DOI:
10.1039/C4MB00548A
(Paper)
Mol. BioSyst., 2015,
11, 307-316
Elucidating the interaction of limonene with bovine serum albumin: a multi-technique approach†
Received
19th September 2014
, Accepted 23rd October 2014
First published on 23rd October 2014
Abstract
The interaction of Bovine Serum Albumin (BSA) with limonene has been studied by UV-visible spectroscopy, fluorescence spectroscopy and molecular docking, and its effects on protein conformation, topology and stability were determined by Circular Dichroism (CD), Dynamic Light Scattering (DLS) and Differential Scanning Calorimetry (DSC). A gradual decrease in Stern–Volmer quenching constants with the increase in temperature showed the static mode of fluorescence quenching. The obtained binding constant (Kb) was ∼104 M−1. The temperature dependent Kb, Gibbs free energy (ΔG), enthalpy (ΔH) and entropy (ΔS) changes were calculated, which revealed that the reaction is spontaneous and exothermic. The UV-visible spectra showed a change in the peaks within the aromatic region indicating hydrophobic interactions with Trp, Tyr and Phe in the protein. Moreover, limonene induced an increase in α-helical contents probably on the cost of random coils or/and β-sheets of BSA, as observed from the far-UV CD spectra. The topology of BSA in the presence of limonene was slightly altered, as obtained from DLS results. The stability was also enhanced as revealed through thermal denaturation study by DSC and CD. Molecular docking study depicted that limonene fits into the hydrophobic pocket close to Sudlow site I in domain IIA of BSA. The present study will be helpful in understanding the binding mechanism of limonene and associated stability and conformational changes.
1. Introduction
Plant derived products have always been preferred over commercial ones for medicinal purposes. Flavonoids, polyphenols and medicinal products are extensively used for antioxidant, anti-inflammatory, anticancer, antimicrobial, antidermatophytic and hepatoprotective activities.1,2 Serum albumins are the most abundant proteins in the circulatory system of a wide variety of organisms, which play a dominant key role in the binding and transport of numerous endogenous and exogenous ligands.3 Serum albumin often enhances the apparent solubility of hydrophobic drugs in plasma and modulates their delivery, disposition, efficacy and distribution to cells in vivo and in vitro.4,5 The drug–protein interaction may result in the formation of a stable protein–drug complex, which has significant effect on the delivery, distribution, free concentration and metabolism of drugs in the blood circulatory system. Thus, the drug–albumin complex may be considered as a model to gain fundamental insights into drug–protein interactions and explore its applications. In this study, BSA has been chosen because of its structural homology with human serum albumin (HSA), low cost, easy availability and unusual ligand-binding properties.6–10
BSA is a non glycated and globular protein composed of 583 amino acids. It is made up of three homologous domains (I, II, III), which are separated into nine loops (L1–L9) by 17 disulphide bridges.11 The loops in each domain are designed of a sequence of large–small–large loops forming a triplet. Each domain in turn is the resultant of two subdomains. BSA has two tryptophans, Trp-134 and Trp-212, which are embedded in the sub domain IB and sub domain IIA, respectively.12,13
The molecular interactions among proteins and many compounds such as drugs and some organic small molecules have been investigated successfully.14–21 However, the binding of plant derived products to serum albumins is now an emerging field to explore the role of plant derivatives in biological systems of herbivores and their mode of action in various diseases.22,23
Limonene exists as two optical isomers, D- and L-limonene, and the racemic mixture dipentene. Limonene, like other monoterpenes, occurs naturally in certain trees, bushes, mainly in peel from citrus fruits, in dill, caraway, fennel, turpentine and in celery. Limonene is used as a flavour and fragrance additive in food, household cleaning products, cosmetics and perfumes.24 Thus, it is also consumed by the human body in several ways. The hydrolytic half-life of D-limonene is >1000 days, as described earlier.25 In addition, limonene has been shown to prevent mammary, liver, lung, and other cancers. It has also been focused to treat a class of rodent cancers, including breast and pancreatic carcinomas. Being a good solvent of cholesterol, D-limonene has been exploited clinically to dissolve cholesterol-containing gallstones.26 Owing to its gastric acid neutralizing effect and its support of normal peristalsis, it has also been utilized for relief of heartburn and gastroesophageal reflux (GERD).
Limonene is a constituent in cosmetics, pharmaceuticals, and solvents. Thus, this study has significant importance with human health concern. In the present work, we demonstrated the binding of limonene to BSA by employing fluorescence, CD, and UV-visible spectroscopic methods, and the effect of limonene on the conformation and stability of BSA was further checked by the help of DLS and DSC, respectively. In addition, molecular docking and displacement studies were also carried out to reveal the binding site of limonene. BSA and limonene are both antioxidants, and this property may be synergistically increased upon BSA–limonene complex formation. The present study may be helpful to postulate how herbivores combat free radical scavenging by exploring the binding efficacy of limonene to BSA.
2. Materials and methods
2.1 Materials
Essentially fatty acid free bovine serum albumin (A7030) and (R)-(+)-limonene (183164) were products of Sigma-Aldrich, India. All other reagents were of analytical grade.
2.2 Preparation of solutions
All experiments were carried out in 20 mM phosphate buffer, pH 7.4. BSA was used without further purification as its purity was checked by SDS-PAGE at high concentration. BSA was dialyzed properly against the respective buffer. Protein stock solutions (5 mg ml−1) were prepared in 20 mM phosphate buffer, pH 7.4. The concentration of native proteins in 20 mM phosphate buffer was determined spectrophotometrically from the extinction coefficient reported at 280 nm.
2.3 UV-visible spectroscopic measurements
Absorption measurements were performed at 37 °C using a Perkin-Elmer Lambda 25 double beam UV-vis spectrophotometer attached with a peltier temperature programmer-1 (PTP-1). Fixed concentrations of BSA (6 μM) and limonene (6 μM) (molar ratio of P![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) L = 1
L = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were taken and spectra were measured.
1) were taken and spectra were measured.
