Research highlights: microfluidic analysis of antimicrobial susceptibility
Coleman
Murray
,
Oladunni
Adeyiga
,
Keegan
Owsley
and
Dino
Di Carlo
*
Department of Bioengineering, California NanoSystems Institute, Jonsson Comprehensive Cancer Center, University of California Los Angeles, 420 Westwood Plaza, 5121 Engineering V, Box 951600, Los Angeles, California 90095, USA. E-mail: dicarlo@ucla.edu
First published on 28th January 2015
Abstract
Antimicrobials remain an integral part of the treatment of patients with an infection. New microfluidic technologies are poised to help clinicians prescribe the right antimicrobials, sooner, reducing long hospital stays and improving outcomes. Given that current microbiologic diagnostic testing methods require a significant turnaround time (days), clinicians, in general, initially empirically determine a suitable therapy. After review of laboratory data, including information regarding the susceptibility of the microbial pathogen to specific anti-infectives, a clinician will then make alterations in therapy as appropriate, to direct therapy toward the pathogen involved in the illness. Important steps needed to quickly ascertain this information include the timely isolation of the microorganism, followed by direct antibiotic susceptibility tests (ASTs) or determination of the presence within the microorganism of any resistance genes or proteins that will impair the activity of a potential therapy. Recent microfluidic technologies highlighted here that can intrinsically interface at the scale of the organisms are starting to address these challenges by improving the speed and accuracy of tests aimed at helping physicians to give the right antimicrobials sooner.
The increased prevalence of antibiotic-resistant bacterial strains represents a major global health challenge. Furthermore, pathogen identification and subsequent antibiotic susceptibility tests (ASTs) are crucial in order to determine the proper therapeutic route as quickly as possible. The current gold standard AST, known as a broth microdilution test (BMD), involves co-culture of patient-isolated bacteria with antibiotic doped media of varying concentrations. The sample turbidity is then quantified via optical density measurement over the course of 16 to 20 hours. However, sample turbidity can yield false positives if, for example, the bacteria form filamentous structures without dividing when treated with antibiotic. While effective, the traditional AST has a substantial lead time and a limited sensitivity of approximately 107 cfu mL−1. The test usually follows an initial 1–2 day culture period to expand the initial microbial population isolated from the patient and enable testing of a variety of antibiotic classes.
Single-cell morphological analysis
To address these challenges, Kwon et al.1 developed a high throughput microfluidic approach to monitor bacterial morphology as a metric for rapid identification of susceptibility. As shown in Fig. 1a, the microfluidic component consists of a simple fluidic channel connected to a loading well, integrated into a 96-well plate format. Bacterial samples are first suspended in 37 °C agarose gel broth and added to the microfluidic channel. When cooled to room temperature, the broth gels and immobilizes the bacteria within the microfluidic channel. Antibiotic solution is added to the loading well and completely diffuses through the gelled broth after ~5 minutes (Fig. 1b). The immobilized bacteria are then imaged under bright field microscopy with a 60× objective. Automated image analysis quantifies single bacteria in terms of division, filamentary formation and swelling, which are then used to determine susceptibility or resistance (Fig. 1c).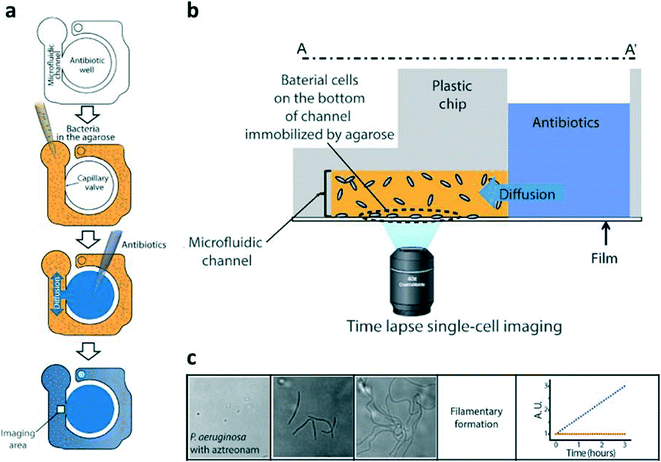 | ||
