Affinity proteomics led identification of vimentin as a potential biomarker in colon cancers: insights from serological screening and computational modelling†
Shoiab
Bukhari
a,
Taseem A.
Mokhdomi
a,
Naveed A.
Chikan
a,
Asif
Amin
a,
Hilal
Qazi
a,
Sajad H.
Wani
a,
Asrar H.
Wafai
a,
Sumira
Tyub
b,
Farhat
Mustafa
c,
Masood S.
Mir
d,
Nisar A.
Chowdri
e and
Raies A.
Qadri
*a
aDepartment of Biotechnology, University of Kashmir, Srinagar, J & K-190006, India. E-mail: raies@kashmiruniversity.ac.in; Fax: +91-1942428723; Tel: +91-9419001315
bCenter of Research and Development, University of Kashmir, Srinagar, J & K-190006, India
cDepartment of Pathology, Sher-i-Kashmir Institute of Medical Sciences, Srinagar, J & K-190011, India
dRabbit House Unit, Department of Pathology, SKAUST-K, Srinagar, J & K-190011, India
eDepartment of General Surgery, Sher-i-Kashmir Institute of Medical Sciences, Srinagar, J & K-190011, India
First published on 2nd October 2014
Abstract
Proteomic analysis using multiplex affinity reagents is perhaps the most reliable strategy to capture differentially expressed proteins that are slightly or immensely modified. In addition to expressional variation, it is comprehensively evident that the immunogenicity of a protein can be a deciding factor for instigating an inflammation afflicted-carcinogenesis. Considering both these factors, a simple and systematic strategy was designed to capture the immunogenic cancer biomarkers from sera of colorectal cancer patients. The affinity reagent, in the form of an antibody repertoire against the secretome of the HT29 cell line was used to grade the sera samples on the basis of the degree of immuno-reactivity and to capture differentially expressed antigens from the patient sera. Following affinity based 2DE-MALDI-TOF; the proteins were identified as (1) soluble vimentin; and (2) TGF-beta-inhibited membrane-associated protein (PP16B), in colon cancer sera and (3) keratin, type II cytoskeletal protein in rectal cancer sera. Pathway reconstruction and protein–protein networking of identified proteins predicted only Vimentin to be physically and genetically engaged in close proximity with the most established colorectal cancer associated tumorigenic pathways. Furthermore, our findings suggest that a possible surface stoichiometric shift in the structure of protein could be due to mutations in the coding sequence of Vimentin that may elicit its enhanced secretion possibly due to protein-hyperphosphorylation. Of the three proteins identified, only Vimentin showed higher expression in sera of colon cancer patients alone. Thus, it could be argued that vimentin might help in predicting individuals at higher risk of developing colon cancers. Our data are therefore suggestive of using vimentin as an antigen for tumor vaccination in an autologous set-up for colon cancers.
Introduction
In the application for biomarker discovery in colorectal cancer (CRC), both two dimensional gel electrophoresis (2DE) coupled with mass spectrometry (MS) as well as label free quantitative MS have helped to develop a database of candidate cancer biomarkers.1,2 But affinity based proteomics is gathering increased attention as an indispensable tool to be applied within the interdisciplinary network of cancer research.3 This is mainly due to the fact that affinity reagents, commonly antibodies serve as a precise tool for the specific strategy that involves immuno-capturing of relevant cancer biomarkers. Affinity-based proteomics involves the development of affinity reagents to be used for systematic exploration of the proteome. Creating affinity-based reagents to the human proteome theoretically means having a wide range of binders (antibodies) that would be needed in order to achieve complete coverage of the proteome.3–5 Large numbers of Protein Affinity Reagents (PAR) are available commercially for cancer diagnosis.6,7 Immunotherapy has dramatically changed the outcome of many tumors. The theory behind specific immunotherapy is to use vaccines to activate a unique lymphocyte (such as a B cell or T cell) response, which will have an immediate anti-tumor effect and also a “memory response” to help fight future tumor challenges.8,9 The source of vaccine in principle is either the “whole cell lysate” of autologous/allogenic tumor cells or the tumor associated antigen itself.10–13 Autologous tumor cells are an obvious source of tumor associated antigens (TAA) for vaccination purposes, since, by definition, all relevant candidates TAA should be contained within them. Some of these “active” specific vaccines are made either from whole tumor cell preparations or tumor cell membranes.13–17 Based on this concept, Hoover et al. conducted a clinical trial in which patients with stage II/III colorectal cancer were vaccinated with irradiated autologous tumor cells and BCG, randomized versus surgery alone.18–22 Since the repertoire of polyclonal antibodies generated in a process makes the 1st line of defense as an anti-tumor response when “whole cell lysates” are used for vaccination purpose, it was therefore realized to make use of these antibodies to capture the immunogenic TAA present in the sera of CRC patients. While putting forward the strategy to establish an effective platform for coverage of candidate biomarkers essentially TAA from sera of CRC patients, we utilized the “anti-secretory fraction” antibody repertoire raised in a rabbit against the secretome of the human colon adenocarcinoma cell line, HT29 (instead of whole cell lysate) as an affinity reagent for immuno-capturing possible TAA present in the sera of CRC patients followed by their characterization using 2DE and MS/MALDI-TOF.Materials and methods
Cell lines, media selection and secretome collection