2.4 Steady state fluorescence quenching measurements
The Shimadzu 5301 PC fluorescence spectrophotometer is equipped with a constant temperature holder and the temperatures (15, 25 and 37 °C) were maintained by a constant temperature water circulator (Julabo Eyela). The excitation and emission slit widths were set at 3 nm. The titration of the limonene (0–30 μM) to 2 μM BSA solutions was carried out in a dual-path length fluorescence cuvette (10 × 3.5 mm). The shorter path length was oriented towards the emission side. Such a low concentration of BSA (2 μM) with absorbance value of ∼0.07 was used throughout the fluorescence experiments to minimize the inner filter effect. Intrinsic fluorescence was measured by exciting at 280 nm. The emission spectra were recorded in the range of 300–450 nm and the data were plotted at 339 nm. The decrease in fluorescence intensity at 339 nm was analyzed according to the Stern–Volmer eqn (1):| | | F0/F = Ksv[Q] + 1 = kqτ0[Q] + 1 | (1) |
where, F0 and F were the fluorescence intensities in absence and presence of quencher (limonene), Ksv is the Stern–Volmer quenching constant, kq is the bimolecular rate constant of the quenching reaction and τ0 the average integral fluorescence life time of tryptophan, which is ∼10−9 s. Binding constants and binding sites were obtained from eqn (2):| | log(F0/F − 1) = log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kb + n Kb + n![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log[Q] log[Q] | (2) |
where, Kb is the binding constant and n is the number of binding sites. The change in free energy was calculated from eqn (3) whereas change in enthalpy and entropy at different temperatures were analysed from the van't Hoff equation as given in eqn (4):| | ΔG° = −RT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kb Kb | (3) |
| |  | (4) |
where, ΔG° is the free energy change, ΔH° is the enthalpy change, ΔS° is the entropy change, R (1.987 cal mol−1 K−1) is a gas constant and T is the absolute temperature (K).
The synchronous fluorescence spectra were recorded at Δλ 15 (for tyrosine) and 60 nm (for tryptophan) in the absence and presence of limonene over a wavelength range of 290–350 nm.
The three-dimensional fluorescence spectra were measured under the following conditions: the emission wavelength was recorded between 200 and 500 nm, the initial excitation wavelength was set at 200 nm with an increment of 5 nm, the number of scanning curves was 31, and the other scanning parameters were the same as those of the fluorescence emission spectra.
2.5 Fluorescence resonance energy transfer (FRET) to the limonene
The fluorescence spectra of BSA (2 μM) and absorption spectra of limonene (2 μM) between 300 to 400 nm were scanned in similar way as given in the method sections ‘Fluorescence Quenching’ and ‘UV-visible’ experiments at 25 °C. If the emission spectrum of the donor (BSA) significantly overlaps with the absorption spectrum of acceptor (limonene), these donor–acceptor pairs will be considered to be within Förster distance, and the possibility of energy transfer between them could be ascertained.27 Therefore, the degree of energy transfer depends upon the area of overlap and the distance between these donor–acceptor molecules. The efficiency of energy transfer (E) is calculated using the following eqn (5):| |  | (5) |
where, F0 and F are the fluorescence intensities of BSA in the absence and presence of limonene, respectively; r is the distance between donor and acceptor and R0 is the critical distance at which transfer efficiency equals 50%, which can be calculated from the following equation:| | | R06 = 8.79 × 10−25 K2n−4ϕJ | (6) |
where K2 is the orientation factor related to the geometry of the donor and acceptor of dipoles, n is the refractive index of the medium, ϕ is the fluorescence quantum yield of the donor in the absence of an acceptor, and J expresses the degree of spectral overlap between the donor emission and the acceptor absorption, which can be evaluated by integrating the overlap spectral area between 300 to 400 nm from the following equation:| |  | (7) |
where, F(λ) is the fluorescence intensity of the donor at wavelength range λ, which is dimensionless, and ε(λ) is the molar absorptivity (extinction coefficient) of the acceptor wavelength λ in M−1 cm−1. In our present study K2, φ and n were taken as 2/3, 0.118 and 1.336, respectively.28
2.6 Circular dichroic measurements
The isothermal wavelength scan studies of BSA in the absence and presence of limonene were carried out with a JASCO-J815 spectropolarimeter equipped with a Peltier-type temperature controller. The instrument was calibrated with D-10-camphorsulfonic acid. All the isothermal CD measurements were made at 25 °C. Spectra were collected with a 50 nm min−1 scan speed, 0.1 nm data pitch and a response time of 2 s. Each spectrum was the average of 2 scans. For far-UV CD spectra (190–250 nm) and near-UV CD (250–300 nm) the cells of 0.1 cm and 1 cm path length were taken respectively. Helical content was calculated using online available K2D software. All spectra were smoothed by the Savitzky–Golay method with 25 convolution width. BSA concentrations used for far-UV CD and near-UV CD were 2 μM and 15 μM, respectively.
2.7 Differential scanning calorimetry
The differential scanning calorimetric measurements were carried out using a VP-DSC microcalorimeter (Micro Cal, Northampton, MA). The buffer and protein solutions were degassed under mild vacuum prior to the experiment. Samples were prepared in 20 mM sodium phosphate buffer, pH 7.4. The DSC measurements of BSA (18 μM) in the presence of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 ratio of limonene were performed from 25 to 90 °C at a scan rate of 0.5 °C min−1. Data was analyzed using the Origin software provided with the instrument to obtain the temperature at the midpoint of the unfolding transition (Tm) and calorimetric enthalpy (ΔH°).
15 ratio of limonene were performed from 25 to 90 °C at a scan rate of 0.5 °C min−1. Data was analyzed using the Origin software provided with the instrument to obtain the temperature at the midpoint of the unfolding transition (Tm) and calorimetric enthalpy (ΔH°).
2.8 Dynamic light scattering (DLS) measurements
DLS measurements were carried out at 830 nm using DynaPro-TC-04 dynamic light scattering equipment (Protein Solutions, Wyatt Technology, Santa Barbara, CA) equipped with a temperature-controlled microsampler. BSA (2 mg ml−1) was incubated with the limonene for 8 hours. The samples were spun at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min and were filtered serially through 0.22 and 0.02 μm Whatman syringe filters directly into a 12 μl quartz cuvette. For each experiment, 20 measurements were taken. Mean hydrodynamic radius (Rh) and polydispersity were analysed using Dynamics 6.10.0.10 software at optimized resolution. The Rh was estimated on the basis of an autocorrelation analysis of scattered light intensity data based on translation diffusion coefficient by the Stokes–Einstein relationship:
000 rpm for 10 min and were filtered serially through 0.22 and 0.02 μm Whatman syringe filters directly into a 12 μl quartz cuvette. For each experiment, 20 measurements were taken. Mean hydrodynamic radius (Rh) and polydispersity were analysed using Dynamics 6.10.0.10 software at optimized resolution. The Rh was estimated on the basis of an autocorrelation analysis of scattered light intensity data based on translation diffusion coefficient by the Stokes–Einstein relationship:| |  | (8) |
where Rh is the hydrodynamic radius, k is the Boltzmann constant, T is the temperature, η is the viscosity of water and D is the diffusion coefficient.29
2.9 Molecular docking and binding displacement measurement study using site markers
To determine the amino acid residues involved in the binding site of limonene on BSA the docking studies were performed by auto dock 4.2.0 software (http://autodock.scripps.edu) as reported earlier.30 Lamarckian genetic algorithm (LGA) implemented with an adaptive local method search was applied to determine the possible conformation of the drug that binds to the protein.31 The crystal structure of BSA was obtained from Brookhaven Protein Data Bank having PDB id (4F5S), and the 3d sdf file of limonene (CID 22311) was obtained from PubChem. Water molecules and hydrogen atoms were removed from the protein. Then, partial Kollman charges were assigned to BSA. The protein was held rigid and all torsional bonds are taken as being free during docking calculations.