| Fig. 1 (a) Bacterial cells were mixed with agarose and loaded into a microfluidic channel, where the cells were immobilized by gel solidification. Liquid nutrients, some spiked with antimicrobials, were then loaded into the wells. These liquid samples diffuse into the agarose through openings between the channels and the wells. (b) Time-lapse imaging was performed in the imaging region (yellow box). (c) Quantification of P. aeruginosa treated with β lactam antibiotics over the course of three hours showed no bacterial division but filamentary formation, which can yield a false positive result due to increased turbidity. Adapted with permission from Choi et al., 2014.1 | ||
To calibrate the system, four clinically-relevant strains were tested with increasing antibiotic concentrations and categorized as susceptible or resistant. The system was able to match the standard minimum inhibitory concentration (MIC) for each strain with high confidence. After calibrating with known strains, 189 clinical samples were tested and compared to gold standard BMD tests. The system demonstrated a 91.5% agreement with the BMD method, with a minor discrepancy value of 7.01% and a major discrepancy value of 1.49%. Furthermore, the technology presented achieved results rapidly, demonstrating a turnaround time reduction of 25% since only microscopic changes in cell growth can provide a signal (as compared to turbidity), thereby decreasing the time to determine proper patient treatment.
Gradient antibiotic susceptibility
A similar microfluidic approach published by Wu et al.2 again takes advantage of smaller scales to reduce assay time and increase the number of data points collected. However, instead of integration in a 96-well plate format, they make use of gradient generation to test a range of antibiotic concentrations in a microfluidic format. The system consists of a commercially-available microfluidic device with a single 6 μL chamber, containing a 3D suspension of bacteria immobilized in agarose (Fig. 2). Two feed channels on either side of the chamber establish a linear concentration gradient of the antibiotic under consideration across the culture chamber. A simple PID system built from a microcontroller, thermometer, and heating element maintains the system at 37 °C. The system is then observed under phase contrast microscopy for 3–4 hours, with image intensity used as a measure of bacterial growth. From these images, the authors are able to obtain the minimum inhibitory concentration (MIC), as well as growth curves for a range of well-controlled antibiotic concentrations.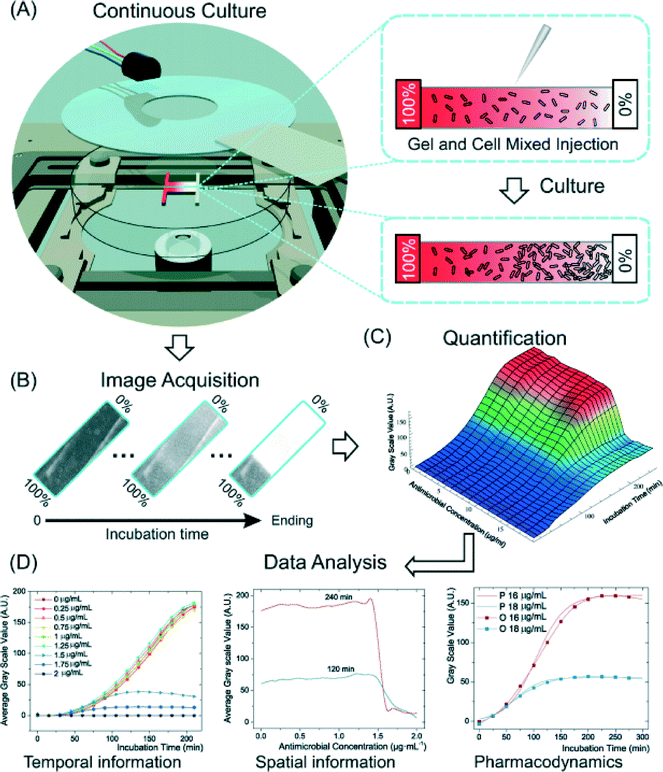 | ||
| Fig. 2 (a) A schematic representation of the device used by Wu et al.2 An agarose gel–bacteria mixture is loaded into the device and allowed to solidify. An antibiotic gradient is established across the device, resulting in a gradient of cell density after a brief culture period. (b) Phase contrast images of the device track bacteria growth over time, providing growth curves as functions of time and antibiotic concentration (c). From these images, quantitative pharmacodynamic data can be extracted (d). (Hou et al., 2014)2 Reproduced by permission of The Royal Society of Chemistry. | ||