Colon cancer cell line cultures of HT29, Colo 205, Colo 320DM, and HCT-15 (all of them are colon adenocarcinoma) were maintained in DMEM/RPMI (Gibco) supplemented with 10% FBS (Gibco) at 5% CO2 under standard cell culture conditions. To allow media selection, cells were alternatively cultured in various media preparation viz. 10% FBS supplemented media, 1% BSA supplemented media, serum free media (SFM), transferrin supplemented SFM, or insulin supplemented SFM. The best media were the ones which caused optimal growth and minimal cell death as determined by LDH assays.23 For secretome analysis, cells were initially grown in standard culture medium (supplemented with 10% FBS) and after reaching 70% confluency, they were shifted to supplemented or serum free medium and allowed to grow for 24–48 hours depending upon the experimental design. After 24 or 48 hours, the culture supernatant was collected, centrifuged, filtered to remove debris and stored at −20 °C until use.Immunization and purification of polyclonal antibodies
New Zealand rabbits were immunized with conditioned and concentrated secretory fractions (pre-cleared from abundant proteins using Agilent Multi Affinity Resin, HU-7 Plasma) of HT29 cells in accordance with the established guidelines (see ethical statement). The primary immunization consisted of 300 mg of the secretory fraction of HT29 cells in Freund's complete adjuvant administered subcutaneously and followed by three booster injections of 100 mg each in Freund's incomplete adjuvant with 4 week intervals. Polyclonal fractions from antisera of immunized animals were purified on a protein-G column using established protocols. Briefly, 5 ml of total antisera was buffered with PBS and filtered using a 0.22 mm pore-size filter (Millipore). The precleared antisera were allowed to flow through the protein-G column. The unbound material was washed away with 1 × PBST (16PBS, 0.1% Tween20, pH 7.25) and the captured antibodies were eluted using a low pH glycine buffer (0.2 M glycine, pH 2.5). The eluted antibody fraction was collected and neutralized with TBS BUFFER, pH 7.4, supplemented with glycerol and NaN3 to a final concentration of 50% and 0.02%, respectively, for long-term storage at −20 °C.Immunoprecipitation of sera antigens
Immunoprecipitation of antigens from a patient sera and normal sera (pre-cleared from abundant proteins using Agilent Multi Affinity Resin, HU-7 Plasma) was carried out as follows: 20 mg of proteins from patient and normal sera was pre-cleared by incubating with 200 μl of protein G beads in microfuge tubes with gentle rocking for 1 hour at room temperature to remove proteins bound nonspecifically to protein G. Beads were pelleted down and the supernatant (pre-cleared sera) was collected for further use. Meanwhile the polyclonal fraction of anti-secretome antibodies (anti-Sc antibody repertoire) was immobilized on 50 μl of protein G beads, by adding 20 μl patient sera and normal sera separately in two microfuge tubes and kept at room temperature for 2 hours with gentle rocking. Beads were then washed thrice with wash buffer and were subsequently incubated with pre-cleared sera overnight at 4 °C with gentle rocking. Following incubation, the supernatant was removed. Beads were washed three times with 1 ml wash buffer each. Antibodies and bound proteins were mildly eluted three times consecutively using 40 μl elution buffer (100 mM Glycine, pH 2.5) and neutralized with 10 μl of 1 M Tris pH 8.8. The eluates were then stored at −80 °C until further use.Two-dimensional differential gel electrophoretic analysis of immunocaptured antigens
50 μg of protein antigens (immunocaptured from patient/normal sera) were resuspended in 140 μl of two-dimensional sample buffer (8 M urea, 2 M thiourea, 4% (w/v) CHAPS, 65 mM DTT, 1% pharmalytes 3–10 pH, pH 8.5) and incubated for 30 min at room temperature. The protein solution was loaded onto the IEF tray and active rehydration was carried out at 20 °C overnight using immobilized nonlinear strip gels (pH 3–10, 11 cm) on Biorad IEF CELL under a layer of mineral oil. IEF was performed using stepwise increase in voltage and running times: 500 V for 1 hour, then 2000 V for 2 h, 4000 V for 4 h and finally 8000 V for 4 h. Focused strips were equilibrated for 15 min in 50 mM Tris-HCl, pH 8.8, containing 6 M urea, 30% (v/v) glycerol, 2% (w/v) SDS, and 1% (w/v) DTT, followed by incubation for 15 min in the same buffer lacking DTT, but containing 2.5% (w/v) iodoacetamide. Equilibrated IPG strips were placed on 10% SDS-gels, and sealed with a 1% (w/v) agarose solution. 2nd dimension was carried out at 20 mA for 1 h and 40 mA per gel until the run was complete, using the Hoefer midi gel format. The gels were captured using a GS-800 gel scanner (BioRad, Hertfordshire, UK). The differential spots were identified manually and sent to CDFD, Hyderabad, and the Biophysics Center, IISc, Bangalore (India) individually for protein characterization using mass spectrometry. Briefly, manually excised spots were subjected to in-gel trypsination (Promega) as described.25 Peptides were desalted as described33 and mixed with a-cyano-4-hydroxycinnamic acid (CHCA) prior to MALD-TOF/TOF mass spectrometry (Ultraflex III, Bruker). MS and MS/MS data were used in subsequent searches by using the MSDB protein sequence database for human proteins.Bioinformatic tools for protein search and network reconstruction
The 3D model of proteins was built by using an i-tasser server. For protein search, SNP hits and pathway networking following major databases were used like NCBI, UniProtKB/Swiss-Prot, ClinVar, dbSNP, GenomeNet, CaSNP and COSMIC. In addition, the functional protein association networks or protein interactions were searched using STRING database (http://string-db.org/). The combining pathway databases of BioCarta (http://www.biocarta.com/genes/index.asp), and the PubMed literature (http://www.ncbi.nlm.nih.gov/pubmed/) were also used to search the correlated regulatory pathways. The GeneMania 3.1.2 (http://www.genemania.org/) was applied thoroughly for compiling protein functional classification, physical and genetic interaction networks.Molecular dynamics simulation