Moreover, the protein was set to be rigid and there is no consideration of solvent molecules on docking. To determine the binding site on BSA, blind docking was carried out and the grid size was set to be 126, 126 and 126 along the X, Y and Z axes with 0.564 Angstrom grid spacing. Auto dock parameters were used with GA population size: 150 and maximum number of energy evolutions: 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500
500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. The 10 best solutions based on docking score were retained for further analysis. Discovery studio 3.5 was used for visualization and for the identification of residues involved in binding. Binding displacement studies between limonene and BSA in the presence of two site markers, warfarin (for site I) and diazepam (for site II) were measured using the fluorescence titration method. The titration of limonene was carried out on the solution having protein and site markers in the ratio of 1
000. The 10 best solutions based on docking score were retained for further analysis. Discovery studio 3.5 was used for visualization and for the identification of residues involved in binding. Binding displacement studies between limonene and BSA in the presence of two site markers, warfarin (for site I) and diazepam (for site II) were measured using the fluorescence titration method. The titration of limonene was carried out on the solution having protein and site markers in the ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and the Ksv values were calculated by using eqn (1).
1 and the Ksv values were calculated by using eqn (1).
3. Results and discussion
3.1 UV-visible absorption spectroscopy
Ultraviolet-visible absorption spectroscopy is an influential tool for steady-state studies of protein–drug interaction. Changes in the far and near UV regions correspond to the secondary and tertiary structure, respectively. In proteins, we discriminate various internal chromophoric groups that give rise to electronic absorption bands. The aromatic amino acids contribute to bands in the range of 255–300 nm. Fig. 1A shows that the absorption peak of BSA centres at ∼280 nm mainly due to the tryptophan residue. However, after addition of the limonene, the maximal absorption peak as well as the absorption intensity of BSA is increased. This indicates that the interaction of limonene leads to conformational change in BSA, which is primarily near to the tryptophan residue. In Fig. 1A, the maximum change was observed at 280 nm where the increase in absorbance with 6 μM limonene indicates complex formation between BSA and limonene.
 |
| | Fig. 1 (A) Absorption spectra of BSA gradually titrated with limonene at 25 °C. (A) Limonene only, (B) BSA = 6 μM and (C) BSA/limonene = 1. (B) Fluorescence quenching of BSA by limonene at 25 °C. [BSA = 2 μM; limonene = 0–30 μM]. The inset corresponds to molecular structure of Limonene. | |
3.2 Tryptophan fluorescence quenching by limonene
Tryptophan fluorescence quenching was performed for the determination of the interaction between limonene and BSA by titration of limonene against protein at 25 °C. BSA has a strong fluorescence emission peak at ∼339 nm on excitation at 280 nm; addition of the limonene caused reduction in the emission spectra of BSA as shown in Fig. 1B. The values of emission intensity at 339 nm were used to measure drug-binding affinity. The fluorescence intensity of tryptophan fluorescence emission decreases continuously but at higher concentration of limonene the decreasing pattern of emission gets saturated, which is a clear indication of binding of limonene to a specific binding site on BSA. The same experimental procedures were also followed at 15 and 37 °C where we found that on increasing the temperature, the quenching also decreases, or in other words, the extent of lowering in fluorescence emission was higher at lower temperature. The decrease in fluorescence intensity upon addition of limonene was analysed according to the Stern–Volmer equation (as shown in Fig. 2A). There is a linear dependence between F0/F and the molar concentration of limonene in the Stern–Volmer plot. The slopes decrease with increasing temperature, indicating that the ligand binding to the protein occurred by ‘static quenching’. When the value of kq was calculated it was greater than the maximum scatter collision quenching constant, i.e. 2.0 × 1010 mol−1 s−1.32 This shows that quenching is not initiated by dynamic diffusion but occurs by formation of a strong complex between BSA and limonene. Moreover, the absorption spectra of BSA–limonene (Fig. 1A) were transparently different from those of BSA or limonene alone, giving clear proof of forming a protein drug complex with a new structure. From the obtained results, it is to be noted that a static type of quenching occurs during the binding of limonene with BSA. The Ksv values for limonene at different temperatures are given in Table 1.
Table 1 Binding parameters of limonene interaction to BSA in 20 mM Phosphate buffer pH 7.4 at different temperatures obtained and calculated from fluorescence quenching results
|
T (K) |
K
sv (×104 M−1) |
k
q (×1013 M−1 s−1) |
n
|
K
b (×104 M−1) |
ΔG (kcal mol−1) |
ΔH (kcal mol−1) |
ΔS (cal mol−1 K−1) |
| 288 |
1.146 |
1.146 |
1.00 |
1.171 |
−5.34 |
|
|
| 298 |
0.798 |
0.798 |
0.99 |
0.765 |
−5.27 |
−7.2 |
−6.53 |
| 310 |
0.541 |
0.541 |
0.98 |
0.473 |
−5.19 |
|
|
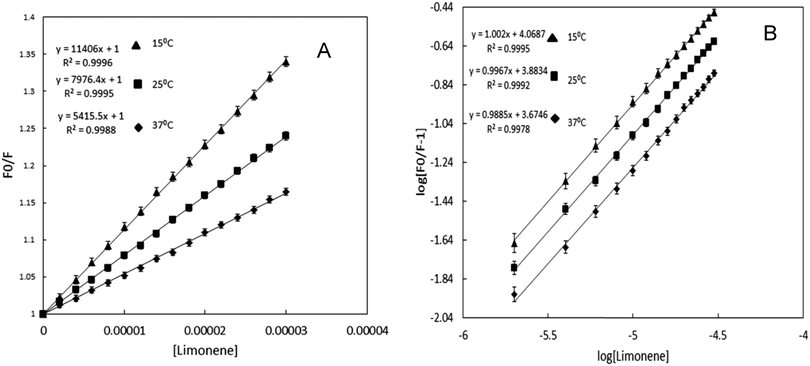 |
| | Fig. 2 (A) The Stern–Volmer plots for the binding of Limonene with BSA at 288 (▲), 298 (■) and 310 (◆) K. Excitation wavelength was 280 nm, [BSA = 2 μM; limonene = 0–30 μM]. (B) Plot between log[(F0/F) − 1] and log[limonene] for BSA–limonene interaction at 288 (▲), 298 (■) and 310 (◆) K. [BSA = 2 μM; limonene = 0–30 μM]. | |
3.3 Determination of binding constant and number of binding sites
For the determination of the binding constant and number of binding sites, log[(F0/F) − 1] vs. log[limonene] was plotted (Fig. 2B). Using eqn (2) from the slopes and intercepts of modified Stern–Volmer plots, the number of binding sites (n) and the value of binding constant were calculated, respectively. For limonene, the values of Kb and n were calculated at different temperatures and the observed values are listed in Table 1. The data shows that a decrease in Ksv or a decrease in Kb on increasing the temperature is a clear indication of static quenching.33
3.4 Thermodynamics of BSA–limonene interaction
Generally, a small molecule binds to a macromolecule by the following four binding modes: hydrogen bonds, van der Waals attractions, electrostatic interactions, and hydrophobic interactions. The thermodynamic parameters, enthalpy change (ΔH) and entropy change (ΔS) of the reaction are very important for confirming binding modes. The temperature-dependence of the binding constant was investigated at three different temperatures (15, 25 and 37 °C) by considering that BSA could not undergo any structural degradation.