The authors validate their device by comparing MICs and growth curves for common antibiotics and bacterial strains against the values obtained using the traditional methods. For all antibiotics tested (ampicillin, vancomycin, spectinomycin, streptomycin, tetracycline), the obtained MIC was within 15% of the value obtained using the gold standard. Additionally, the growth curves provided by the experiment provide insight into the mechanism of action of the antibiotics. The latter three antibiotics (spectinomycin, streptomycin, and tetracycline) are known inhibitors of translation, and demonstrated a reduction in growth rate dependent on antibiotic concentration, with a large drop at the MIC. The former two are inhibitors of cell wall synthesis, and have distinctive growth curves, ampicillin showing characteristic cell swelling (and thus larger apparent growth) before lysis, and vancomycin having no apparent concentration dependence until the MIC is reached. All pharmacodynamic results match expected behavior for these antibiotics, obtained from literature.
This simple microfluidic approach provides a flexible solution to studying the pharmacodynamic effects of antibiotics on bacteria, on much shorter timescales than existing techniques, while maintaining comparable results. In both the work of Kwon et al.1 and Wu et al.2 encapsulation in gel and confinement of microorganisms is employed and appears to be key to enabling high quality imaging and image analysis for robust assays.
Rapid bacterial detection in blood
As an alternative to growing and directly testing cell killing using added antibiotics, direct detection of bacteria with unique genomic features associated with a particular strain may show promise in picking the right antibiotics sooner. In the work of Zhao et al., the authors introduce a technology called Integrated Comprehensive Droplet Digital Detection (IC 3D)3 to detect single cells of specific strains of bacteria from diluted whole blood within 1.5–4 h in a single step, and without the use of bacterial culture or other amplification methods. The technology was developed by integrating these elements: detection of specific bacterial lysate with DNAzyme fluorescence-coupled sensors, droplet formation using droplet microfluidics, and microscopic fluorescence quantification using a 3D particle-counting prototype instrument.First, the authors describe the DNAzyme sensors used in their technology, which have been described previously.4 The sensors used are short, synthetic, single-strand catalytic DNA oligonucleotides that react specifically with lysates of the bacteria of interest to initiate an RNase activity. This specific reaction results in the release of a quenched fluorophore. The authors first demonstrate the ability of these molecules to differentiate the lysates of several E. coli strains from other bacterial strains and lysates of two mammalian cell lines in vitro with high specificity. However, comparison of clinically-isolated E. coli strains with other clinically-relevant gram negative organisms isolated from patients in 10% blood, such as C. freundii and K. oxytoca, shows the potential for cross reactivity, in that the magnitude difference in fluorescence intensity is smaller, although still statistically significant. Importantly, the bacteria used in these experiments were lysed with a solution optimized for DNAzyme sensor function and droplet generation and stability.
Next, the authors designed a droplet microfluidic system so that the sensors used would have increased sensitivity (through a higher concentration of lysate in the small volume) and a shorter detection time for detecting the bacteria confined within generated droplets. This system was fabricated using standard soft lithography with PDMS as the material. It has three inlets: one for oil, and two aqueous inlets (one for bacteria containing buffer or 10% diluted blood, the other for DNAzymes mixed with the bacterial lysis reagent). Fluids are introduced into the chip using a standard syringe pump, and DNAzymes are co-localized with bacteria within droplets. Using a multi-layer device, the goal is to achieve high throughput processing ability; the ability to process 1 mL of blood in under 40 min.