The MDS studies were performed using the Gromacs 4.5.3 package.26 For wt VIM, the modeled structure was used as a starting structure for MDS. The Accelrys Discovery Studio27 was used to make two single point mutations on the wild type structure. All structures were applied with the GROMOS96 43a1 force field and then placed in a model of a pre-equilibrated water bath, and counter-ions were added to achieve a neutral box using the “genion” tool that accompanies the gromacs package. The solvent molecules were restrained to the original position with a force constraint of 100 kcal mol−1 for 5000 steps before being subjected to energy minimization for 5000 iterations. For regulating the temperature inside the box Berendsen temperature coupling method was used.28 Electrostatic interactions were computed using the Particle Mesh Ewald method.29The ionizing state of the residues, the pressure and other parameters were set in the standard range. The non-bonded pair list was updated every 10 steps and conformations were stored every 2 picoseconds (ps). Position restraint simulation for 500 ps was implemented to allow solvent molecules to enter the cavity region of the structure. Finally, the system was subjected to MDS for 30 nanoseconds (ns). Root mean square deviation (RMSD), root mean square fluctuation (RMSF), solvent accessible surface area (SASA), radius of gyration (Rg) and PCA were carried out by using inbuilt gromacs tools. g-hbond was used to calculate the number of distinct hydrogen bonds formed by specific residues to other amino acids within the protein during the simulation (NH bond). g_sham was used extensively to obtain a free energy landscape. The graphs were plotted using the Grace GUI toolkit 5.1.22 version. All the visualizations were carried out using Pymol, Ligplus, VMD30,31 and graphs were plotted using the Grace Program and GNUPlot.32 The trajectories were analyzed using the inbuilt tool in the GROMACS distribution.Ethical statement
The protocols/experiments involving the use of human specimens were duly examined and approved by Institutional Ethics Committee (IEC), Sher-i-Kashmir Institute of Medical Sciences, Srinagar (SIMS 1 24/IEC-SKIMS/2013). The experimentation involving the use of laboratory animals was approved by Institutional Animal Ethics Committee, Faculty of Veterinary Sciences and Animal Husbandry, SKUAST-K and conforms to the Guidelines for Care and Use of Laboratory Animals in Scientific Research (INSA 2000), Committee for the Purpose of Control and Supervision on Experiments on Animals (ICMR, 2000).Results
Secretome selection for development of an affinity reagent
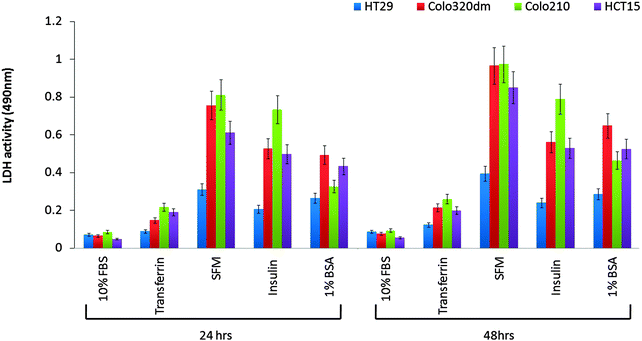
A key determinant for our selection of the secretory protein fraction from various Human adenocarcinoma cell lines was to obtain the secretome fractions with no or little cytoplasmic protein contaminants. The cytoplasmic proteins would leak out in the media only in the case of cell death or when cells are under extreme stress. Therefore the crucial step in secretome preparation was the selection of the cell line and media. Human colon cancer cell lines like HT29, HCT-15, COLO320 DM and COLO210 were grown in 10% FBS-Media or serum free media with additive factors like transferrin, insulin or 1% BSA separately. In order to allow cells adapt to serum free media, the percentage of the serum in the conditioned medium was gradually reduced and it was found that transferrin supplemented DMEM was the best for cell lines over a period of 48 hours compared to 1% BSA supplemented DMEM, insulin supplemented DMEM or serum free media. Cells were allowed to grow for 48 h and subsequently secretary fractions were collected at 24 h and 48 h from all the media preparations. LDH levels measured in all the secretome preparations indicated that in the case of the HT29 cell line there was no or little cell death in 10% FBS supplemented media followed by transferrin supplemented media at 24 h as well as 48 h (Fig. 1). Since FBS in principle is also considered as a contaminant protein mixture in a representative secretome, therefore we selected the transferrin supplemented media for secretome preparation that would only contain transferrin and the fraction of secretory proteins of a particular cell line. Following selection of media, all the cell secretomes were analyzed by SDS-PAGE to examine the band profile across different cell lines (Fig. S1, ESI†). The band profiles correlated with the LDH assay based cell death patterns, indicating leakage of cytoplasmic protein. We could understand that those secretory protein preparations that had previously displayed higher levels of LDH had more number of protein bands on the gel. Therefore, based on the LDH levels and secretome analysis, the secretory fraction of HT29 collected at 24 h was concentrated and used for immunization purpose for the development of the affinity reagent.Evaluation of affinity reagent