According to the binding constants of limonene to BSA at the three different temperatures, the thermodynamic parameters were determined from the linear second law of thermodynamics plot (Fig. 3) and the observed values are presented in Table 1. For the determination of the enthalpy–entropy relation in the BSA–limonene interaction, three temperatures, viz. 15, 25 and 37 °C, are considered only, because during the binding process, the structure of the protein is assumed to be structurally unaltered as major conformational changes gives the false reading of thermodynamic parameters for interaction studies. In other words, the obtained enthalpy–entropy changes are mainly caused by the binding of the limonene molecules to BSA. As shown in Table 1, ΔG in every condition is negative, which suggests that the interaction process is spontaneous, ΔH and ΔS for the complex formation between limonene and BSA are found to be −7.2 kcal mol−1 and −6.53 cal mol−1 K−1, respectively. Thus, the interaction of limonene with BSA is an exothermic reaction accompanied by negative ΔS value. Negative ΔS value suggests that the bound water to the protein molecule in or near the binding pockets was not disturbed and BSA is stabilized in the presence of limonene as further confirmed by CD and DSC.
 |
| | Fig. 3 van't Hoff plot for temperature dependence of Kb. Obtained from BSA fluorescence quenching by limonene at 15, 25 and 37 °C. | |
3.5 Energy transfer between BSA and limonene
A possibility of energy transfer between BSA and limonene was investigated for further confirmation of the proximity of binding. Fig. 4 shows the spectral overlap between the emission spectrum of BSA and the UV-absorption spectra of the limonene with the molar ratio of BSA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) limonene (donor
limonene (donor![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acceptor) as 1. R0 and r were calculated by using the J value of 7.74 × 10−16 cm3 M−1 and values obtained for the BSA–limonene complex are 1.6 and 2.3 nm, respectively. The energy transfer took place from BSA to limonene with significant possibility. The distance between the donor and the acceptor was on the scale of 2–8 nm that satisfies 0.5R0 < r < 1.5R0 in accordance with Förster’s non-radiative energy transfer theory.34,35 The range of r values does not exceed the dimensions of the protein (8 × 8 × 3 nm), which shows that the energy transfer from BSA to limonene is possible. This further justifies that the energy transfer between BSA and limonene contributes to the noticeable decrease of protein fluorescence intensity through static quenching mechanism.36 FRET results are listed in Table 4.
acceptor) as 1. R0 and r were calculated by using the J value of 7.74 × 10−16 cm3 M−1 and values obtained for the BSA–limonene complex are 1.6 and 2.3 nm, respectively. The energy transfer took place from BSA to limonene with significant possibility. The distance between the donor and the acceptor was on the scale of 2–8 nm that satisfies 0.5R0 < r < 1.5R0 in accordance with Förster’s non-radiative energy transfer theory.34,35 The range of r values does not exceed the dimensions of the protein (8 × 8 × 3 nm), which shows that the energy transfer from BSA to limonene is possible. This further justifies that the energy transfer between BSA and limonene contributes to the noticeable decrease of protein fluorescence intensity through static quenching mechanism.36 FRET results are listed in Table 4.
 |
| | Fig. 4 Fluorescence resonance energy transfer. Spectral overlap of the fluorescence emission of BSA and absorption spectra of limonene [BSA = limonene = 2 μM]. | |
3.6 Synchronous fluorescence spectroscopy studies
Synchronous fluorescence spectroscopy was used to measure the fluorescence quenching and also provide information about the conformational changes in the protein. The possible shift of the maximum emission wavelength λmax is related to the alteration of the polarity around the chromophore micro-environment,35 representing the value of difference between the excitation and emission wavelengths. When the values of Δλ are stabilized at 15 or 60 nm, the synchronous fluorescence gives the characteristic information of tyrosine and tryptophan residues, respectively.37 The synchronous fluorescence spectra of the BSA–limonene system are shown in Fig. 5. The maximum emission wavelength has negligible shift when Δλ is fixed at 15 nm. This expresses that the conformation of BSA was unchanged around tyrosine residue.35 However, in Fig. 5B, the maximum emission wavelength has a red shift in the presence of limonene when Δλ is fixed at 60 nm. This implied that limonene has more probability to cause conformational changes close to tryptophan residues than tyrosine.37 Moreover, the fluorescence intensity decreased regularly with the addition of limonene in both systems (Fig. 5A and B), which further depicts the occurrence of fluorescence quenching in the binding process.
