M. Burslem, H. F. Kyle, A. L. Breeze, T. A. Edwards, A. Nelson, S. L. Warriner and A. J. Wilson, ChemBioChem, 2014, 15, 1083–1087 CrossRef CAS PubMed.
- S. Kushal, B. B. Lao, L. K. Henchey, R. Dubey, H. Mesallati, N. J. Traaseth, B. Z. Olenyuk and P. S. Arora, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 15602–15607 CrossRef CAS PubMed.
- B. B. Lao, I. Grishagin, H. Mesallati, T. F. Brewer, B. Z. Olenyuk and P. S. Arora, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 7531–7536 CrossRef CAS PubMed.
- R. Dubey, M. D. Levin, L. Z. Szabo, C. F. Laszlo, S. Kushal, J. B. Singh, P. Oh, J. E. Schnitzer and B. Z. Olenyuk, J. Am. Chem. Soc., 2013, 135, 4537–4549 CrossRef CAS PubMed.
- S. Yin, S. Kaluz, N. S. Devi, A. A. Jabbar, R. G. de Noronha, J. Mun, Z. Zhang, P. R. Boreddy, W. Wang, Z. Wang, T. Abbruscato, Z. Chen, J. J. Olson, R. Zhang, M. M. Goodman, K. C. Nicolaou and E. G. Van Meir, Clin. Cancer Res., 2012, 18, 6623–6633 CrossRef CAS PubMed.
- Q. Shi, S. Yin, S. Kaluz, N. Ni, N. S. Devi, J. Mun, D. Wang, K. Damera, W. Chen, S. Burroughs, S. R. Mooring, M. M. Goodman, E. G. Van Meir, B. Wang and J. P. Snyder, ACS Med. Chem. Lett., 2012, 3, 620–625 CrossRef CAS PubMed.
- C. A. Lipinski, J. Pharmacol. Toxicol. Methods, 2000, 44, 235–249 CrossRef CAS.
- J. L. Ruas, L. Poellinger and T. Pereira, J. Biol. Chem., 2002, 277, 38723–38730 CrossRef CAS PubMed.
- J. Gu, J. Milligan and L. E. Huang, J. Biol. Chem., 2001, 276, 3550–3554 CrossRef CAS PubMed.
- D. Lando, D. J. Peet, D. A. Whelan, J. J. Gorman and M. L. Whitelaw, Science, 2002, 295, 858–861 CrossRef CAS PubMed.
- H. Cho, D. R. Ahn, H. Park and E. G. Yang, FEBS Lett., 2007, 581, 1542–1548 CrossRef CAS PubMed.
- S. S. Sidhu, Curr. Opin. Biotechnol., 2000, 11, 610–616 CrossRef CAS.
- J. K. Scott and G. P. Smith, Science, 1990, 249, 386–390 CAS.
- S. E. Cwirla, E. A. Peters, R. W. Barrett and W. J. Dower, Proc. Natl. Acad. Sci. U. S. A., 1990, 87, 6378–6382 CrossRef CAS.
- J. J. Devlin, L. C. Panganiban and P. E. Devlin, Science, 1990, 249, 404–406 CAS.
- C. Tiede, A. A. Tang, S. E. Deacon, U. Mandal, J. E. Nettleship, R. L. Owen, S. E. George, D. J. Harrison, R. J. Owens, D. C. Tomlinson and M. J. McPherson, Protein Eng., Des. Sel., 2014, 27, 145–155 CrossRef CAS PubMed.
- T. Hoffmann, L. K. Stadler, M. Busby, Q. Song, A. T. Buxton, S. D. Wagner, J. J. Davis and P. Ko Ferrigno, Protein Eng., Des. Sel., 2010, 23, 403–413 CrossRef CAS PubMed.
- L. K. Stadler, T. Hoffmann, D. C. Tomlinson, Q. Song, T. Lee, M. Busby, Y. Nyathi, E. Gendra, C. Tiede, K. Flanagan, S. J. Cockell, A. Wipat, C. Harwood, S. D. Wagner, M. A. Knowles, J. J. Davis, N. Keegan and P. K. Ferrigno, Protein Eng., Des. Sel., 2011, 24, 751–763 CrossRef CAS PubMed.