For the development of the affinity reagent, antibodies were raised against the HT29 secretome harvested after 24 h. Since the cancer cell secretome is considered to be the reservoir of tumor associated antigens (TAA), theoretically we attempted to generate the repertoire of anti-TAA antibodies that could be further used to affinity-capture the immunogenic cancer biomarker from the sera of the patients. One of the limiting factor while adopting this methodical approach is the specificity of generated antibody repertoire. As a proof of concept, the affinity reagent (anti-Sc) was probed with cell lysates and secretomes of different cell lines to make sure that the immune response generated against the secretory proteins of HT29 cells was at least partially discriminative. It was observed that the antibody in an unbiased manner picked up bands from all the cell line lysates and secretomes, however, relatively more prominent bands were picked up from the HT29 cell lysate and secretome compared to A549 and THP-1 lysates (Fig. 2). Keeping in view that all cell lysates would be having similar domains to a certain extent, we expected less specificity for our affinity reagent, however, surprisingly the qualitative observation demonstrated that the level of specificity was much more synchronized with the HT92 secretome as depicted from the intensity and the number of bands that were visualized on the blot.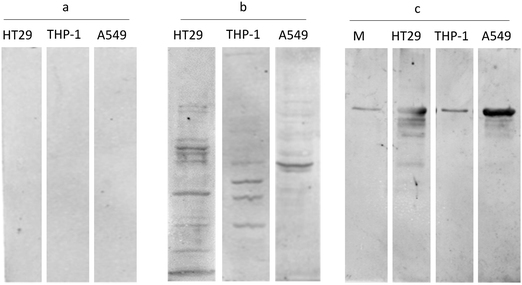 | ||
Fig. 2 Western blot analysis24 for evaluating the affinity reagent specificity: HT29, THP-1 and A549 cell lysates probed with (a) pre-immune sera, (b) anti-secretome antisera at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 dilution, and (c) HT29, THP-1 and A549 cell secretome probed with anti-secretome antisera at 1 200 dilution, and (c) HT29, THP-1 and A549 cell secretome probed with anti-secretome antisera at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 dilution. HT29 cell lysate and secretome showing prominent and more number of bands compared to other cell lysates, indicating that the antibody fraction has specificity and sensitivity towards HT29 cell lysate. M indicates the media control showing a single band of transferrin (added as a supplement in the media). 200 dilution. HT29 cell lysate and secretome showing prominent and more number of bands compared to other cell lysates, indicating that the antibody fraction has specificity and sensitivity towards HT29 cell lysate. M indicates the media control showing a single band of transferrin (added as a supplement in the media). | ||
Selection of immunocompetent patient sera as a source of tumor associated antigen
The selection of patient sera was determined by quantifying the reactivity of patient sera while probing with anti-Sc antibody. As hypothesized, the repertoire of polyclonal antibodies (anti-Sc antibody) that were raised against the secretome of the HT29 cell line may give us the signature pattern of highly immunogenic tumor associated antigens or that might have been altered/over expressed in tumor cells. Keeping this in view, we screened the sera of colorectal patients (n = 41), controls (n = 20), and sera from patients with gut-associated diseases other than cancer [mesenteric lymphadenitis (n = 3), ulcerative colitis (n = 2)] for measuring their respective reactivity when probed with anti-Sc antibody. Based on the reactivity of sera, we were able to catalogue sera into three discrete groups, i.e. TS1 (all tumor sera) showing the highest seroreactivity, followed by, TS2 (all tumor sera) and NST (normal/diseased as well as tumor sera) groups reflecting a low sero-reactivity (Fig. S2, ESI†). Surprisingly, all the tumor sera in the TS1 group had raised CEA levels (>20 ng ml−1), while in the case of the NST group the CEA levels varied from 2–8 ng ml−1. It was evident from our results that all the tumor sera samples having CEA levels more than 20 ng ml−1 showed the highest-reactivity while probed with affinity reagent (anti-Sc antibody.). It would be worth mentioning that TS2 and NST groups had none of the tumor samples (sera) that were clinically reported as metastatic in nature. Therefore, it was concluded that sero-reactivity varied due to the presence of antibodies in affinity reagent (anti-Sc) against certain antigens/cancer biomarkers that were present in aggressive types of colon/rectal serum samples. In order to characterize these antigens, serum samples of the TS1 group comprising of 10 Tumor serum samples were selected for immuno-capturing.Immuno-capturing and identification of antigens from the sera of colorectal cancer patients
In order to capture the immunogenic tumor associated proteins, we implemented an immuno-precipitation methodology to identify those antigens that would only be present in tumor sera compared to the normal sera. We kept a constant immuno-capturing tool as immobilized anti-Sc antibody that was previously raised against the secretome of HT29 cells. This platform provided us a logical strategy for effective implementation of antibody based proteomics wherein the capturing tool (affinity reagent- anti-Sc antibody) would remain same for normal as well as tumor samples. In practice, we incubated pre-cleared normal and tumor sera separately with this affinity reagent followed by the elution of the antibody–antigen complex. Later on, 2DE was applied to eluted fractions to establish a comparative profile of immunogenic tumor associated antigens present in tumor sera as well as in normal sera. Differential protein spots were picked and sent for identification by peptide mass fingerprinting (PMF). Though there were many spots that were differentially expressed, but to make this study error free and target oriented, only those protein spots were further analyzed that were exclusively present in tumor sera but absent in normal sera (Fig. 3). These spots were excised and subjected to in-gel tryptic digestion and the resulting peptides were analyzed by MALDI-TOF-MS/MS. The acquired spectra were processed and searched against a nonredundant SwissProt protein sequence database using the Mascot and other search engines (Fig. S3–S5, ESI†). Subsequently, 3 proteins were identified- (1) soluble vimentin (53.6 kDa, pI 5.06) and (2) Keratin, type II cytoskeletal protein (67 kDa, pI 8.1) in colon cancer sera; and (3) TGF-beta-inhibited membrane-associated protein (PP16B; 63,551 Da, PI 6.06) in rectal cancer sera.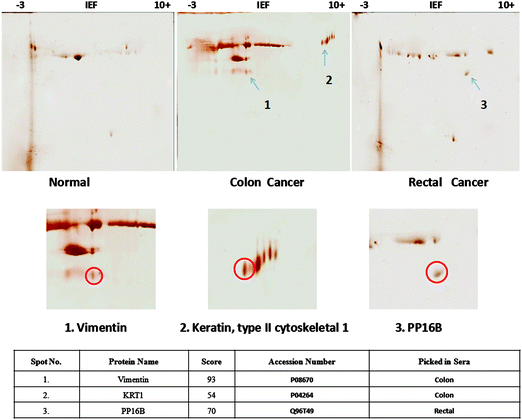 | ||