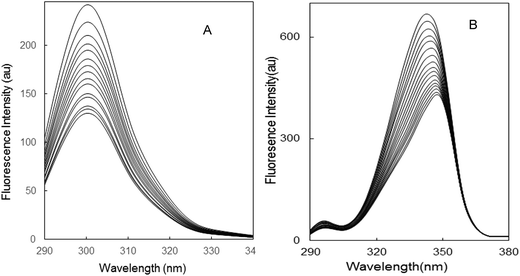 |
| | Fig. 5 Synchronous fluorescence spectra of BSA: (A) Δλ = 15 nm; (B) Δλ = 60 nm; [BSA = 2 μM; limonene = 0–30 μM]. | |
3.7 Three dimensional fluorescence spectroscopic analysis
To study the conformational changes in BSA upon addition of limonene (Fig. 6) three dimensional fluorescence spectra were measured and related parameters are shown in Table 2. Peak 1 is the Rayleigh scattering peak (λex = λem), peak 4 is the second order scattering peak (λex = 2λem). Peak 2 and peak 3 are the two typical fluorescence peaks. It is shown in the figure that both peak 2 (333.5 → 313.2 nm) and peak 3 (261 → 252.9 nm) of BSA were quenched by limonene with no shift of emission wavelength whereas Rayleigh scattering peaks were enhanced giving clear evidence of complex formation.32 Peak 2 mainly reflects the spectral behaviour of Trp, which is mainly caused by the transition of n → π* of aromatic amino acids in BSA. Peak 3 exhibits the spectral characteristics of polypeptide backbone (π → π* transition).38 All these results and analysis deciphered that the binding of limonene to BSA induced conformational and micro environmental changes.
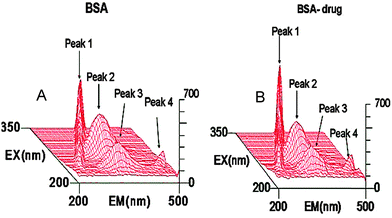 |
| | Fig. 6 Three-dimensional fluorescence spectra of BSA and the BSA–limonene system (A) BSA = 2 μM, limonene = 0; (B) BSA = 2 μM, limonene = 2 μM. | |
Table 2 Three dimensional fluorescence spectra characteristic parameters of the BSA and limonene–BSA system
| System |
Peak 2 (λex/λem) (nm/nm) |
Δλ (nm) |
Peak 2 (intensity) |
Peak 3 (λex/λem) (nm/nm) |
Δλ (nm) |
Peak 3 (intensity) |
| (A) BSA |
280/340 |
60 |
333.5 |
225/345 |
120 |
261 |
(B) BSA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) limonene (1 limonene (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
280/340 |
60 |
313.2 |
225/345 |
120 |
252.9 |
3.8 Circular dichroism measurement
Circular dichroism (CD) is an imperative technique in biological chemistry and structural biology especially for secondary structure determination.39 Spectra in the far-ultraviolet wavelength range (typically from ∼200 to 250 nm) provide information on the polypeptide backbone conformations of proteins. Secondary structural elements, such as α-helices, β-sheets, β turns, and random coil structures, all make bands of individual shapes and magnitudes in the far ultraviolet region. Due to the binding of ligands to globular proteins, the intermolecular forces liable for sustaining the secondary and tertiary structures can be rehabilitated triggering a conformational alteration of the protein.
In order to obtain an insight into the structure of BSA, the far-UV CD spectra were recorded in the presence and absence of limonene and are shown in Fig. 7. The CD spectrum of BSA exhibited two negative minima in the UV region at 208 and 222 nm, which is characteristic of the α-helix structure of the protein. The similar spectral features were noticed in our previous report.40 The binding of limonene to BSA leads to increase both (208 and 222 nm) of these negative minima peaks, clearly indicating the induction of the α-helix structure of the protein upon interaction with the limonene. Further, the CD spectra of BSA in the presence of limonene were found to be similar in shape, revealing that the structure of BSA is predominantly α-helix even after the addition of limonene. Using K2D software, the α-helicity of BSA was calculated. It increased from 62% to 66.80% in the presence of 30 μM limonene. Overall, it is clear from the spectra that BSA is more stabilized in the presence of 30 μM of limonene rather than 10 μM. Near UV CD spectra (250–320 nm) exhibited changes around 263 nm on the addition of limonene to BSA [Fig. 7B]. This indicated that both the secondary as well as tertiary structures of BSA are changed due to limonene binding.
 |
| | Fig. 7 (A) Secondary structural rearrangements. The far-UV CD spectra of [2 μM BSA only, 2 μM BSA + 10 μM limonene and 2 μM BSA + 30 μM limonene]. (B) Near-UV CD spectra of BSA (15 μM) with increasing concentration of limonene (0–225 μM). | |
3.9 Thermo stability study of limonene–BSA interaction by differential scanning calorimetry
Generally, ligand binding either stabilizes or destabilizes the proteins. DSC was employed to investigate the effect of limonene on the thermal stability of BSA. ΔTm and ΔH are the two main parameters obtained from DSC that give information about the effect of ligand binding on the thermal stability of the protein. Fig. 9 shows the DSC thermograms for BSA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) limonene in the molar ratios of 1
limonene in the molar ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 and 1
0 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15. BSA unfolds cooperatively and gives a single endothermic peak with melting temperature of 61.14 °C.41 It is observed that the thermal denaturation of BSA was found to be only partially reversible under the conditions of this study. Under saturating conditions limonene stabilized the BSA as evidenced by the escalation in melting temperature ΔTm by 3.0 °C also accompanied by an increase in enthalpy value. These results indicated that the binding stabilizes the protein structure linearly with the CD results.
15. BSA unfolds cooperatively and gives a single endothermic peak with melting temperature of 61.14 °C.41 It is observed that the thermal denaturation of BSA was found to be only partially reversible under the conditions of this study. Under saturating conditions limonene stabilized the BSA as evidenced by the escalation in melting temperature ΔTm by 3.0 °C also accompanied by an increase in enthalpy value. These results indicated that the binding stabilizes the protein structure linearly with the CD results.
3.10 Dynamic light scattering study
It was clear from the above mentioned investigations that upon interaction with limonene, BSA undergoes conformational changes. Conformational changes might be affected by the size of the protein molecules. Dynamic light scattering was used to examine the hydrodynamic radius of native BSA and the BSA–limonene complex. In Fig. 8, the hydrodynamic radii of native BSA and BSA in the presence of limonene were plotted and the observed data are shown in Table 3. Polydispersity is the parameter that describes the homogeneity of a solution.42 The value of hydrodynamic radius (3.7 nm) for native BSA is satisfactory with earlier reports.43 The hydrodynamic radii of BSA-limonene complex found to be lower than the BSA alone. The reduction in hydrodynamic radii upon ligand binding may be due to the “collapsing” of the protein as limonene binds with BSA. This response may result in shrinkage in the molecular volume due to a conformational alteration; similar results were reported for HSA in the presence of pollutants.44 The possible mechanism for the drop in protein hydrodynamic radius is that limonene disrupts the solvent shell around the BSA.