Finally, generated droplets that were collected into a cuvette were analyzed with a 3D particle counter that uses a custom-built prototype instrument. The prototype consists of a small, portable microscope able to hold and position a cylindrical cuvette so that excitation light from a diode laser is focused on the cuvette contents. After emission from the sample is collected through the same objective, it is transmitted through a set of dichroic filters, focused by another lens into a pinhole, and then collimated by an additional lens to the photomultiplier tube (PMT). In this case, particles (e.g. fluorescence of droplets) are recognized using a pattern recognition algorithm rather than threshold intensity, so as to decrease false positive/negative rates. The authors integrate the above described elements to achieve their final technology (Fig. 3). In a proof of concept experiment, in buffer or diluted blood DNAzymes were able to detect target E. coli K12 down to single digits within hours. Future work in combining with other fluorogenic DNAzymes or detectors specific for beta-lactamases and carbapenemases could aid in rapid analysis of specific antibiotic resistance phenotypes as well.
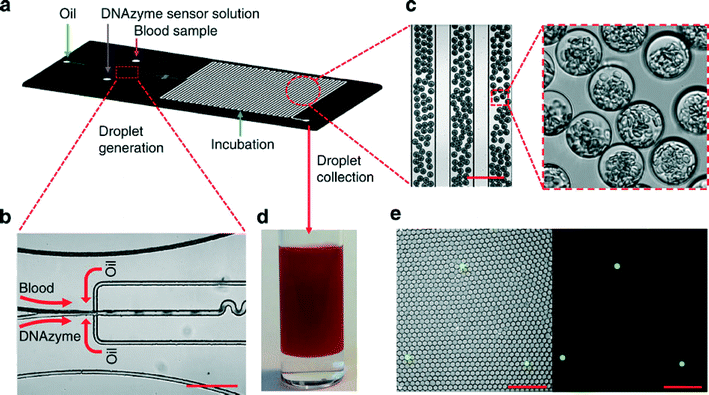 | ||
| Fig. 3 (a) Device layout for the droplet-based microfluidic device. The device has 3 inlets: one oil inlet, and two inlets for blood samples and the DNAzyme/bacterial lysis buffer. (b, c) Microscopy images showing the formation of uniform 30 μm droplets that consist of 10% blood and DNAzyme/bacterial lysis buffer solution. Scale bar, 200 μm. (d) Collected droplets in a cuvette for counting with a 3D particle counter. (e) Fluorescence microscopy images indicating that after a 3 h reaction, DNAzyme sensors light up droplets containing single E. coli K12 bacterium in 10% blood. Left panel: overlay of fluorescence and bright field images. Right panel: fluorescence image. Scale bar, 200 μm. (Kang et al., 2014)3 Reproduced by permission of the Nature Publishing Group. | ||
References
- J. Choi, J. Yoo, M. Lee, E.-G. Kim, J. S. Lee, S. Lee and S. Kwon, A rapid antimicrobial susceptibility test based on single-cell morphological analysis, Sci. Transl. Med., 2014, 6(267), 267ra174, DOI:10.1126/scitranslmed.3009650.
- Z. Hou, Y. An, K. Hjort, K. Hjort, L. Sandegren and Z. Wu, Time lapse investigation of antibiotic susceptibility using a microfluidic linear gradient 3D culture device, Lab Chip, 2014, 14(17), 3409–3418, 10.1039/c4lc00451e.
- D.-K. Kang, M. M. Ali, K. Zhang, S. S. Huang, E. Peterson, M. A. Digman and W. Zhao, Rapid detection of single bacteria in unprocessed blood using Integrated Comprehensive Droplet Digital Detection, Nat. Commun., 2014, 5, 5427, DOI:10.1038/ncomms6427.
- M. M. Ali, S. D. Aguirre, H. Lazim and Y. Li, Fluorogenic DNAzyme probes as bacterial indicators, Angew. Chem., Int. Ed., 2011, 50(16), 3751–3754, DOI:10.1002/anie.201100477.
| This journal is © The Royal Society of Chemistry 2015 |