| Fig. 3 Two-dimensional gels display differential protein spots precipitated after incubation of pooled sera with anti-Sc antibody (affinity reagent). Spots encircled were identified as vimentin, KRT1 and PP16B by MALDI-TOF MS/MS. See Fig. S7 (ESI†) for the 2DE gel display of anti-Sc antibody only (reagent control). | ||
Pathway reconstruction and network analysis
Pathways and network interaction profiles that involved proteins derived from 2-DE experiments were analyzed using various software but representative and detailed pathways were reconstructed using GeneMania database, a web-based integrative software. The network architecture represents the connections between the individual query proteins such as vimentin, PP16B and KRT1 and in combination with APC and KRAS proteins. Protein networks for analyzing shortest pathways between the identified proteins were built by STRING and cytoscape (data not shown). Sub networks were built from the master network to focus on activated pathways and genetic interaction profiles. In this analysis, hubs (KRAS, APC) of protein networks are proposed to be the key regulatory proteins involved in the major colorectal cancer pathway. Due to the high complexity of the network, we only examined the physical and genetic interaction plots of vimentin, PP16B and KRT1 with highest significance and found that only vimentin originated as one of the only hits having highest degrees of connectivity with nodal proteins as well as with hubs. Vimentin shows an equidistant dependency on both the genes viz. APC and KRAS (Fig. S6.1, ESI†) considering the genetic interaction network while KRT1 shows a distant network dependency via NEF1 (Fig. S6.2, ESI†) and PP16B did not show any network dependency separately as well as with KRAS and APC. Moreover PP16B was not connected to any of the hubs and shown as unconnected in the network (Fig. S6.3, ESI†). The most significant sub networks derived from vimentin connected with APC and KRAS hubs are associated with cellular component disassembly involved in the execution phase of apoptosis. This network is associated with the canonical Wnt receptor signaling pathway and the beta-catenin destruction complex via DMD, NEB, GSK3B and CTNNB1. The vimentin derived sub network involves many known proteins supposed to be linked with colorectal cancer.Evaluation of soluble vimentin in serum samples of CRC patients
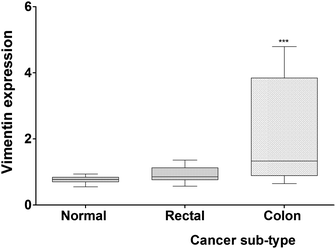
While evaluating the capacity of vimentin as a cancer biomarker, soluble vimentin levels were quantified in 63 serum samples comprising 43 pre-operative serum samples from colorectal patients and 20 serum samples as healthy controls. Soluble vimentin present in the sera of CRC patients varied greatly as compared to that of healthy controls. Vimentin showed a significant correlation with colon cancers (p < 0.001; 99% CI −1.998 to −0.7242) compared to rectal (p > 0.01; 99% CI −0.7120 to 0.4088) vs. healthy controls. It was found that vimentin expression was marginally higher (∼5-fold increased) in the colon cancer patients compared either to rectal cancer patients or healthy controls (Fig. 4).Evaluation of the CRC associated SNP effect on the vimentin structure using MDS
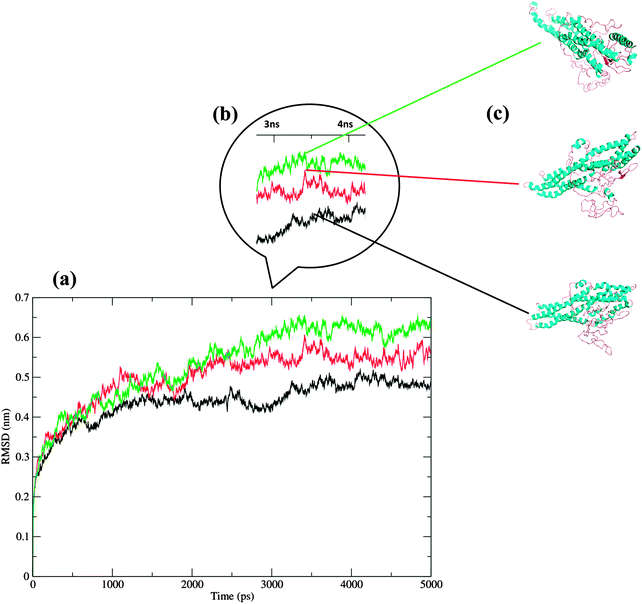
Two somatic SNPs were selected, Q190L – colon cancer associated and R345C – rectal cancer associated; after screening the various databases for colorectal cancer associated mutations in vimentin(data not shown). These SNPs were found to be in the amino acid stretch of the vimentin ecto-domain against which a therapeutic anti-cancer antibody named as pritumumab is used to treat brain tumors.49 We used MDS to investigate the effect of vimentin mutations on the dynamics of the protein structure. The protein was modeled using i-tasser and the model that was selected for further analysis fell in the range. Fig. 5 shows the picturesque happenings at different intervals of time during the simulations on the protein structure, thus outlining the effect of mutations in general. To investigate the structural events further, Gromacs inbuilt tools g_rms, g_rmsf, g_ gyrate, g-covar and g_anaeig were used. g_rmsf gave us the RMSD (root mean square deviation) value curves of VIM in the mutant and wild-type systems during the simulations (Fig. S8, ESI†), indicating that both the mutations are affecting the conformation of the vimentin. The inset in Fig. 6 presents the picture (Fig. 6b) retrieved at a point in the simulations where both the mutant structures show maximum deviation. The deviations can be a result of changes in the secondary structure architecture as seen in Fig. 6c. g_rmsf was used to calculate the atomic standard deviation and the deviations were plotted against the head region of the vimentin containing multiple phosphorylation sites, evidently associated with the regulation of the vimentin structure (Fig. 7). On observing RMSF values it is evident that mutation at codon 345 is making the structure more fluctuating and the mutation at codon 190 is making the structure a bit rigid. In the r_rmsf tool the option -oq was used to convert the RMSF values into B factor values and implicit them on the average structure (blue representing the most stable and red the most fluctuating). The comparative B factor projection (Fig. S8a, ESI†) on Wt and both the mutant structures primarily indicate the variations within the average structure, giving us an insight into the change in the fluctuation pattern between the structures. The coloring pattern set as default ranging between blue to red, blue representing most stable and red most fluctuating. The significant change in fluctuation can be observed in the mutant structure along with this change in the average secondary structure layout in Fig. S8b (ESI†); the figure also illustrates the average secondary structure layout of the three. The g_ gyrate tool was used to analyze the shape of the protein over time (Fig. S9, ESI†). The results are in accordance to our earlier findings of RMSF showing 345 mutations making the structure slightly expanded and 190 mutations effecting the structure other way around by making it more compact. g-covar and g_anaeig tools were used for understanding the effect of this mutation on global correlated motions in atomic simulations. In Fig. S10 (ESI†) the projections PC 1 vs. PC 2 of both structures are projected (black – Wt/red – Mu, green – Mu), the cluster obtained from the wt structure is stable; whereas the projection of first two PCs of both mutants covers a large area.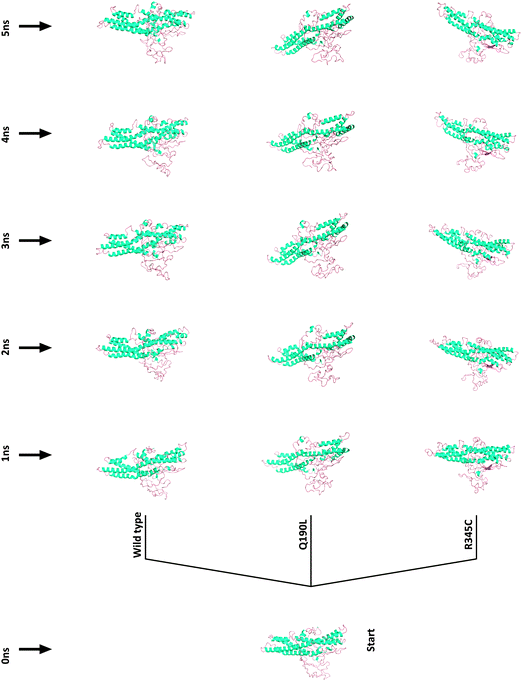 | ||