 |
| | Fig. 8 Hydrodynamic radii pattern of BSA in the absence and presence of limonene. | |
 |
| | Fig. 9 DSC thermograms of BSA and its complex with limonene (BSA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) limonene = 1 limonene = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15). 15). | |
Table 3 Hydrodynamic radii and polydispersity of BSA in the absence and presence of limonene
| Conditions |
R
h (nm) |
Pd (%) |
| (A) BSA |
3.7 ± 0.04 |
17.0 |
(B) BSA + limonene (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) 5) |
3.5 ± 0.03, 19.3 ± 0.35 |
11.7 |
(C) BSA + limonene (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15) 15) |
3.4 ± 0.03, 10.9 ± 0.2 |
11.0 |
Table 4 FRET parameters obtained from BSA–limonene binding
|
J (cm3 M−1) |
R
0 (nm) |
r (nm) |
E
FRET
|
| 7.74 × 10−16 |
1.6 |
2.3 |
0.1 |
Table 5 Molecular docking parameters obtained from BSA–limonene binding
| Binding site |
Amino acid residues |
Forces involved |
ΔG (kcal mol−1) |
K
b (M−1) |
| Site I |
Tyr149, Arg217 |
Hydrophobic |
−4.6 |
2.4 × 103 |
| Leu237, Arg256 |
| Leu259, Ala260 |
| Ile263, Ser286 |
| Ile289, Ala290 |
Two molar ratios of BSA and limonene, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 and 1
5 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15, were taken to observe the effect of limonene on protein dynamics, and the results indicated a continuous shrinkage in the hydrodynamic radius. As CD data suggested, limonene affects the secondary and tertiary structure of BSA. This also might be a reason that conformationally altered secondary structural components tends to decrease hydrodynamic radius.
15, were taken to observe the effect of limonene on protein dynamics, and the results indicated a continuous shrinkage in the hydrodynamic radius. As CD data suggested, limonene affects the secondary and tertiary structure of BSA. This also might be a reason that conformationally altered secondary structural components tends to decrease hydrodynamic radius.
3.11 Molecular docking study and binding displacement measurement using site markers
The molecular docking study was performed to further reveal the interaction of limonene with BSA. BSA comprises of three homologous domains, each domain made up of subdomains that possess common structural motifs. The principal regions of ligand binding to BSA are located in hydrophobic cavities in subdomains IIA and IIIA, which are consistent with Sudlow sites I and II, respectively.12 In the present study, Autodock 4.2.0 program was applied to calculate the possible conformation of the limonene that binds to the BSA. The best energy ranked results are summarized in Table 5. Fig. 10A and B show that limonene more favorably fits in the hydrophobic pocket close to Sudlow site I in domain IIA with ΔG and Kb of −4.64 kcal mol−1 and 2.4 × 103 M−1, respectively. Using the eqn (3), the binding constant (Kb) for protein–ligand interactions was calculated from the obtained free energy change of docking. The Tyr149, Arg217, Leu237, Arg256, Leu259, Ala260, Ile263, Ser286, Ile289, and Ala290 of site I are involved in hydrophobic interaction.45,46 Interaction parameters, including Kb and ΔG, agreed well with the results from the fluorescence quenching measurements. Therefore, molecular docking in this study yields useful information about the specific residues of BSA involved in the interactions with limonene for better understanding of the protein–ligand interaction at the molecular level. For further confirmation of the site involved in the binding of limonene with BSA, a displacement study was done by exploiting standard site markers, warfarin for site I47 and diazepam for site II. The Ksv value of BSA–limonene was 1.14 × 104 that decreases to 1.0 × 104 and 2.03 × 103 in the presence of diazepam and warfarin, respectively. These differences in Ksv values in the absence and presence of site markers are significant enough to deduce the binding site locations as reported in the literature.48,49 As evident from the above mentioned values, the Ksv of BSA–limonene decreased markedly in the presence of warfarin. It indicates that limonene binds to close to Sudlow site I in domain IIA of BSA as Ksv remained the same in the case of diazepam and decreased with warfarin.
 |
| | Fig. 10 Molecular docking results of BSA complexed with limonene (A) limonene is shown in a stick representation, and BSA represented with ribbon model (B) detailed view of the docking poses of BSA–limonene complex. | |
4. Conclusions
In the present work, we have computed the binding parameters and conformational alterations leading to an enhancement in the stability of the BSA–limonene complex using different spectroscopic, calorimetric and molecular docking methods. Putting all results together, it is to be concluded that limonene binds with BSA via a static quenching manner and the binding process is spontaneous and exothermic. DSC and CD results enlighten the limonene as a stabilizer of BSA, while molecular docking and displacement study reveal the binding site of limonene close to Sudlow site I in domain IIA of BSA. BSA as a drug carrier may aid in the delivery of limonene to an inflamed region and facilitate drug access.
Acknowledgements
Facilities provided by I. B. U, Aligarh Muslim University are gratefully acknowledged. S. K. C, E. A., J. M. K and P. A are highly thankful to the Council of Scientific and Industrial Research, New Delhi, for financial assistance in the form of senior research fellowship (SRF) and junior research fellowship (JRF).
References
- B. Eyheraguibel, C. Richard, G. Ledoigt and A. Ter Halle, J. Agric. Food Chem., 2011, 59, 4868–4873 CrossRef CAS PubMed.
- T. Kaur, K. Hussain, S. Koul, R. Vishwakarma and D. Vyas, PLoS One, 2013, 8, e69112 CAS.
- G. Zolese, G. Falcioni, E. Bertoli, R. Galeazzi, M. Wozniak, Z. Wypych, E. Gratton and A. Ambrosini, Proteins, 2000, 40, 39–48 CrossRef CAS.
- H. Xu, Q. Liu and Y. Zuo, J. Solution Chem., 2009, 38, 15–25 CrossRef CAS.
- H.-X. Zhang and P. Mei, J. Solution Chem., 2009, 38, 351–361 CrossRef CAS.
- Y. Q. Wang, H. M. Zhang, G. C. Zhang, W. H. Tao, Z. H. Fei and Z. T. Liu, J. Pharm. Biomed. Anal., 2007, 43, 1869–1875 CrossRef CAS PubMed.
- N. Zhou, Y.-Z. Liang and P. Wang, J. Photochem. Photobiol., A, 2007, 185, 271–276 CrossRef CAS PubMed.
- C. Bertucci, S. Cimitan, A. Riva and P. Morazzoni, J. Pharm. Biomed. Anal., 2006, 42, 81–87 CrossRef CAS PubMed.