- V. Azzarito, K. Long, N. S. Murphy and A. J. Wilson, Nat. Chem., 2013, 5, 161–173 CrossRef CAS PubMed.
- T. A. Edwards and A. J. Wilson, Amino Acids, 2011, 41, 743–754 CrossRef CAS PubMed.
- B. N. Bullock, A. L. Jochim and P. S. Arora, J. Am. Chem. Soc., 2011, 133, 14220–14223 CrossRef CAS PubMed.
- D. J. Williamson, M. A. Fascione, M. E. Webb and W. B. Turnbull, Angew. Chem., Int. Ed., 2012, 51, 9377–9380 CrossRef CAS PubMed.
- W. L. Matochko, K. Chu, B. Jin, S. W. Lee, G. M. Whitesides and R. Derda, Methods, 2012, 58, 47–55 CrossRef CAS PubMed.
- V. Munoz and L. Serrano, Nat. Struct. Biol., 1994, 1, 399–409 CrossRef CAS.
- P. A. t Hoen, S. M. Jirka, B. R. Ten Broeke, E. A. Schultes, B. Aguilera, K. H. Pang, H. Heemskerk, A. Aartsma-Rus, G. J. van Ommen and J. T. den Dunnen, Anal. Biochem., 2012, 421, 622–631 CrossRef CAS PubMed.
- P. Molek, B. Strukelj and T. Bratkovic, Molecules, 2011, 16, 857–887 CrossRef CAS PubMed.
- R. N. De Guzman, J. M. Wojciak, M. A. Martinez-Yamout, H. J. Dyson and P. E. Wright, Biochemistry, 2005, 44, 490–497 CrossRef CAS PubMed.
- M. Gebauer and A. Skerra, Curr. Opin. Chem. Biol., 2009, 13, 245–255 CrossRef CAS PubMed.
- S. E. Hufton, N. van Neer, T. van den Beuken, J. Desmet, E. Sablon and H. R. Hoogenboom, FEBS Lett., 2000, 475, 225–231 CrossRef CAS.
- S. J. McConnell and R. H. Hoess, J. Mol. Biol., 1995, 250, 460–470 CrossRef CAS PubMed.
- B. Heyd, F. Pecorari, B. Collinet, E. Adjadj, M. Desmadril and P. Minard, Biochemistry, 2003, 42, 5674–5683 CrossRef CAS PubMed.
- K. Nord, E. Gunneriusson, J. Ringdahl, S. Stahl, M. Uhlen and P. A. Nygren, Nat. Biotechnol., 1997, 15, 772–777 CrossRef CAS PubMed.
- E. Bianchi, A. Folgori, A. Wallace, M. Nicotra, S. Acali, A. Phalipon, G. Barbato, R. Bazzo, R. Cortese and F. Felici, et al. , J. Mol. Biol., 1995, 247, 154–160 CrossRef CAS.
- P. A. Dalby, R. H. Hoess and W. F. DeGrado, Protein Sci., 2000, 9, 2366–2376 CrossRef CAS PubMed.
- C. Souriau, L. Chiche, R. Irving and P. Hudson, Biochemistry, 2005, 44, 7143–7155 CrossRef CAS PubMed.
- C. Dominguez, R. Boelens and A. M. Bonvin, J. Am. Chem. Soc., 2003, 125, 1731–1737 CrossRef CAS PubMed.
- F. Long, A. A. Vagin, P. Young and G. N. Murshudov, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2008, 64, 125–132 CrossRef CAS PubMed.
- K. Cowtan, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2006, 62, 1002–1011 CrossRef PubMed.
- P. D. Adams, R. W. Grosse-Kunstleve, L. W. Hung, T. R. Ioerger, A. J. McCoy, N. W. Moriarty, R. J. Read, J. C. Sacchettini, N. K. Sauter and T. C. Terwilliger, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2002, 58, 1948–1954 CrossRef PubMed.
- G. N. Murshudov, A. A. Vagin and E. J. Dodson, Acta Crystallogr., Sect. D: Biol. Crystallogr., 1997, 53, 240–255 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5mb00284b |
| ‡ Adhiron crystal structure PDB accession code: 5a0o. |
| This journal is © The Royal Society of Chemistry 2015 |