| Fig. 5 Pictorial representation of vimentin structural variation due to SNPs retrieved at 1 ns intervals of time during simulations extending from 1 to 5. | ||
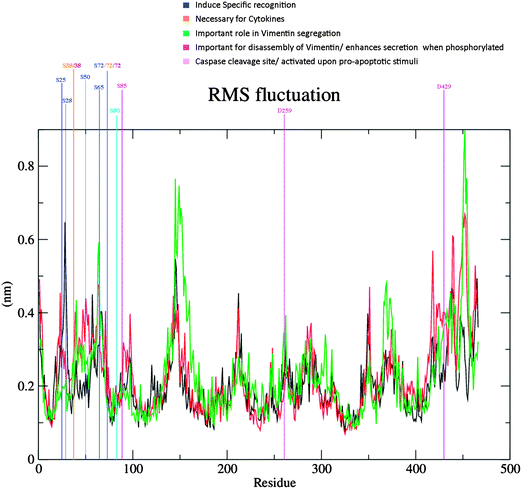 | ||
| Fig. 7 RMSF Plot of the three runs, incorporated with the color scheme representing different important functional aspects of concerned amino acids. | ||
Discussion
Tumor cell secretome consists of secretory proteins and tumor peptides shed by tumors that are regarded as exogenous byproducts systematically phagocytosed by antigen presenting cells (APCs), in particular by dendritic cells. Within this context of the immune synapse, alternative/additional co-stimulatory ligands would also be interacting. This immune assault against the tumor cells or secretory proteins initiates the physiologically significant, and naturally occurring anti-tumor response. Cancer vaccines are developed with an intend of boosting the body's natural ability to generate a potent immune response against the cancer cells. “Whole cell lysate” based cancer vaccines34 are more complex yet a complete dose can elicit better anti-tumor response. Some of these “active” specific vaccines are made from membrane preparations of tumor cells, autologous as well as allogenic.34,35 Melacine, (manufactured by Corixa Corp.), is an example of allogeneic melanoma tumor cell lysate combined with the adjuvant DETOX used as an active specific immunogenic cancer vaccine.36–38 Elsewhere a fractionated tumor-derived chaperone-rich cell lysate was used as an anticancer vaccine. This approach led to the identification of many tumor associated antigens, of which a peptidome was constituted and used as a vaccine.37 We utilized a fairly similar strategy, but not to develop anti- cancer vaccine, rather to immuno-capture the tumor associated antigens against which the anti-tumor responses were generated. To appreciate the cellular events that help in generation of multivalent affinity-matured antibodies, it is useful, as we tried to do so, to mimic the actual physiological scenario under which the multiple immunogen entry into lymphoid tissues result in generation of the repertoire of polyclonal antibodies with, on average orders of magnitude, higher affinity for multiple immunogenic antigens and increasing affinity heterogeneity that accompanies the increasing average affinity of antibodies compared to antigen specific monoclonal antibodies. Different strategies have been followed for disease biomarker mining by applying affinity proteomics.38–40 However almost every approach has mildly underestimated the strategy of immunizing the host with multiple proteins in a non-selective manner to have a repertoire of polyclonal antibodies generated against the immunogenic antigens in an increasing order. We utilized an immuno-affinity based antigen evaluation approach to assist us in capturing the tumor antigens wherein we generated a repertoire of polyclonal antibodies against the non-fractionated pure secretome preparations of HT29 cells in an unbiased manner. We identified three proteins viz., vimentin, PP16B and keratin, type II cytoskeletal protein. These proteins were captured by the affinity reagent (anti-Sc) from the pooled of sera of 5 colon cancer patients, 5 rectal cancer patients and a pool of 5 normal volunteers, separately. Protein–Protein interaction network based plotting revealed that vimentin was the only protein out of three that showed the bilateral genetic dependence directly related to the potential oncogenic signal axis shared between the K-ras and APC. Furthermore, vimentin was found to be picked from the pool of colon cancer patient sera. It was found that the levels of vimentin were elevated in the colon cancer sera compared to rectal cancer and normal sera. Furthermore, patients who had CEA > 20 ng ml−1, stage II or stage III, irrespective of being colon or rectal cancer sera, were found to have higher levels vimentin. Our data suggest that vimentin could be regarded as a predictive cancer biomarker for colon cancers with diagnostic potential. Vimentin belongs to the family of intermediate filament (IF) proteins, mostly expressed in normal mesenchymal cells and various other cellular phenotypes. In terms of functional classification, vimentin is one of those proteins that help in cellular integrity and molecular movement inside the cell. Over-expression of cellular vimentin has been reported in various epithelial cancers. Vimentin is also considered as a marker for epithelial-mesenchymal transition (EMT). However, in our study vimentin was picked from the sera of patients and the expression levels as well were recorded from the sera of colorectal cancer patients therefore indicating that vimentin could be selectively secreted by cancer cells. The unique property of vimentin secretion can be attributed to the spatial reorganization and dynamic assembly/disassembly of vimentin in response to several stimuli.41 Phosphorylation, as an important post translational event, is thought to regulate the dynamic states of vimentin.42–44 Vimentin has been recognized as a substrate for several