- Y.-J. Hu, Y. Liu, R.-M. Zhao, J.-X. Dong and S.-S. Qu, J. Photochem. Photobiol., A, 2006, 179, 324–329 CrossRef CAS PubMed.
- Y.-P. Wang, Y.-L. Wei and C. Dong, J. Photochem. Photobiol., A, 2006, 177, 6–11 CrossRef CAS PubMed.
- U. Kragh-Hansen, Pharmacol. Rev., 1981, 33, 17–53 CAS.
- D. C. Carter, B. Chang, J. X. Ho, K. Keeling and Z. Krishnasami, Eur. J. Biochem., 1994, 226, 1049–1052 CrossRef CAS PubMed.
-
J. R. Lakowicz, Principles of Fluorescence Spectroscopy, Springer, New York, 3rd edn, 2008 Search PubMed.
- Y. J. Hu, W. Li, Y. Liu, J. X. Dong and S. S. Qu, J. Pharm. Biomed. Anal., 2005, 39, 740–745 CrossRef CAS PubMed.
- C. Dufour and O. Dangles, Biochim. Biophys. Acta, 2005, 1721, 164–173 CrossRef CAS PubMed.
- S. Bi, D. Song, Y. Tian, X. Zhou, Z. Liu and H. Zhang, Spectrochim. Acta, Part A, 2005, 61, 629–636 CrossRef PubMed.
- Y. Q. Wang, H. M. Zhang and G. C. Zhang, J. Pharm. Biomed. Anal., 2006, 41, 1041–1046 CrossRef CAS PubMed.
- X. F. Liu, Y. M. Xia and Y. Fang, J. Inorg. Biochem., 2005, 99, 1449–1457 CrossRef CAS PubMed.
- M. I. Kaldas, U. K. Walle, H. van der Woude, J. M. McMillan and T. Walle, J. Agric. Food Chem., 2005, 53, 4194–4197 CrossRef CAS PubMed.
- X. Long, S. Liu, L. Kong, Z. Liu and S. Bi, Talanta, 2004, 63, 279–286 CrossRef CAS PubMed.
- A. Papadopoulou, R. J. Green and R. A. Frazier, J. Agric. Food Chem., 2005, 53, 158–163 CrossRef CAS PubMed.
- J. Namiesnik, K. Vearasilp, A. Nemirovski, H. Leontowicz, M. Leontowicz, P. Pasko, A. L. Martinez-Ayala, G. A. Gonzalez-Aguilar, M. Suhaj and S. Gorinstein, Appl. Biochem. Biotechnol., 2014, 172, 2849–2865 CrossRef CAS PubMed.
- F. Sheng, Y. Wang, X. Zhao, N. Tian, H. Hu and P. Li, J. Agric. Food Chem., 2014, 62, 6813–6819 CrossRef CAS PubMed.
- Y. W. Kim, M. J. Kim, B. Y. Chung, Y. Bang du, S. K. Lim, S. M. Choi, D. S. Lim, M. C. Cho, K. Yoon, H. S. Kim, K. B. Kim, Y. S. Kim, S. J. Kwack and B. M. Lee, J. Toxicol. Environ. Health, Part B, 2013, 16, 17–38 CAS.
- W. F. Hink and B. J. Fee, J. Med. Entomol., 1986, 23, 400–404 CAS.
- H. Igimi, T. Hisatsugu and M. Nishimura, Am. J. Dig. Dis., 1976, 21, 926–939 CrossRef CAS.
- T. Förster, Ann. Phys., 2, 1948, 473, 55–75 CrossRef.
- Q. Wang, X. Liu, M. Su, Z. Shi and H. Sun, New J. Chem., 2014 10.1039/c4nj00327f.
- E. Ahmad, P. Sen and R. H. Khan, Cell Biochem. Biophys., 2011, 61, 313–325 CrossRef CAS PubMed.
- N. Jiang, C. Yang, X. Dong, X. Sun, D. Zhang and C. Liu, Org. Biomol. Chem., 2014, 12, 5250–5259 CAS.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, J. Comput. Chem., 2009, 30, 2785–2791 CrossRef CAS PubMed.
- M. Ishtikhar, S. Khan, G. Badr, A. Osama Mohamed and R. Hasan Khan, Mol. BioSyst., 2014, 10, 2954–2964 RSC.
- S. R. Feroz, S. B. Mohamad, N. Bujang, S. N. A. Malek and S. Tayyab, J. Agric. Food Chem., 2012, 60, 5899–5908 CrossRef CAS PubMed.
- Y.-J. Hu, Y. Liu, L.-X. Zhang, R.-M. Zhao and S.-S. Qu, J. Mol. Struct., 2005, 750, 174–178 CrossRef CAS PubMed.
- A. Varlan and M. Hillebrand, Molecules, 2010, 15, 3905–3919 CrossRef CAS PubMed.
- Z. Chi, R. Liu, Y. Teng, X. Fang and C. Gao, J. Agric. Food Chem., 2010, 58, 10262–10269 CrossRef CAS PubMed.
- A. Bhogale, N. Patel, J. Mariam, P. M. Dongre, A. Miotello and D. C. Kothari, Colloids Surf., B, 2014, 113, 276–284 CrossRef CAS PubMed.
- S. Sugio, A. Kashima, S. Mochizuki, M. Noda and K. Kobayashi, Protein Eng., 1999, 12, 439–446 CrossRef CAS PubMed.
- P. Kumar, B. Baidya, S. K. Chaturvedi, R. H. Khan, D. Manna and B. Mondal, Inorg. Chim. Acta, 2011, 376, 264–270 CrossRef CAS PubMed.
- J. M. Khan, S. A. Abdulrehman, F. K. Zaidi, S. Gourinath and R. H. Khan, Phys. Chem. Chem. Phys., 2014, 16, 5150–5161 RSC.
- A. Y. Khan, M. Hossain and G. S. Kumar, Mol. Biol. Rep., 2013, 40, 553–566 CrossRef CAS PubMed.
- M. Yu, Z. Ding, F. Jiang, X. Ding, J. Sun, S. Chen and G. Lv, Spectrochim. Acta, Part A, 2011, 83, 453–460 CrossRef CAS PubMed.
- J. M. Khan, S. K. Chaturvedi and R. H. Khan, Biochem. Biophys. Res. Commun., 2014, 441, 681–688 CrossRef PubMed.
- E. Ahmad, G. Rabbani, N. Zaidi, B. Ahmad and R. H. Khan, PLoS One, 2012, 7, e38372 CAS.
- B. Yang, F. Hao, J. Li, K. Wei, W. Wang and R. Liu, Food Chem. Toxicol., 2014, 65, 227–232 CrossRef CAS PubMed.
- X. Tan and Z. Song, RSC Adv., 2014, 4, 3263–3271 RSC.
- J. Tian, J. Liu, Z. Hu and X. Chen, Bioorg. Med. Chem., 2005, 13, 4124–4129 CrossRef CAS PubMed.
- N. Zaidi, E. Ahmad, M. Rehan, G. Rabbani, M. R. Ajmal, Y. Zaidi, N. Subbarao and R. H. Khan, J. Phys. Chem. B, 2013, 117, 2595–2604 CrossRef CAS PubMed.
- Y. Song, Y. Liu, W. Liu, F. A. Villamena and J. L. Zweier, RSC Adv., 2014, 4, 47649–47656 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4mb00548a |
|
| This journal is © The Royal Society of Chemistry 2015 |
Click here to see how this site uses Cookies. View our privacy policy here. ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) L = 1
L = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were taken and spectra were measured.
1) were taken and spectra were measured.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kb + n
Kb + n![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log[Q]
log[Q]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln
ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kb
Kb


![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 ratio of limonene were performed from 25 to 90 °C at a scan rate of 0.5 °C min−1. Data was analyzed using the Origin software provided with the instrument to obtain the temperature at the midpoint of the unfolding transition (Tm) and calorimetric enthalpy (ΔH°).
15 ratio of limonene were performed from 25 to 90 °C at a scan rate of 0.5 °C min−1. Data was analyzed using the Origin software provided with the instrument to obtain the temperature at the midpoint of the unfolding transition (Tm) and calorimetric enthalpy (ΔH°).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min and were filtered serially through 0.22 and 0.02 μm Whatman syringe filters directly into a 12 μl quartz cuvette. For each experiment, 20 measurements were taken. Mean hydrodynamic radius (Rh) and polydispersity were analysed using Dynamics 6.10.0.10 software at optimized resolution. The Rh was estimated on the basis of an autocorrelation analysis of scattered light intensity data based on translation diffusion coefficient by the Stokes–Einstein relationship:
000 rpm for 10 min and were filtered serially through 0.22 and 0.02 μm Whatman syringe filters directly into a 12 μl quartz cuvette. For each experiment, 20 measurements were taken. Mean hydrodynamic radius (Rh) and polydispersity were analysed using Dynamics 6.10.0.10 software at optimized resolution. The Rh was estimated on the basis of an autocorrelation analysis of scattered light intensity data based on translation diffusion coefficient by the Stokes–Einstein relationship:
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500
500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. The 10 best solutions based on docking score were retained for further analysis. Discovery studio 3.5 was used for visualization and for the identification of residues involved in binding. Binding displacement studies between limonene and BSA in the presence of two site markers, warfarin (for site I) and diazepam (for site II) were measured using the fluorescence titration method. The titration of limonene was carried out on the solution having protein and site markers in the ratio of 1
000. The 10 best solutions based on docking score were retained for further analysis. Discovery studio 3.5 was used for visualization and for the identification of residues involved in binding. Binding displacement studies between limonene and BSA in the presence of two site markers, warfarin (for site I) and diazepam (for site II) were measured using the fluorescence titration method. The titration of limonene was carried out on the solution having protein and site markers in the ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and the Ksv values were calculated by using eqn (1).
1 and the Ksv values were calculated by using eqn (1).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) limonene (donor
limonene (donor![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acceptor) as 1. R0 and r were calculated by using the J value of 7.74 × 10−16 cm3 M−1 and values obtained for the BSA–limonene complex are 1.6 and 2.3 nm, respectively. The energy transfer took place from BSA to limonene with significant possibility. The distance between the donor and the acceptor was on the scale of 2–8 nm that satisfies 0.5R0 < r < 1.5R0 in accordance with Förster’s non-radiative energy transfer theory.34,35 The range of r values does not exceed the dimensions of the protein (8 × 8 × 3 nm), which shows that the energy transfer from BSA to limonene is possible. This further justifies that the energy transfer between BSA and limonene contributes to the noticeable decrease of protein fluorescence intensity through static quenching mechanism.36 FRET results are listed in Table 4.
acceptor) as 1. R0 and r were calculated by using the J value of 7.74 × 10−16 cm3 M−1 and values obtained for the BSA–limonene complex are 1.6 and 2.3 nm, respectively. The energy transfer took place from BSA to limonene with significant possibility. The distance between the donor and the acceptor was on the scale of 2–8 nm that satisfies 0.5R0 < r < 1.5R0 in accordance with Förster’s non-radiative energy transfer theory.34,35 The range of r values does not exceed the dimensions of the protein (8 × 8 × 3 nm), which shows that the energy transfer from BSA to limonene is possible. This further justifies that the energy transfer between BSA and limonene contributes to the noticeable decrease of protein fluorescence intensity through static quenching mechanism.36 FRET results are listed in Table 4.



![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) limonene (1
limonene (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1)
1)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) limonene in the molar ratios of 1
limonene in the molar ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 and 1
0 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15. BSA unfolds cooperatively and gives a single endothermic peak with melting temperature of 61.14 °C.41 It is observed that the thermal denaturation of BSA was found to be only partially reversible under the conditions of this study. Under saturating conditions limonene stabilized the BSA as evidenced by the escalation in melting temperature ΔTm by 3.0 °C also accompanied by an increase in enthalpy value. These results indicated that the binding stabilizes the protein structure linearly with the CD results.
15. BSA unfolds cooperatively and gives a single endothermic peak with melting temperature of 61.14 °C.41 It is observed that the thermal denaturation of BSA was found to be only partially reversible under the conditions of this study. Under saturating conditions limonene stabilized the BSA as evidenced by the escalation in melting temperature ΔTm by 3.0 °C also accompanied by an increase in enthalpy value. These results indicated that the binding stabilizes the protein structure linearly with the CD results.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5)
5)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15)
15)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 and 1
5 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15, were taken to observe the effect of limonene on protein dynamics, and the results indicated a continuous shrinkage in the hydrodynamic radius. As CD data suggested, limonene affects the secondary and tertiary structure of BSA. This also might be a reason that conformationally altered secondary structural components tends to decrease hydrodynamic radius.
15, were taken to observe the effect of limonene on protein dynamics, and the results indicated a continuous shrinkage in the hydrodynamic radius. As CD data suggested, limonene affects the secondary and tertiary structure of BSA. This also might be a reason that conformationally altered secondary structural components tends to decrease hydrodynamic radius.