kinases owing to its highly complex phosphorylation pattern.45,46 The disassembly of vimentin is enhanced by phosphorylation that splits vimentin into nonfilamentous (monomeric, dimeric, and tetrameric) particles, shifting the equilibrium between polymeric and depolymerized vimentin.47 Consequent to the equilibrium shift, the dissembled vimentin is secreted out in the cellular microenvironment and the soluble vimentin is thought to expedite the pathological state of the cell. Phosphorylated disassembled vimentin has been demonstrated to enhance the recycling of integrins subjected to endocytosis to the plasma membrane during cell migration.48 While trying to find the possible mechanism behind the secretion of vimentin, it is evident from our study that the serum levels of vimentin were higher in colon cancer sera compared to rectal and normal sera. Our simulation data demonstrate the structural variation imposed by mutations (colorectal cancer associated) in some of the important regions of vimentin particularly over the span of approximately 30 serine residues located in the first 100 amino acids at the N terminus of vimentin. Our findings suggest that the mutations at 190 and 345 codons, associated with colorectal cancer expand the overall vimentin structure. Incidentally, this mutation lies in the EDV stretch located in the C2 domain of vimentin against which a therapeutic antibody (Pritumumab) has been designed to limit the growth of brain tumors.49,50 However both the mutations have a profound effect on the atomic motions of full protein as well as the backbone of the protein. Accordingly, it can be hypothesized that excessive phosphorylation of vimentin, as a result of the expanded and destabilized structure due to colorectal cancer associated mutations, may lead to the secretion of vimentin subunits. This dissociated and secreted vimentin may underlie important steps in migratory control, that in the case of tumors are able to initiate the metastatic behavior of the cells.51Conclusions
In conclusion, the current study lays down a strategy to capture the potential immunogenic cancer biomarkers that might be responsible for colorectal cancer development and/or progression such as Vim, PP16B and KRT1 of which vimentin has been validated through clinical sample screening and proteomic data using an Affinity-2DE-MS-System's biology approach. Our preliminary study indicate that vimentin in its soluble form may act as a potential ligand that in turn might be responsible for activating the oncogenic pathways for the progression of cancers, though this needs to be further established. Clinical and functional validation of all the three biomarkers will further substantiate their role in the pathophysiology of colorectal carcinogenesis and/or as therapeutic targets.Abbreviations
| 2DE-MALDI TOF | Two dimensional gel electrophoresis-matrix-assisted laser desorption/ionization-time-of-flight |
| TGF-β | Transforming growth factor beta |
| IEF | Isoelectric focusing |
| LDH | Lactate dehydrogenase |
Acknowledgements
The work is supported by research grant from Science and Engineering Research Board, Department of Science and Technology, Ministry of Science and Technology (SR/SO/HS-90/2009). The authors are much thankful to Dr Ayub Qadri, National Institute of Immunology, Delhi, for providing A549, colon and THP-1 cell lines and Mr Raghu Tadala, Molecular Biophysics Unit, proteomics facility, IISC, Bangalore, for MS/MS data validation.References
- M. J. Taussig, O. Stoevesandt, C. A. K. Borrebaeck, A. R. Bradbury, D. Cahill, C. Cambillau and M. Uhlén, Nat. Methods, 2007, 4(2), 187, DOI:10.1038/nmeth0207-187a
.
- L. Berglund, E. Björling, K. Jonasson, J. Rockberg, L. Fagerberg, C. Al-Khalili Szigyarto and M. Uhlén, Proteomics, 2008, 8(14), 2832–2839, DOI:10.1002/pmic.200800203
.
- M. Uhlén and S. Hober, J. Mol. Recognit., 2009, 22(2), 57–64, DOI:10.1002/jmr.891
.
- M. Uhlen, Mol. Cell. Proteomics, 2005, 4(12), 1920–1932, DOI:10.1074/mcp.m500279-mcp200
.
- C. Agaton, Mol. Cell. Proteomics, 2003, 2, 405–414, DOI:10.1074/mcp.m300022-mcp200
.
- B. B. Haab, A. G. Paulovich, N. L. Anderson, A. M. Clark, G. J. Downing, H. Hermjakob and M. Uhlen, Mol. Cell. Proteomics, 2006, 5(10), 1996–2007, DOI:10.1074/mcp.t600020-mcp200
.
- O. Stoevesandt and M. J. Taussig, Proteomics, 2007, 7(16), 2738–2750, DOI:10.1002/pmic.200700155
, ahead of publication.
- K. A. Foon and M. M. Safa, Oncology Issues, 2008, 18–24 Search PubMed
.
- K. Ali, D. R. Soond, R. Piñeiro, T. Hagemann, W. Pearce, E. L. Lim and B. Vanhaesebroeck, Nature, 2014 DOI:10.1038/nature13444
.
- L. N. Liu, R. Shivakumar, C. Allen and J. C. Fratantoni, Electroporation Protocols, 2008, 139–153, DOI:10.1007/978-1-59745-194-9_9
.
- D. Avigan, Clin. Cancer Res., 2004, 10(18), 6347S–6352S, DOI:10.1158/1078-0432.ccr-050005
.
- M. Buckwalter and P. Srivastava, J. Immunol., 2007, 178, 48.16 Search PubMed
.
- J. D. Eaton, M. J. A. Perry, S. Nicholson, M. Guckian, N. Russell, M. Whelan and R. S. Kirby, BJU Int., 2008, 89(1), 19–26, DOI:10.1046/j.1464-410x.2002.02572.x
.
- R. C. Fields, K. Shimizu and J. J. Mule, Proc. Natl. Acad. Sci. U. S. A., 1998, 95(16), 9482–9487, DOI:10.1073/pnas.95.16.9482
.
- Y. Wu, G. Wu, L. Wang, Y.-Y. Zhang, Z. Li and D.-C. Li, Med. Oncol., 2010, 27(3), 736–742, DOI:10.1007/s12032-009-9277-x
.
- P. G. Lokhov, J. Cancer, 2010, 230, DOI:10.7150/jca.1.230
.
- M. L. Disis, J. R. Gralow, H. Bernhard, S. L. Hand, W. D. Rubin and M. A. Cheever, J. Immunol., 1996, 156(9), 3151–3158 CAS
.
- M. G. Hanna, H. C. Hoover, J. B. Vermorken, J. E. Harris and H. M. Pinedo, Vaccine, 2001, 19(17–19), 2576–2582, DOI:10.1016/s0264-410x(00)00485-0
.
- T. Maier, Blood, 2003, 102(7), 2338–2344, DOI:10.1182/blood-2002-08-2455
.
- D. Jager, E. Jager and A. Knuth, J. Clin. Pathol., 2001, 54(9), 669–674, DOI:10.1136/jcp.54.9.669
.
- K. Tani, M. Azuma, Y. Nakazaki, N. Oyaizu, H. Hase, J. Ohata and Y. Wakumoto, Mol. Ther., 2004, 10(4), 799–816, DOI:10.1016/j.ymthe.2004.07.001
.
- M. G. Hanna
Jr. and L. C. Peters, Cancer Res., 1978, 38, 204–209 Search PubMed
.
- J. Weyermann, D. Lochmann and A. Zimmer, Int. J. Pharm., 2005, 288(2), 369–376, DOI:10.1016/j.ijpharm.2004.09.018
.
- H. Towbin, T. Staehelin and J. Gordon, Proc. Natl. Acad. Sci. U. S. A., 1979, 76(9), 4350–4354, DOI:10.1073/pnas.76.9.4350
.
- A. Shevchenko, M. Wilm, O. Vorm and M. Mann, Anal. Chem., 1996, 68(5), 850–858, DOI:10.1021/ac950914h
.
- B. Hess, C. Kutzner, D. van der Spoel and E. Lindahl, J. Chem. Theory Comput., 2008, 4(3), 435–447, DOI:10.1021/ct700301q
.
-
Accelrys Software Inc., Discovery Studio Modeling Environment, Release 3.5, Accelrys Software Inc., San Diego, 2012 Search PubMed
.
- H. J. C. Berendsen, J. P. M. Postma, W. F. van Gunsteren, A. DiNola and J. R. Haak, J. Chem. Phys., 1984, 81(8), 3684, DOI:10.1063/1.448118
.
- T. E. I. Cheatham, J. L. Miller, T. Fox, T. A. Darden and P. A. Kollman, J. Am. Chem. Soc., 1995, 117(14), 4193–4194, DOI:10.1021/ja00119a045
.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14(1), 33–38, DOI:10.1016/0263-7855(96)00018-5
.
- B. Knapp, N. Lederer, U. Omasits and W. Schreiner, J. Comput. Chem., 2010, 31(16), 2868–2873, DOI:10.1002/jcc.21581
.
-
P. J. Turner, XMGRACE, Version 5.1. 19, Center for Coastal and Land-Margin Research, Oregon Graduate Institute of Science and Technology, Beaverton, OR, 2005 Search PubMed
.
- J. Rappsilber, Y. Ishihama and M. Mann, Anal. Chem., 2003, 75(3), 663–670, DOI:10.1021/ac026117i
.
- P. Reichardt, B. Dornbach and M. Gunzer, Curr. Top. Microbiol. Immunol., 2010, 229–249, DOI:10.1007/978-3-642-03858-7_12
.
- M. G. Hanna, H. C. Hoover, J. B. Vermorken, J. E. Harris and H. M. Pinedo, Vaccine, 2001, 19(17-19), 2576–2582, DOI:10.1016/s0264-410x(00)00485-0
.
- P. G. Lokhov and E. E. Balashova, J. Cancer, 2010, 1, 230–241, DOI:10.7150/jca.1.230
.
- J. Copier and A. Dalgleish, Int. Rev. Immunol., 2006, 25(5–6), 297–319, DOI:10.1080/08830180600992472
.
- J. A. Sosman and V. K. Sondak, Expert Rev. Vaccines, 2003, 2(3), 353–368, DOI:10.1586/14760584.2.3.353
.
- M. W. Graner, A. Romanoski and E. Katsanis, Int. J. Hyperthermia, 2013, 29(5), 380–389, DOI:10.3109/02656736.2013.793406
.
- D. J. Brennan, D. P. O'Connor, E. Rexhepaj, F. Ponten and W. M. Gallagher, Nat. Rev. Cancer, 2010, 10(9), 605–617, DOI:10.1038/nrc2902
.
- W. W. Franke, C. Grund, C. Kuhn, B. W. Jackson and K. Illmensee, Formation of Cytoskeletal Elements During Mouse Embryogenesis, Differentiation, 1982, 23(1–3), 43–59, DOI:10.1111/j.1432-0436.1982.tb01266.x
.
- O. Carpén, I. Virtanen, V. P. Lehto and E. Saksela, Polarization of NK cell cytoskeleton upon conjugation with sensitive target cells, J. Immunol., 1983, 131(6), 2695–2698 Search PubMed
.
- L. Palmberg, M. Sjolund and J. Thyberg, Differentiation, 1985, 29(3), 275–283, DOI:10.1111/j.1432-0436.1985.tb00327.x
.
- Y.-H. Chou, J. R. Bischoff, D. Beach and R. D. Goldman, Cell, 1990, 62(6), 1063–1071, DOI:10.1016/0092-8674(90)90384-q
.
- C. M. O'Connor, D. L. Gard and E. Lazarides, Cell, 1981, 23(1), 135–143, DOI:10.1016/0092-8674(81)90278-6
.
- Y. H. Chou, E. Rosevear and R. D. Goldman, Proc. Natl. Acad. Sci. U. S. A., 1989, 86(6), 1885–1889, DOI:10.1073/pnas.86.6.1885
.
- J. E. Eriksson, T. He, A. V. Trejo-Skalli, A. S. Harmala-Brasken and J. Hellman,
et al.
, J. Cell Sci., 2004, 117(6), 919–932, DOI:10.1242/jcs.00906
.
- J. Ivaska, K. Vuoriluoto, T. Huovinen, I. Izawa, M. Inagaki and P. J. Parker, EMBO J., 2005, 24(22), 3834–3845, DOI:10.1038/sj.emboj.7600847
.
- M. C. Glassy and H. Hagiwara, Hum. Antibodies, 2009, 18, 127–137 CAS
.
- D. M. Muzny, M. N. Bainbridge, K. Chang, H. H. Dinh, J. A. Drummond, G. Fowler and I. F. Newsham,
et al.
, Nature, 2012, 487(7407), 330–337, DOI:10.1038/nature11252
.
- A. Bouamrani, C. Ramus, E. Gay, L. Pelletier, M. Cubizolles, S. Brugière and J.-P. Issartel, PLoS One, 2010, 5(2), e9238, DOI:10.1371/journal.pone.0009238
.
Footnote |
| † Electronic supplementary information (ESI) available: Supplementary Data file 1. See DOI: 10.1039/c4mb00506f |
| This journal is © The Royal Society of Chemistry 2015 |